Abstract
Adenoviruses often cause lethal infections in immunocompromised individuals. Adoptive transfer of immune T cells offers a therapeutic option, but this strategy has been hindered by the paucity of information on molecular targets of cellular immunity and by the immunologic heterogeneity of the 51 human adenoviruses, which are grouped from A to F on the basis of genome size, composition, homology, and organization. Clonal analysis of the adenovirus-specific cytotoxic T lymphocyte (CTL) responses of seropositive individuals identified 5 novel CD8+ T-cell epitopes, all located in conserved regions of the capsid protein hexon. Reactive T cells were cross-reactive between 2 to 4 groups, while no T cells specific for a single subgroup were detected. Thus, by exploiting these peptide targets, it is possible to prepare a T-cell population capable of reacting with most adenoviruses that cause disease in immunocompromised patients.
Introduction
In the immunocompromised human host, adenoviruses may cause hepatitis, pneumonitis, encephalitis, and hemorrhagic cystitis.1-3 Such infections, now a leading cause of morbidity and mortality in transplantation patients, have been attributed to many of the 51 known human adenoviral (Ad) serotypes distributed among 6 subgroups (A-F)4,5 but most commonly involving viruses of subgroups B and C.3,6-9 Severity of lymphocytopenia after transplantation and continuation of immunosuppression have proven to be the most important risk factors for progressive Ad disease.3,10 Thus, in the absence of effective antiviral drugs, immunotherapy may offer an attractive option for the treatment of adenovirus infections.11-13 However, such an approach will be feasible only with an improved understanding of the molecular targets for cellular immunity. Because many serotypes can cause disease, adoptively transferred CTLs must be serotype and subgroup cross-reactive if this strategy is to be of practical value for the widest range of recipients.14-16
In contrast to the immunocompromised host, immunocompetent persons have a high frequency of adenovirus-specific cytotoxic T lymphocytes (CTLs) and generally suffer only mild and limited disease from adenoviruses.15 For this group an improved understanding of the immunodominant T-cell target antigens may lead to the development of less immunogenic Ad vectors, whose current value in human gene transfer studies is limited, in part, by the potent immune response they elicit.17,18
Little is known about human T-cell immunity to adenoviruses.18-21 Immunodominant antigens have yet to be identified, and only one CD4+ epitope has been mapped.22 Indeed, most published studies have characterized immune responses to adenoviruses in mouse models, in which the immediate early proteins, E1A and E1B, appear to be favored targets for specific T cells.23,24 However, adenovirus is a species-specific virus, making it unwise to rely solely on experimental data generated from animal models with limited permissiveness to human adenoviruses.
We have established a system for generating adenovirus-specific, cytotoxic, and helper T-cell responses from healthy human seropositive donors in vitro using replication-incompetent Ad vectors to stimulate viral specificity.15 These CTLs recognize and kill target cells infected with multiple wild-type Ad isolates.15,16 Here we describe how this strategy can be used to identify the molecular targets and HLA restriction elements involved in the adenovirus-specific T-cell response. Four of the new target epitopes we identified induce T cells that are cross-reactive among at least 4 subgroups and may therefore be useful in developing immunotherapies that will be active against any subgroup of adenovirus. Reactivity to another epitope is specific to only 2 subgroups, demonstrating that adenovirus-specific T cells can be either broadly cross-reactive or reactive within particular subgroups. Identification of these epitopes may facilitate the restoration or enhancement of immunity to adenoviruses in the immunocompromised host and help guide efforts to reduce the immunogenicity of Ad vectors, thereby improving the efficacy of gene therapy studies.
Materials and methods
Donors and cell lines
Peripheral blood mononuclear cells (PBMCs) and skin biopsies were taken from adenovirus-seropositive volunteers with their informed consent. The PBMCs were used to generate T-cell lines and dendritic cells (DCs). Epstein-Barr virus (EBV)–transformed B-lymphoblastoid cell lines (LCLs) were generated with concentrated EBV-containing supernatants of cultured B95-8 cells.25 Both LCLs and fibroblasts were maintained in RPMI 1640 (Hyclone, Logan, UT) with 5% and 10% fetal bovine serum (FBS; Hyclone), respectively, and 2 mM l-glutamine (GlutaMAX, Invitrogen, Carlsbad, CA).
Viruses and vectors
The Ad5f35GFP and Ad5f35null vectors were supplied by Dr Alan Davis (Baylor College of Medicine, Houston, TX). Ad5f35 has an Ad5 backbone with a chimeric Ad5/Ad35 fiber, selected for its ability to transduce hematopoietic cells.26-28 All multiplicities of infection (MOIs) are based on virus particles (vp; 1 infectious unit [iu] = 100 vp). The wild-type adenