Abstract
To reveal the relationship between hypodiploidy with 30 to 39 chromosomes and near-triploidy in acute lymphoblastic leukemia (ALL), we studied 24 patients presenting with one of these aneuploidies among 623 adults with ALL registered in the Leucémie Aigüe Lymphoblastique de l'Adulte (LALA) protocols. The 2 ploidy groups presented a striking similarity of their cytogenetic profiles: chromosomes 2, 3, 4, 7, 13, 15, 16, and 17, significantly monosomic in hypodiploidy 30 to 39, were also frequently disomic in near-triploidy, whereas those retained in pairs in hypodiploidy 30 to 39 were frequently tetrasomic in near-triploidy. DNA content data revealed the simultaneous presence of 2 aneuploid peaks in most tested cases (DNA indexes: 0.72-0.87/1.39-1.89) and a multiple correspondence analysis applied on cytogenetic profiles ascertained their strong relationship. We thus assumed that near-triploidy derives from the duplication of hypodiploidy with 30 to 39 chromosomes and that both aneuploid groups are 2 expressions of the same disease. These 24 patients presented with B-cell phenotype, low leukocytoses (median white blood cell count, 4.2 × 109/L), and poor prognosis (complete remission, 57%; median disease-free-survival, 8 months; median survival, 10.4 months) comparable to that of Ph+ patients treated according to the same protocol. We suggest that hypodiploidy with 30 to 39 chromosomes or near-triploidy should be regarded as a new high-risk factor in the risk stratification of adult ALL protocols.
Introduction
Identification of recurring chromosomal abnormalities has had a major impact on risk assignment in acute lymphoblastic leukemia (ALL).1-5 Current therapeutic protocols consider a number of these abnormalities when assigning patients to the low- or high-risk categories.3,5,6
Ploidy is recognized as a prognostic factor in childhood ALL.1,7 Hyperdiploidy more than 50 chromosomes (51-65 chromosomes with nonrandom gains of chromosomes X, 4, 6, 10, 14, 17, 18, and 21) confers the most favorable prognosis and most patients in this ploidy group can be also identified by the DNA content of blast cells (DNA index > 1.16).8,9 Hypodiploidy (modal number lower than 46 chromosomes) has long been associated with poor prognosis.10 However, patients with 45 chromosomes fare better than those with hypodiploidy of fewer than 45 chromosomes,11 and patients with near-haploidy (fewer than 30 chromosomes) have the poorest response to therapy. Near-triploidy is uncommon in childhood ALL (0.3% in Pui's series) and has been reported to be predictive of adverse outcome.12
Unlike in childhood ALL, few large series with cytogenetic studies have been reported in adults and the prognostic significance of ploidy has seldom been documented.2,6,13-15 In the Groupe Français de Cytogénétique Hématologique (GFCH) and United Kingdom Adult Lymphoblastic Leukaemia (UKALL) XA studies,13,14 hypodiploidy with fewer than 46 chromosomes conferred poor prognosis, whereas high hyperdiploidy (> 50 chromosomes) was associated with favorable outcomes. In the GFCH study, near-triploidy and near-tetraploidy differed in outcomes because these 2 groups had the shortest and the longest survivals, respectively.13
Hypodiploidy with modal numbers ranging from 30 to 39 chromosomes and with a characteristic pattern of chromosome loss has been described by Callen in a study including both children and adults.16 In this report, patients with hypodiploidy of 30 to 39 chromosomes were generally older than 40, mainly men, and had a common B-cell phenotype. Because of the few cases, no prognostic significance was related to this ploidy group. The GFCH study on adult ALL has confirmed that this type of hypodiploidy was found among adults, but no sex predominance was observed.13 The GFCH study has also revealed that all patients had a B-cell phenotype and that prognosis was as poor as in Ph+ ALLs. Furthermore, a relationship between the 2 groups was suggested in the GFCH study because frequent disomies in the hypodiploid group with 30 to 39 chromosomes and frequent tetrasomies (or trisomies) in the near-triploid group affected the same pairs of chromosomes (chromosomes 1, 6, 8, 10, 18, 21, and 22). The incidence of hypodiploidy with 30 to 39 chromosomes has not been assessed so far in large series of childhood ALL.
In this study, we compared the clinical, hematologic, and cytogenetic features of adult ALL with hypodiploidy with 30 to 39 chromosomes and with near-triploidy, recorded in the ongoing Leucémie Aigüe Lymphoblastique de l'Adulte (LALA) protocols.17,18 We confirmed that the 2 ploidy groups are strongly related and showed that they are 2 possible expressions of the same entity. For identifying this entity, we specified the clues that are based on the cytogenetic profile of chromosome losses and gains and on DNA content. We emphasize the poor prognostic impact conferred by these abnormalities and we recommend to consider them as high-risk factors in the risk assignment of B-cell lineage ALL.
Patients, materials, and methods
Patients
From January 1994 to July 2000, 809 adults with de novo ALL were registered in the LALA-94 and LALA-SA protocols. Diagnosis of ALL was based on the morphologic and cytochemical criteria of the French-American-British (FAB) classification on bone marrow lymphoblasts. Leukemic cells were tested for the presence of B-cell (CD19, CD20, cytCD79a, cytμ chains, sIgM), T-cell (CD2, CD5, CD 7, CD1a, CD3, CD4, CD8, cytCD3), and myeloid (CD13, CD33) markers, according to the classification used for LALA-87 or proposed by the European Group for the Immunological Characterization of Leukemia (EGIL).19 The study was approved by the Ethics Committee, Hospices Civils de Lyon (Lyon, France), and all patients gave informed consent.
Cytogenetic analysis
Prior to initiation of treatment, cytogenetic investigations were carried out on bone marrow cells or occasionally on short unstimulated peripheral blood cultures (24 or 48 hours or both) by local cytogenetic laboratories. Metaphases were treated for RHG or GTG banding according to the practice of each center, and karyotypes were centrally reviewed by cytogeneticists of the centers participating in the LALA group, in annual workshops. Chromosomal abnormalities were classified using the recommendations of the International System for Human Cytogenetic Nomenclature.20 Designation as normal karyotype required a complete analysis of at least 20 metaphases with good-quality banding. Patients were classified according to the groups classically used for ALL,5,6 normal karyotypes, pseudodiploidy (abnormal karyotype with 46 chromosomes), hypodiploidy (fewer than 46 chromosomes), hyperdiploidy with 47 to 50 chromosomes, hyperdiploidy with more than 50 chromosomes (51-65 chromosomes), near-triploidy (66-80 chromosomes), and near-tetraploidy (81-102 chromosomes). To isolate and better define the hypodiploidy with 30 to 39 chromosomes, the hypodiploid group was further divided into 3 categories according to modal numbers: near-haploidy (fewer than 30 chromosomes), hypodiploidy with 30 to 39 chromosomes, and hypodiploidy with 40 to 45 chromosomes. For hyperdiploidies with modal numbers ranging from 60 to 65, we used the profiles of chromosome gains to include a patient either in the hyperdiploidy with more than 50 group (preferential gains of chromosome X, 4, 6, 10, 14, 17, 18, and 21) or in the near-triploid group (specific profile described in this article).
In Table 1, which lists the karyotypes, the numerical abnormalities observed in patients with hypodiploidy of 30 to 39 chromosomes were described with respect to a haploid karyotype <n>, whereas in patients with near-triploidy, they were described with respect to a triploid karyotype <3n>.
Karyotypes and DNA indexes of patients with hypodiploidy with 30 to 39 chromosomes and with near-triploidy
Patient no. . | Modal no.* . | DNA index (% of leukemic cells in G0/G1) . | Karyotype . |
|---|---|---|---|
| 1 | 32 | ND | 32<n>,X,+Y,+1,+6,+8,+10,+11,+19,+21,+22[7]°/46,XY[5] |
| 2 | 34 | 0.72 (43)/1.43 (39) | 33-35<n>,X,+X,+1,+5,+6,+8,+10,+13,+14,+18,+19,+21[cp5]/64-68,idemx2[cp6]/46,XX[14] |
| 3 | 34 | ND | 34<n>,X,+1,+6,+8,+10,+11,+12,+14,+16,+18,+19,+21[6]/46,XX[6] |
| 4 | 34 | 0.78 (22.4)/1.89 (3.8) | 34<n>,X,+1,+5,+6,+7,+9,+10,+11,+18,+19,+20,+21[2]/69-74,idemx2,-7,+13,+14,-19,+22[cp8]/46,XY[11] |
| 5 | 35 | 0.78 (56)/1.55 (26) | 35<n>,X,del(1)(q32q43),+1,del(5)(q24q34),+5,+6,+8,+9,+10,+11,+14,+18,+19,+21,+22[14]/70,idemx2[2]/46,XY[4] |
| 6 | 37 | ND | 37<n>,X+Y,+1,+4,+del(5)q23q35),+6,+8,+10,+11,+12,+14,+18,+19,+21,+22[5]/74,idemx2[3] |
| 7 | 36 | 0.82 (2.3)† 1.58 (2.5) Relapse: 1.43 (93) | 36<n>,X,+Y,+1,+2,+4,+5,+6,+11,+16,+18,+19,+20,+21,+mar[7]/46,XY[12] Relapse: 62-64<3n>,XXY,add(3)(p21),-3,-7,-8,-13,-14,+16,-17,-28,+mar[21] |
| 8 | 37 | ND | 37<n>,X,+1,+3,+5,+7,+8,+9,+10,+11,+14,+15,+16,+19,+20,+22,inc[14]/46,XY[21] |
| 9 | 38 | 0.87 (44.7)/1.67 (29.2) | 38<n>,X,+1,+2,+4,+5,+6,+8,+9,+10,+11,+12,+13,+14,+20,+21,+22[8]/76<3n>,idemx2[9]/46,XX[1] |
| 10 | 39 | 0.82 (53.1) | 39<n>,X,+1,+4,+5,+6,del(6)(q23),+7,+8,+9,+10,+11,+14,+17,+18,+19,+20,+2mar[11]/72-75,idemx2[2]/46,XX[18] |
| 11 | 39 | ND | 39<n>,X,+Y,+1,+2,+5,+6,+8,+9,+10,+11,+12,+13,+14,+19,+20,+21,+22[6]/46,XY[14] |
| 12 | 64 | ND | 64<3n>,XX,-2,-3,+5,i(7)(q10),+9,-12,del(12)(p11),-13,-14,-15,-17,+18,-19,+mar,inc[3]/46,XX[5] |
| 13 | 70 | ND | 69-71<3n>,XY,+1,-3,-4,+6,-7,-13,-15,+22,+3-5mar[cp12]/46,XY[3] |
| 14 | 68 | ND | 68-70<3n>,XXX,+1,-3,add(4)(q10),-7,+8,+11,+12,-13,-15,-16,-17,+18,-19,+21[cp11]/46,XX[5] |
| 15 | 65 | 1.41 (8) | 63-66<3n>,XXY,+1,-2,-3,-4,+5,-7,+8,-10,-12,-13,+14,-15,-16,-17,+18,-20,+21,+22[cp4]/46,XY[7] |
| 16 | 65 | 1.39 (7) | 64-66<3n>,XXX,+1,-3,-4,-5,+6,-7,-9,-10,+11,-13,+14,-15,-16,-17,+19,-20,+21,+22[cp20] |
| 17 | 67 | 0.72 (12)/1.44 (57) | 66-67,<3n>,XX,+1,-3,-5,-7,-9,+11,-14,-15,-16,-17,+19,+21,+22,+2mar[cp11]/46,XX[3] |
| 18 | 70 | 0.8 (12)/1.56 (10) | 70<3n>,XXY,+Y,+1,-2,-3,+4,+5,+6,-7,+8,-9,+10,+11,-12,-13,-14,-15,-16,-17,+18,+19,-20,+21,+22[4]/46,XY[11] |
| 19 | 68 | 0.8 (25)/1.53 (52) | 68<3n>,XXY,+1,-3,-4,+6,-7,-15,+22[12]/46,XY[3] |
| 20 | 73 | 0.82 (41)/1.59 (10) | 73<3n>,XXY,+Y,+1,-3,-4,add(5)(q13),+6,-9,+11,+12,+14,-15,-17,+18,+21,+22,inc[11) |
| 21 | 69 | ND | 69-73<3n>,XXY,-3,-4,-5,+6,-7,+8,-9,+10,+11,+12,-13,+14,-15,-16,-17,+21,+22,+mar[cp7]/46,XY[12] |
| 22 | 69 | 0.88 (18)/1.53 (25) | 66-70,<3n>XXX,add(2)(q37),-3,-4,-7,del(7)(q21q35),-9,-14,-15,-17,+19,+21,+22,+1-5mar[cp15]/46,XX[7] |
| 23 | 74 | 0.78 (23)/1.49 (76) | 68-74<3n>,XXY,+Y,-3,-4,-5,+6,-7,+8,-9,+11,-15,+18,+19,+21×2,+22×2,+mar[cp10] |
| 24 | 74 | 0.82 (2)/1.56 (21) | 72-74<3n>,XXY,+Y,+1,-2,-3,-4,+6×2,-9,+10,+11,-12,+14,-15,-17,+18×2,+19×2,-20,+21,+22,+mar[cp20] |
Patient no. . | Modal no.* . | DNA index (% of leukemic cells in G0/G1) . | Karyotype . |
|---|---|---|---|
| 1 | 32 | ND | 32<n>,X,+Y,+1,+6,+8,+10,+11,+19,+21,+22[7]°/46,XY[5] |
| 2 | 34 | 0.72 (43)/1.43 (39) | 33-35<n>,X,+X,+1,+5,+6,+8,+10,+13,+14,+18,+19,+21[cp5]/64-68,idemx2[cp6]/46,XX[14] |
| 3 | 34 | ND | 34<n>,X,+1,+6,+8,+10,+11,+12,+14,+16,+18,+19,+21[6]/46,XX[6] |
| 4 | 34 | 0.78 (22.4)/1.89 (3.8) | 34<n>,X,+1,+5,+6,+7,+9,+10,+11,+18,+19,+20,+21[2]/69-74,idemx2,-7,+13,+14,-19,+22[cp8]/46,XY[11] |
| 5 | 35 | 0.78 (56)/1.55 (26) | 35<n>,X,del(1)(q32q43),+1,del(5)(q24q34),+5,+6,+8,+9,+10,+11,+14,+18,+19,+21,+22[14]/70,idemx2[2]/46,XY[4] |
| 6 | 37 | ND | 37<n>,X+Y,+1,+4,+del(5)q23q35),+6,+8,+10,+11,+12,+14,+18,+19,+21,+22[5]/74,idemx2[3] |
| 7 | 36 | 0.82 (2.3)† 1.58 (2.5) Relapse: 1.43 (93) | 36<n>,X,+Y,+1,+2,+4,+5,+6,+11,+16,+18,+19,+20,+21,+mar[7]/46,XY[12] Relapse: 62-64<3n>,XXY,add(3)(p21),-3,-7,-8,-13,-14,+16,-17,-28,+mar[21] |
| 8 | 37 | ND | 37<n>,X,+1,+3,+5,+7,+8,+9,+10,+11,+14,+15,+16,+19,+20,+22,inc[14]/46,XY[21] |
| 9 | 38 | 0.87 (44.7)/1.67 (29.2) | 38<n>,X,+1,+2,+4,+5,+6,+8,+9,+10,+11,+12,+13,+14,+20,+21,+22[8]/76<3n>,idemx2[9]/46,XX[1] |
| 10 | 39 | 0.82 (53.1) | 39<n>,X,+1,+4,+5,+6,del(6)(q23),+7,+8,+9,+10,+11,+14,+17,+18,+19,+20,+2mar[11]/72-75,idemx2[2]/46,XX[18] |
| 11 | 39 | ND | 39<n>,X,+Y,+1,+2,+5,+6,+8,+9,+10,+11,+12,+13,+14,+19,+20,+21,+22[6]/46,XY[14] |
| 12 | 64 | ND | 64<3n>,XX,-2,-3,+5,i(7)(q10),+9,-12,del(12)(p11),-13,-14,-15,-17,+18,-19,+mar,inc[3]/46,XX[5] |
| 13 | 70 | ND | 69-71<3n>,XY,+1,-3,-4,+6,-7,-13,-15,+22,+3-5mar[cp12]/46,XY[3] |
| 14 | 68 | ND | 68-70<3n>,XXX,+1,-3,add(4)(q10),-7,+8,+11,+12,-13,-15,-16,-17,+18,-19,+21[cp11]/46,XX[5] |
| 15 | 65 | 1.41 (8) | 63-66<3n>,XXY,+1,-2,-3,-4,+5,-7,+8,-10,-12,-13,+14,-15,-16,-17,+18,-20,+21,+22[cp4]/46,XY[7] |
| 16 | 65 | 1.39 (7) | 64-66<3n>,XXX,+1,-3,-4,-5,+6,-7,-9,-10,+11,-13,+14,-15,-16,-17,+19,-20,+21,+22[cp20] |
| 17 | 67 | 0.72 (12)/1.44 (57) | 66-67,<3n>,XX,+1,-3,-5,-7,-9,+11,-14,-15,-16,-17,+19,+21,+22,+2mar[cp11]/46,XX[3] |
| 18 | 70 | 0.8 (12)/1.56 (10) | 70<3n>,XXY,+Y,+1,-2,-3,+4,+5,+6,-7,+8,-9,+10,+11,-12,-13,-14,-15,-16,-17,+18,+19,-20,+21,+22[4]/46,XY[11] |
| 19 | 68 | 0.8 (25)/1.53 (52) | 68<3n>,XXY,+1,-3,-4,+6,-7,-15,+22[12]/46,XY[3] |
| 20 | 73 | 0.82 (41)/1.59 (10) | 73<3n>,XXY,+Y,+1,-3,-4,add(5)(q13),+6,-9,+11,+12,+14,-15,-17,+18,+21,+22,inc[11) |
| 21 | 69 | ND | 69-73<3n>,XXY,-3,-4,-5,+6,-7,+8,-9,+10,+11,+12,-13,+14,-15,-16,-17,+21,+22,+mar[cp7]/46,XY[12] |
| 22 | 69 | 0.88 (18)/1.53 (25) | 66-70,<3n>XXX,add(2)(q37),-3,-4,-7,del(7)(q21q35),-9,-14,-15,-17,+19,+21,+22,+1-5mar[cp15]/46,XX[7] |
| 23 | 74 | 0.78 (23)/1.49 (76) | 68-74<3n>,XXY,+Y,-3,-4,-5,+6,-7,+8,-9,+11,-15,+18,+19,+21×2,+22×2,+mar[cp10] |
| 24 | 74 | 0.82 (2)/1.56 (21) | 72-74<3n>,XXY,+Y,+1,-2,-3,-4,+6×2,-9,+10,+11,-12,+14,-15,-17,+18×2,+19×2,-20,+21,+22,+mar[cp20] |
ND indicates not done.
Model number is the most common chromosome number in a tumor cell population.20
At diagnosis, as analyzed by flow cytometry, the hypodiploid peak was slightly higher than the sensitivity threshold, whereas the near-triploid peak was clearly detected.
Molecular studies
In patients with a B-cell phenotype, a systematic screening for fusion transcripts MLL-AF4, BCR-ABL, or E2A-PBX1 was performed from bone marrow or peripheral blood samples or both at diagnosis.
DNA content by flow cytometry
Cytogenetic pellets were used for this study because methanol/acetic acid fixation allows a good preservation of cellular DNA as previously published.21 After 2 washes in phosphate-buffered saline (PBS), DNA staining with propidium iodide was carried out with the “DNA-prep3” kit (Beckman Coulter, Marseille, France) according to the manufacturer's recommendations. Diploid lymphocyte samples fixed according to the same protocol were used as controls. The DNA index was determined as the ratio of DNA content in leukemic G0/G1 cells versus normal G0/G1 lymphocytes.
Treatment
Patients were treated according to 2 concomitant protocols depending on their age. Fourteen patients, aged between 18 and 55 years, were treated according to the LALA-94 protocol, whereas 10 patients, older than 55, were included in the LALA-SA protocol.17,18
In the LALA-94, an induction course was administered over a 4-week period and consisted of a 4-drug standard combination. Marrow response status was determined by bone marrow aspirates at about day 28 of induction chemotherapy. B-cell lineage ALL without high-risk criteria according to Hoelzer et al22 were randomized on day 35 and received either intensive consolidation chemotherapy combining mitoxantrone with intermediate-dose cytarabine, or a standard dose consolidation course combining cyclophosphamide with cytarabine, and 6-mercaptopurine. Then for 2 years, the patients followed a chemotherapy program involving consolidation and maintenance courses. On day 35, all patients with a diagnosis of high-risk ALL22 were eligible for a second course of consolidation (or salvage), whatever the response to the induction course. This consolidation/salvage course consisted of mitoxantrone and intermediate-dose cytarabine. Patients who did not achieve complete remission (CR) at that time were not considered for further treatment in the protocol. All patients in CR after this second course were eligible for genetic randomization. Based on an intention-to-treat principle, all patients with Ph+ ALL or with central nervous system–positive (CNS+) ALL eligible for hematopoietic stem cell transplantation (HSCT) were distributed in one of the 2 following HSCT groups: (1) matched related allogeneic bone marrow transplantation if they had a matched related donor or (2) autologous peripheral blood stem cell transplantation if they did not meet criteria for the first group. In the high-risk ALL group, patients without HLA sibling donors were randomized between the chemotherapy program and the autologous peripheral stem cell transplantation.
In the LALA-SA protocol, remission induction consisted of 4-drug 4-week standard induction regimen. Patients who did not reach CR after the induction phase were offered a rescue therapy, identical to the first consolidation course. The first consolidation course included cytarabine for 3 days and mitoxantrone for 2 days. An interferon (IFN) phase was then started with IFN α-2b. Thereafter, patients entered the second consolidation phase, which consisted of vincristine, adriamycin, and dexamethasone infusions. The maintenance phase included oral 6-mercaptopurine and intramuscular methotrexate for 18 months.
Morphologic response was evaluated using bone marrow aspiration and peripheral blood examination. Responses were classified as CR or failure. Resistant disease and early death were included in failure. Patients were considered to be in CR when the neutrophil count was more than 1.5 × 109/L, platelet count was more than 150 × 109/L, bone marrow examination was normal, and all extramedullary localizations had resolved.
Statistical analysis
The Fisher exact test was used to determine the statistically significant chromosome losses in hypodiploidy with 30 to 39 chromosomes as compared to a normal diploid set of chromosomes. Both the Fisher exact test and Mantel-Haenszel tests were used to determine the statistically significant gains and losses in the near-triploid group, as compared to a triploid set of chromosomes. For the 2 tests, a loss or gain was significant at P < .05.
Comparison between hypodiploid with 30 to 39 chromosomes and near-triploid profiles were performed on a multivariate basis using a multiple correspondence analysis with the Splus (Insightful, Seattle, WA) software.23
To determine the prognostic value of hypodiploidy with 30 to 39 chromosomes/near-triploidy, patients presenting these abnormalities were compared with a cohort of 149 patients with Ph+ ALL (considered as a high-risk group of B-lineage ALL17,24,25 ) and to a second control group involving all other B-cell lineage ALLs, treated during the same period with identical intensive chemotherapy regimens. Because the study group (hypodiploid with 30-39 chromosomes/near-triploid) involves only B-cell lineage ALLs, T-cell lineage ALLs were not taken into account for control groups.
Overall survival (OS) was calculated from the day of initial randomization until death, or date last known to be alive. Disease-free survival (DFS) was calculated from the date of CR until relapse, death, or date last known to be alive. OS and DFS curves were estimated using the Kaplan-Meier method, and their 95% symmetrical confidence interval was calculated according to the method of Greenwood.26 Differences were assessed for statistical significance using the log-rank test. The 95% confidence intervals (CIs) on proportions of patients in CR were calculated using a binomial formula. Associations between categorical factors were performed with the χ2 statistics. For continuously distributed variables other than event time, differences between groups were tested using the Wilcoxon rank-sum test. All the P values indicated are 2-tailed and reported as statistically significant if < .05. Computations were performed using BMDP PC-90 statistical program (BMDP Statistical Software, Los Angeles, CA).
Results
The 24 patients included in the study were selected from 623 successfully karyotyped, of the 809 patients included in LALA trials over a 6-year period (751 patients in the LALA-94 and 58 in the LALA-SA). They presented either a hypodiploidy with 30 to 39 chromosomes (11 patients) or a near-triploidy (13 patients).
Frequency of hypodiploidy with 30 to 39 chromosomes and near-triploidy
Patients were distributed between groups as follows: normal karyotype 23.8%, pseudodiploidy 42.4%, hypodiploidy 9.4%, hyperdiploidy with 47 to 50 chromosomes 13%, hyperdiploidy with more than 50 chromosomes 7.9%, near-triploidy 2.1%, and near-tetraploidy 1.4%.
The hypodiploid group was composed of one patient with a near-haploid karyotype, 11 with modal numbers ranging from 32 to 39 chromosomes, 10 with 40 to 44 chromosomes, and 37 with 45 chromosomes (Table 2).
Distribution of ploidy groups according to age and to immunophenotype
. | Total no. (%) . | Patients 15-55 y old, no. . | Patients older than 55 y, no. . | Immunophenotype . | . | |
|---|---|---|---|---|---|---|
| . | . | . | . | B, % . | T, % . | |
| All patients | 623 | 577 | 46 | 72 | 28 | |
| Normal | 148 (23.8) | 141 | 7 | 55 | 43 | |
| Pseudodiploidy | 264 (42.4) | 248 | 16 | 72 | 28 | |
| Near-haploid | 1 (0.1) | 1 | 0 | 100 | 0 | |
| Hypodiploidy | ||||||
| 30-39 chromosomes | 11 (1.8) | 6 | 5 | 100 | 0 | |
| 40-45 chromosomes | 47* (7.5) | 41 | 6 | 77 | 23 | |
| 47-50 chromosomes | 81 (13) | 76 | 5 | 75 | 25 | |
| More than 50 chromosomes | 49 (7.9) | 47 | 2 | 98 | 2 | |
| Near-triploidy | 13 (2.1) | 8 | 5 | 100 | 0 | |
| Near-tetraploidy | 9 (1.4) | 9 | 0 | 55 | 45 | |
| t(9;22)(q34;q11) | 147 (23.5) | 132 | 15 | 100 | 0 | |
| t(4;11)(q21;q23) | 30 (4.8) | 29 | 1 | 100 | 0 | |
| t(1;19)(q23;p13) | 17 (2.7) | 17 | 0 | 100 | 0 | |
. | Total no. (%) . | Patients 15-55 y old, no. . | Patients older than 55 y, no. . | Immunophenotype . | . | |
|---|---|---|---|---|---|---|
| . | . | . | . | B, % . | T, % . | |
| All patients | 623 | 577 | 46 | 72 | 28 | |
| Normal | 148 (23.8) | 141 | 7 | 55 | 43 | |
| Pseudodiploidy | 264 (42.4) | 248 | 16 | 72 | 28 | |
| Near-haploid | 1 (0.1) | 1 | 0 | 100 | 0 | |
| Hypodiploidy | ||||||
| 30-39 chromosomes | 11 (1.8) | 6 | 5 | 100 | 0 | |
| 40-45 chromosomes | 47* (7.5) | 41 | 6 | 77 | 23 | |
| 47-50 chromosomes | 81 (13) | 76 | 5 | 75 | 25 | |
| More than 50 chromosomes | 49 (7.9) | 47 | 2 | 98 | 2 | |
| Near-triploidy | 13 (2.1) | 8 | 5 | 100 | 0 | |
| Near-tetraploidy | 9 (1.4) | 9 | 0 | 55 | 45 | |
| t(9;22)(q34;q11) | 147 (23.5) | 132 | 15 | 100 | 0 | |
| t(4;11)(q21;q23) | 30 (4.8) | 29 | 1 | 100 | 0 | |
| t(1;19)(q23;p13) | 17 (2.7) | 17 | 0 | 100 | 0 | |
h = 45 chromosomes in 37 patients.
The hyperdiploid group consisted of 81 patients with 47 to 50 chromosomes, 49 with hyperdiploidy with more than 50 chromosomes (modal numbers ranging from 51 to 61), 13 with near-triploidy (modal numbers from 64 to 74 chromosomes), and 9 with near-tetraploidy (modal numbers from 80 to 96).
In the present series of adult ALLs, the frequencies of hypodiploidy with 30 to 39 chromosomes and near-triploidy were 1.8% and 2.1%, respectively.
Cytogenetic description of hypodiploidy with 30 to 39 chromosomes
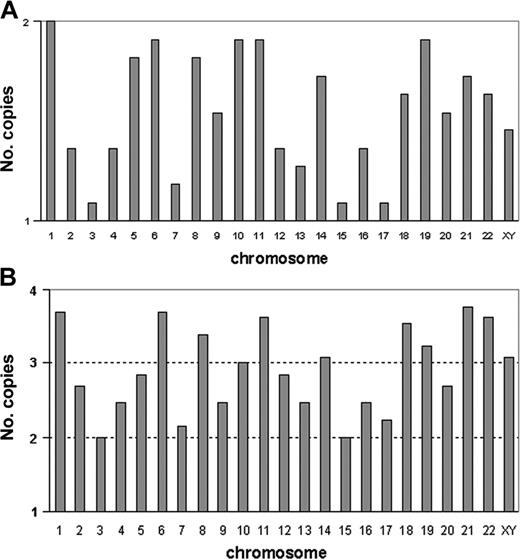
The 11 patients with a modal number ranging from 32 to 39 chromosomes presented similar cytogenetic profiles. In these patients, karyotype could be described either as chromosome losses in a diploid set of chromosomes, or as chromosome gains in a haploid set. Table 1 shows karyotypes with respect to haploidy <n>. This cytogenetic profile is also represented in Figure 1A, which was generated from the cumulated number of copies for each chromosome. All chromosomes, except chromosome 1, were found in one copy. However, the frequencies of monosomies differed according to chromosomes. Compared with a normal diploid set of chromosomes, the Fisher exact test found significant losses of chromosome 2 (P < .023), chromosome 3 (P < .0002), chromosome 4 (P < .023), chromosome 7 (P < .007), chromosome 12 (P < .023), chromosome 13 (P < .007), chromosome 15 (P < .0002), chromosome 16 (P < .023), and chromosome 17 (P < .0002). Loss of sex chromosomes did not reach significance (P > .06). In men, the sex chromosome lost was always chromosome Y. Hypodiploidy with 30 to 39 chromosomes can be therefore defined by the presence of nonrandom monosomies involving in decreasing order, chromosomes 3, 15, and 17, then chromosomes 7 and 13, and less frequently chromosomes 2, 4, 12, and 16.
Profiles of numerical abnormalities characterizing hypodiploidy with 30 to 39 chromosomes and near-triploidy. (A) Each vertical bar graph corresponds to a chromosome (sex chromosomes are analyzed together). For each chromosome, bars indicate the frequencies of monosomies (1 copy) and of disomies (2 copies); the abscissa represents the haploid set (1 copy) and the horizontal line represents the diploid set (2 copies). For each chromosome, frequencies are calculated from the cumulated numbers of copies from the 11 patients with hypodiploidy with 30 to 39 chromosomes. The maximum is observed for chromosome 1, which appears in 2 copies in all patients and the minimum value is observed for chromosomes 3, 15, and 17, which are monosomic in most patients. Therefore, the high values of the bar graph show the chromosomes often retained in pairs, and the low values point to the chromosomes that are frequently haploid. (B) Each vertical bar graph corresponds to a chromosome (sex chromosomes are analyzed together). For each chromosome, bars indicate the frequencies of disomies (2 copies), trisomies (3 copies), and tetrasomies (4 copies). The abscissa represents the haploid (1 copy) and the 3 horizontal lines represent the diploid (2 copies), triploid (3 copies), and tetraploid (4 copies) sets. For each chromosome, frequencies are calculated from the cumulated numbers of copies from the 13 patients with near-triploidy. Therefore, the highest values indicate the frequently trisomic or tetrasomic chromosomes, and the lowest values correspond to the frequently disomic chromosomes.
Profiles of numerical abnormalities characterizing hypodiploidy with 30 to 39 chromosomes and near-triploidy. (A) Each vertical bar graph corresponds to a chromosome (sex chromosomes are analyzed together). For each chromosome, bars indicate the frequencies of monosomies (1 copy) and of disomies (2 copies); the abscissa represents the haploid set (1 copy) and the horizontal line represents the diploid set (2 copies). For each chromosome, frequencies are calculated from the cumulated numbers of copies from the 11 patients with hypodiploidy with 30 to 39 chromosomes. The maximum is observed for chromosome 1, which appears in 2 copies in all patients and the minimum value is observed for chromosomes 3, 15, and 17, which are monosomic in most patients. Therefore, the high values of the bar graph show the chromosomes often retained in pairs, and the low values point to the chromosomes that are frequently haploid. (B) Each vertical bar graph corresponds to a chromosome (sex chromosomes are analyzed together). For each chromosome, bars indicate the frequencies of disomies (2 copies), trisomies (3 copies), and tetrasomies (4 copies). The abscissa represents the haploid (1 copy) and the 3 horizontal lines represent the diploid (2 copies), triploid (3 copies), and tetraploid (4 copies) sets. For each chromosome, frequencies are calculated from the cumulated numbers of copies from the 13 patients with near-triploidy. Therefore, the highest values indicate the frequently trisomic or tetrasomic chromosomes, and the lowest values correspond to the frequently disomic chromosomes.
In addition to the hypodiploid stemline, a hyperdiploid sideline corresponding to a doubling of the hypodiploid line was found in 6 patients (cases 2, 4, 5, 6, 9, and 10). Duplication of the hypodiploid stemlines resulted in near-triploid sidelines. In 3 of these patients (cases 2, 4, and 9), the hyperdiploid outnumbered the hypodiploid cells. These patients were classified as hypodiploidy with 30 to 39 chromosomes because we considered that, when both a hypodiploid stemline and a hyperdiploid sideline were present, the stemline (and not the number of cells in the different clones) determined the inclusion in a given ploidy group.
In case 7, a hyperdiploid clone corresponding to a duplication of the hypodiploid clone initially observed at diagnosis was found as the sole clone at relapse.
Structural abnormalities were seldom observed: 3 deletions, del(1q) (1 patient), del(6q) (1 patient), and del(5q) (2 patients), and 3 marker chromosomes (2 patients). No t(9;22), t(4;11) or t(1;19) was found and the absence of recurrent translocations was ascertained by the negativity of molecular analyses that are systematically carried out in LALA protocols.
Cytogenetic description of near-triploidy
The 13 patients with modal numbers ranging from 64 to 74 presented similarities of cytogenetic profiles. In Table 1, chromosome gains and losses are indicated with regard to triploidy <3n>. The karyotypes of the 13 patients revealed that some chromosomes were more frequently disomic, whereas others were more commonly tetrasomic. This cytogenetic profile of numerical abnormalities is represented in Figure 1B, which was generated from the cumulated number of copies added to the haploid set for each chromosome. Chromosomes 1, 6, 8, 11, 18, and 21 were always trisomic or tetrasomic (never found in 2 copies), and chromosomes 19 and 22 were each found in 2 copies in only 2 cases. Chromosomes more often tetrasomic than trisomic were chromosomes 1, 6, 11, 18, 21, and 22. Conversely, other chromosomes were more often found in 2 copies; chromosomes 3 and 15 were disomic in all patients, whereas chromosomes 4, 7, 9, 13, 16, and 17 were more often disomic than trisomic or tetrasomic. Compared to a triploid set of chromosomes (3 copies of each chromosome), the Fisher exact test found significant losses for chromosomes 2, 3, 7, 9, 13, 15, 16, 17, 20 (P < .002), and 4 (P < .01).
In our patients, the profile of near-triploidy was therefore characterized by frequent disomies of chromosomes 2, 3, 4, 7, 9, 13, 15, 16, 17, and 20. All other chromosomes were either trisomic or tetrasomic, but the most frequent tetrasomies involved chromosomes 1, 6, 11, 18, 21, and 22. All patients with modal numbers ranging from 64 to 74 presented this specific profile.
In these patients, no hypodiploid sideline was detected using the conventional cytogenetic technique.
No recurrent translocations were associated with numerical changes, and markers were found in 6 of the 13 cases. As in the hypodiploid group, the absence of high-risk translocations was confirmed by molecular techniques.
Ploidy assessed by flow cytometry
The DNA content was measured in 6 of the 11 patients with hypodiploidy with 30 to 39 chromosomes and in 9 of the 13 patients with near-triploidy (Table 1; Figure 2).
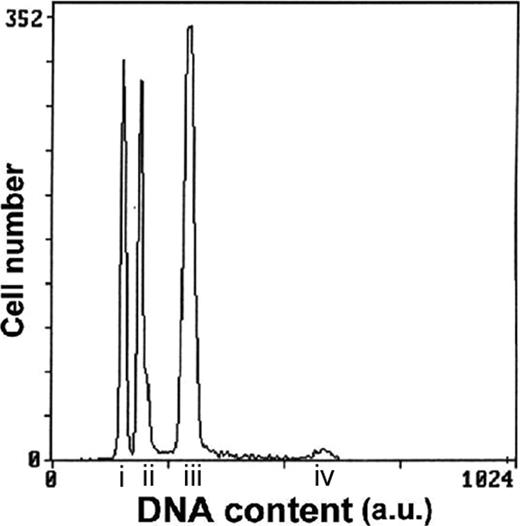
Representative histogram of ALL case number 19 with both hypodiploid with 30 to 39 chromosomes and near-triploid clones. G1 peak of the hypodiploid clone (i); G1 peak of the diploid clone (ii); G1 peak of the near-triploid clone (iii); G2 peak of the near-triploid clone (iv); a.u. indicates arbitrary unit.
Representative histogram of ALL case number 19 with both hypodiploid with 30 to 39 chromosomes and near-triploid clones. G1 peak of the hypodiploid clone (i); G1 peak of the diploid clone (ii); G1 peak of the near-triploid clone (iii); G2 peak of the near-triploid clone (iv); a.u. indicates arbitrary unit.
In the 6 patients with hypodiploidy at karyotype (cases 2, 4, 5, 7, 9, and 10) tested for DNA content, the flow cytometry technique confirmed hypodiploidy with DNA indexes ranging from 0.72 to 0.87. This technique also revealed a second near-triploid clone in 5 of 6 patients, confirming the cytogenetic findings in cases 2, 4, 5, and 9 but disclosing the presence of a near-triploid clone in case 7 where only hypodiploid cells had been detected at karyotype. In 4 of 5 patients, the percentages of hypodiploid cells were higher than those of near-triploid ones. The DNA indexes of the near-triploid clones ranged from 1.43 to 1.89, figures that correspond to nearly twice the value of the DNA indexes of the respective hypodiploid clones (Table 1). In case 7, cells from relapse were also available for DNA content measurement. At relapse, the DNA content analysis detected only the near-triploid clone, in agreement with the cytogenetic data, but this clone displayed a lower DNA index (1.43) than the one found at diagnosis (1.58).
In the 9 patients classified as near-triploid and tested for DNA content, the near-triploid clones were also detected by flow cytometry with DNA indexes ranging from 1.39 to 1.59. In addition to the near-triploid clone, a second hypodiploid clone was also detected in 7 of the 9 samples tested (cases 17, 18, 19, 20, 22, 23, and 24). Their DNA indexes ranged from 0.72 to 0.88 and represented half the value of those of the respective near-triploid clones. When the 2 aneuploid clones were simultaneously present, the most frequent pattern was a predominance of the near-triploid clone. However, in one patient (case 20), the hypodiploid outnumbered the near-triploid cells. Conversely, in case 24, the hypodiploid peak plotted slightly higher than the sensitivity threshold of the technique and was barely detectable.
The 3 hypodiploid, diploid, and near-triploid clones were easily identified in most patients. However, in 3 patients, the sensitivity of flow cytometry did not allow us to detect the expected hypodiploid (cases 15 and 16) or near-triploid (case 10) clones. In case 10, there might be an overlap of the small near-triploid clone with the G2/M peak of the hypodiploid population.
Relationship between hypodiploidy with 30 to 39 chromosomes and near-triploidy
A comparison of the cytogenetic profiles of the 2 groups showed that most of the chromosomes frequently monosomic in hypodiploidy with 30 to 39 chromosomes were also the chromosomes most commonly disomic in near-triploidy. The only exceptions were chromosomes 12 (found as a significant loss only in the hypodiploid group), and chromosomes 9 and 20 (found as significant losses only in near-triploidy). Chromosomes 2, 3, 4, 7, 13, 15, 16, and 17 were significantly lost in both ploidy groups.
Likewise, the chromosomes retained in pairs in hypodiploidy with 30 to 39 chromosomes were also frequently trisomic or tetrasomic in near-triploidy. Chromosomes 1, 5, 6, 8, 10, 11, 14, 18, 19, 21, and 22 were often found in 2 copies in the hypodiploid group, whereas chromosomes 1, 6, 11, 18, 21, and 22 were observed in 4 copies in most near-triploid cases. Chromosomes 5, 8, 10, 14, and 19 were either trisomic or tetrasomic.
Because the most common monosomies in hypodiploidy with 30 to 39 chromosomes corresponded to the most common disomies in near-triploidy, and because the most frequent disomies in hypodiploidy with 30 to 39 corresponded to the most frequent tetrasomies in near-triploidy, we assumed that in ALL, near-triploidy derives from the duplication of hypodiploidy with 30 to 39 chromosomes.
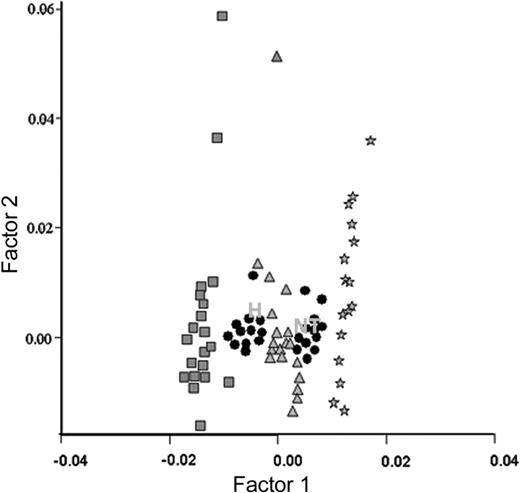
To further analyze the relationship between the 2 groups on a multivariate basis, we performed a multiple correspondence analysis of their cytogenetic profiles (gains and losses). This analysis, illustrated in Figure 3, revealed a striking similarity of the 2 cytogenetic profiles with a near overlap of the plots summarizing the 2 profiles.
Multivariate analysis assessed by multiple correspondence analysis: profiles of patients with hypodiploidy with 30 to 39 chromosomes and near-triploidy. Multiple correspondence analysis or dual scaling represents a qualitative analysis and provides an image of karyotype profiles23 in a less-dimensioned space defined by the factorial axis (factors). Different profiles can be plotted on the same graphic. This multivariate analysis takes into account different levels (interaction). As in component analyses, the distance of a modality from the center of gravity of the cluster reflects the rarity of the corresponding event in the population. The intensity of the association of 2 profiles is inversely proportional to the distance between 2 plots. First the profile for each chromosome (chromosome 1, chromosome 2,..., chromosome 22) was plotted on the same graph according to its monosomic (× 1), or disomic (× 2), or trisomic or tetrasomic (× 3 or × 4) status. Each symbol results from the analysis of the 24 patients. ▪, the profile of < 2n chromosome (monosomic); *, the profile of > 2n chromosomes (trisomic or tetrasomic); ▴, the profile of a diploid chromosomes (2n) (disomic). Neither the number of the chromosome nor the patients they belong to are specified. Then each patient was plotted summarizing the dots that correspond to each of his 22 autosomes as defined (▪ or * or ▴). Each symbol (•) on the graph represents the profile of one patient. Then the points H and NT were plotted. H summarizes the profile of patients (•) with hypodiploidy with 30 to 39 chromosomes. NT summarizes the profile of patients (•) with near-triploidy. The proximity of H and NT indicates their closely associated profiles.
Multivariate analysis assessed by multiple correspondence analysis: profiles of patients with hypodiploidy with 30 to 39 chromosomes and near-triploidy. Multiple correspondence analysis or dual scaling represents a qualitative analysis and provides an image of karyotype profiles23 in a less-dimensioned space defined by the factorial axis (factors). Different profiles can be plotted on the same graphic. This multivariate analysis takes into account different levels (interaction). As in component analyses, the distance of a modality from the center of gravity of the cluster reflects the rarity of the corresponding event in the population. The intensity of the association of 2 profiles is inversely proportional to the distance between 2 plots. First the profile for each chromosome (chromosome 1, chromosome 2,..., chromosome 22) was plotted on the same graph according to its monosomic (× 1), or disomic (× 2), or trisomic or tetrasomic (× 3 or × 4) status. Each symbol results from the analysis of the 24 patients. ▪, the profile of < 2n chromosome (monosomic); *, the profile of > 2n chromosomes (trisomic or tetrasomic); ▴, the profile of a diploid chromosomes (2n) (disomic). Neither the number of the chromosome nor the patients they belong to are specified. Then each patient was plotted summarizing the dots that correspond to each of his 22 autosomes as defined (▪ or * or ▴). Each symbol (•) on the graph represents the profile of one patient. Then the points H and NT were plotted. H summarizes the profile of patients (•) with hypodiploidy with 30 to 39 chromosomes. NT summarizes the profile of patients (•) with near-triploidy. The proximity of H and NT indicates their closely associated profiles.
This nonrandom relationship was also evidenced by the similarities between the near-triploid clones observed as sidelines in the hypodiploid group and the near-triploid clones observed as stemlines in the near-triploid group. In both, the near-triploid clones were characterized by the same profiles of numerical gains. Because in the hypodiploid group, the near-triploid clone unambiguously derived from a doubling of the stemline, we could infer that all near-triploidies presenting the described cytogenetic features derived from a hypodiploid clone. This assumption, based on cytogenetic findings, was further validated by the cell cycle data because the hypodiploid population was found in most near-triploidy cases.
The mechanism of clonal evolution from hypodiploidy with 30 to 39 chromosomes to near-triploidy was also demonstrated by the emergence of only a near-triploid clone at relapse in a patient who presented with a predominant hypodiploid clone at diagnosis (case 7).
Clinical and hematologic presentation
The comparison of clinical and hematologic features did not show any significant difference between the 2 groups (Table 3). Compared to the whole ALL population (median age, 35 years), our patients presented with a relatively high median age (hypodiploidy 30-39: median age, 46 years and range, 15-73 years; near-triploidy: median age, 52 years and range, 21-73 years). Lymph node enlargement was observed in 30% of patients in both groups and hepatosplenomegaly in 30% and 15% of patients in the hypodiploid and the near-triploid groups, respectively. CNS involvement was observed in only 2 patients in the near-triploid group. Both groups had low leukocytoses (4.5 and 4.2 × 109/L, respectively), low hemoglobin levels (97 and 99 g/L, respectively), and low platelet counts (55 and 45 × 109/L, respectively). The 2 cytogenetic groups were exclusively composed of B-lineage ALLs with a predominance of the intermediate CD10+ stage in both. A myeloid marker (CD33) was expressed in only one patient.
Hematologic and clinical features of patients with hypodiploidy with 30 to 39 chromosomes and near-triploidy
. | Hypodiploidy 30-39 chromosomes . | Near-triploidy . |
|---|---|---|
| No. patients | 11 | 13 |
| Median age, y (range) | 46 (15-73) | 52 (21-73) |
| Sex, no. male/no. female | 5/6 | 8/5 |
| Median hemoglobin level, g/L | 97 | 99 |
| Median WBC count, × 109/L (range) | 4.5 (1.6-7.4) | 4.2 (0.9-32.3) |
| Median platelet count, × 109/L | 55 | 45 |
| Immunophenotype* | B lineage | B lineage |
| CD19+CD10-CD20-, % (no. patients) | 27 (3) | 16.6 (2) |
| CD19+CD10+CD20+, % (no. patients) | 73 (8) | 83.3 (10) |
| CR rates, %† | 70 | 46 |
| OS, mos. | 11.1 | 8.1 |
| Median DFS, mos. | 5 | 18.2 |
. | Hypodiploidy 30-39 chromosomes . | Near-triploidy . |
|---|---|---|
| No. patients | 11 | 13 |
| Median age, y (range) | 46 (15-73) | 52 (21-73) |
| Sex, no. male/no. female | 5/6 | 8/5 |
| Median hemoglobin level, g/L | 97 | 99 |
| Median WBC count, × 109/L (range) | 4.5 (1.6-7.4) | 4.2 (0.9-32.3) |
| Median platelet count, × 109/L | 55 | 45 |
| Immunophenotype* | B lineage | B lineage |
| CD19+CD10-CD20-, % (no. patients) | 27 (3) | 16.6 (2) |
| CD19+CD10+CD20+, % (no. patients) | 73 (8) | 83.3 (10) |
| CR rates, %† | 70 | 46 |
| OS, mos. | 11.1 | 8.1 |
| Median DFS, mos. | 5 | 18.2 |
Not done for one patient.
P not significant because of the small number of patients in each group.
Treatment outcome
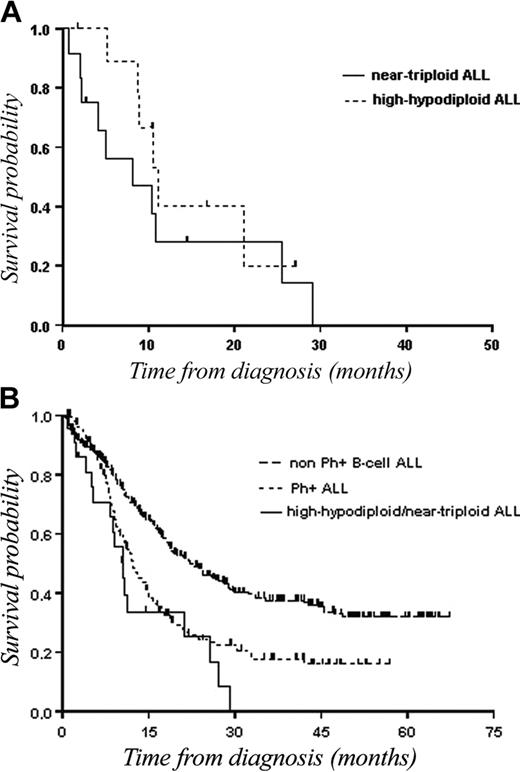
Because we assumed that the 2 ploidy groups belonged to the same entity and because there was no significant difference between patients with hypodiploidy with 30 to 39 chromosomes and those with near-triploidy in terms of CR (70% versus 46%; P = .3) as well as in terms of OS (11.1 months versus 8.1 months; Figure 4A) or DFS (5 months versus 18.2 months), we analyzed the outcome of hypodiploidy with 30 to 39 chromosomes/near-triploidy as a single group. There was also no statistical difference in terms of CR and survival between the 2 ploidy groups when separating patients aged younger than 55 years and those aged over 55 years.
OS of patients with hypodiploidy with 30 to 39 chromosomes and near-triploidy. (A) Kaplan-Meier plot of OS in patients with hypodiploidy with 30 to 39 chromosomes (n = 11) as compared with patients with near-triploidy (n = 13). (B) Kaplan-Meier plot of OS in patients with hypodiploid 30 to 39 chromosomes/near-triploid (n = 24) as compared with patients with non-Ph+ B-cell lineage ALL (n = 364), or patients with Ph+ ALL (n = 147).
OS of patients with hypodiploidy with 30 to 39 chromosomes and near-triploidy. (A) Kaplan-Meier plot of OS in patients with hypodiploidy with 30 to 39 chromosomes (n = 11) as compared with patients with near-triploidy (n = 13). (B) Kaplan-Meier plot of OS in patients with hypodiploid 30 to 39 chromosomes/near-triploid (n = 24) as compared with patients with non-Ph+ B-cell lineage ALL (n = 364), or patients with Ph+ ALL (n = 147).
CR was achieved in 13 (57%; 95% CI, 35%-77%) of 24 patients. Two patients died within 4 weeks following the beginning of treatment. Eight patients were resistant to induction chemotherapy (35%; 95% CI, 16%-57%). Median DFS and median OS were 8 months (95% CI, 4.3-15.1 months) and 10.4 months (95% CI, 8.1-11.1 months), respectively. Ten patients (77% of those who achieved CR) had a relapse.
To evaluate the prognostic value of hypodiploidy with 30 to 39 chromosomes/near-triploidy, the 24 patients were compared with a cohort of 147 cases with Ph+ B-ALL and with another of 364 patients with Ph– B-ALL, treated over the same time period using similar intensive chemotherapy regimens. CR rates were 57%, 54%, and 75% (P < .0001) for patients with hypodiploidy with 30 to 39 chromosomes /near-triploidy, patients with Ph+ ALL, and patients with Ph– ALL, respectively. Median DFSs were 8 months, 8.7 months, and 19.3 months, and median OSs were 10.4 months, 12.2 months, and 22.1 months, respectively (P < .001; Figure 4B).
Discussion
In this study, we show that hypodiploidy with 30 to 39 chromosomes and near-triploidy, defined by specific patterns of numerical abnormalities, characterize the same entity and should be regarded as a new high-risk factor in B-cell ALL.
To be identified, rare cytogenetic entities need large cohorts of patients. Hypodiploidy with 30 to 39 chromosomes with a specific cytogenetic pattern was reported in 1989 in a review of 22 cases of ALL with hypodiploidy,16 but the existence of this subgroup was confirmed only 7 years later in a large series of adult ALL.13 No series of childhood ALL have so far isolated this hypodiploid subgroup. Given the number of large cohorts of childhood ALL published to date, we can infer that this cytogenetic entity might occur more seldom in children than in adults. The frequency of hypodiploidy with 30 to 39 chromosomes with the specific profile of numerical losses can be assessed at 0.2% to 0.4% in Pui's27 and Heerema's11 studies on 2184 and 1880 childhood ALLs, respectively. In adult ALL, the frequency of this group is slightly higher, representing 1.6% in both the GFCH13 and the current study, on 443 and 626 cases, respectively. Near-triploidy also has a lower incidence in childhood ALL (0.3% in Pui's series12 ) than in adult ALL (3% in the GFCH series and 2% in the current study) and the probable relationship between the hypodiploidy with 30 to 39 and near-triploidy has been first established by the GFCH report.13 Our study confirms this relationship because there is no overlap between the 2 French studies, which have registered patients over different time periods (1987-1992 for the GFCH and 1994-2000 for the LALA studies). The frequency of the combined cytogenetic entity can be estimated at 4% in adult ALLs.
The clues that help to identify this entity rely on cytogenetic and cytometric features. Blast cells presented either a hypodiploidy with 30 to 39 chromosome or a near-triploidy or both hypodiploid and near-triploid clones. Actually, the DNA content showed that the 2 clones were present in most patients but that the cytogenetic techniques identified only the proliferative clones. The DNA content histograms displayed 2 aneuploid peaks, the hyperdiploid with a DNA index (1.4-1.6) twice the value of the hypodiploid one (0.7-0.9). Hypodiploidy with 30 to 39 chromosomes was characterized by recurrent losses of chromosomes 2, 3, 4, 7, 12, 13, 15, 16, and 17. All patients with modal numbers ranging from 30 to 39 presented this specific profile. No patient with near-triploidy had a standard triploidy (3 copies of all chromosomes). Patients classified in this group had a near-triploid (64-74) modal number that resulted from the association of frequent disomies involving chromosomes 2, 3, 4, 7, 9, 13, 15, 17, and 20, with frequent tetrasomies of chromosomes 1, 6, 11, 18, 21, and 22. This specific cytogenetic profile also allowed to distinguish near-triploidy from hyperdiploidy with more than 50 chromosomes characterized by nonrandom gains of chromosomes X, 4, 6, 10, 14, 17, 18, and 21. This distinction is extremely important, given the opposed prognostic significance conferred by these 2 cytogenetic groups, especially because treatment decisions may be based on karyotype. This distinction can be made by analyzing both the whole chromosome pattern and the DNA content because patients with hyperdiploidy with more than 50 chromosomes never present an associated hypodiploid clone. Conversely, the presence of a hypodiploid clone in a patient with a profile evocative of near-triploidy allows an unambiguous classification in near-triploidy. However, because in near-triploidy, the hypodiploid clone might involve only a minor population of blasts, its identification requires a thorough examination of the cytogenetic slides and of the DNA content histogram because the hypodiploid peak might be barely detectable. When only karyotypes are available, cytogenetic clues help to make the distinction between near-triploidy and hyperdiploidy more than 50. In near-triploidy, tetrasomy affects several pairs in a metaphase (3 or more than 3 pairs), whereas in hyperdiploidy more than 50, tetrasomy involves fewer pairs of chromosomes (often fewer than 3 pairs) with tetrasomy 21 frequently found as single tetrasomy. Chromosomes 6, 10, 14, and 18, which are gained in both ploidy groups, are not informative to classify a karyotype into one of the 2 groups, except for chromosome 6, which is more often tetrasomic in near-triploidy and more often trisomic in hyperdiploidy more than 50. Other gains are more helpful to make the distinction: tetrasomy 1 is near constant in near-triploidy but it is a very rare event in hyperdiploidy more than 50, tetrasomies 11 and 22 are frequent in near-triploidy, whereas they are rather rare in hyperdiploidy more than 50; trisomy 4 and trisomy 17 are common in hyperdiploidy more than 50 but rare in near-triploidy.
A characteristic feature of this cytogenetic entity was its restriction to B-cell lineage. However, there was no association with a B-differentiation stage. Another salient feature of this disease was a low median leukocytosis, which paralleled the minor tumoral burdens characterizing most patients, and might account for the low proliferative status of leukemic cells. However, the major clinical feature of this cytogenetic group was its prognostic impact. This study, carried out on patients treated according to the same therapeutic regimens, showed that hypodiploidy 30 to 39/near-triploidy conferred as poor an outcome as the Ph chromosome, either because of resistance to therapy or early bone marrow relapses. The median DFS was 8 months and no patients were alive at 3-year follow-up. In most patients, no other high-risk factors could account for this adverse outcome. The other cytogenetic high-risk factors, t(9;22), t(4;11) and t(1;19), were never associated with the numerical abnormalities characterizing our patients. There was no major tumoral syndrome, white blood cell (WBC) counts were low, and a CNS involvement was found in only 9% of patients. However, we cannot rule out that the elevated median age contributed to impair prognosis because our population had a higher median age (46 years) than the overall B-lineage ALLs registered in the LALA (35 years). However, age did not contribute to the poor outcomes of the young patients who had hypodiploidy 30 to 39/near-triploidy as the only high-risk factor. This strong prognostic implication further stresses the need to identify these patients using cytogenetic/cytometric techniques among B-cell lineage ALLs, and to regard them as high-risk subjects.
Based on the similarity of their cytogenetic profiles, we assumed that hypodiploidy with 30 to 39 chromosomes and near-triploidy are different expressions of the same entity and that near-triploidy derives from a doubling of hypodiploidy 30 to 39, with blasts that display either both clones or only the predominant one. A multiple correspondence analysis applied to the cytogenetic profiles of the 2 groups proved their close relationship and further argues in considering them as the same disease. These 2 possible expressions are reminiscent of near-haploidy, which can be occasionally revealed by a hyperdiploid clone, an exact duplicate of the near-haploid one.28-31 The chromosomes commonly retained in pairs in near-haploidy (14 and 21)27,32 also belong to the common disomies in hypodiploidy 30 to 39. So far, no mechanisms have been put forward to account for the recurrent numerical abnormalities that characterize near-haploidy and hypodiploidy 30 to 39, but they might be identical in both cytogenetic groups.33,34 Another similarity between the 2 hypodiploid groups is their adverse prognostic impact.10,11,13 Finally, the distinction between these 2 hypodiploid groups might not be justified but further studies are needed to classify them as a single entity. Their transcriptional profiles might give clues for elucidating the mechanisms underlying their generation.
Appendix
Paritcipants are listed with the name of centers in parentheses: C. Charrin, I. Tigaud, X. Thomas (Lyon); N. Dastugue, F. Huguet (Toulouse); M. Lafage, M. J. Mozziconacci, N. Vey (Marseille); J. L. Lai, P. Fenaux (Lille); C. Bilhou-Nabera, J. M. Boiron (Bordeaux); A. Bernheim, J. M. Vantelon (Villejuif); C. Bastard, A. Stamatoullas (Rouen); A. Hajemeijer, L. Michaux, A. Delannoy (Brussels, Belgium); J. Van Den Akker, C. Perot, M. Kuentz (Paris, Saint Antoine, Creteil); H. Mossafa (Meaux, Argenteuil); O. Maarek, H. Dombret (Paris, Saint Louis).
Prepublished online as Blood First Edition Paper, March 23, 2004; DOI 10.1182/blood-2003-04-1299.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Mariana Titorov for editorial assistance. Participants are listed in the “Appendix.”

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal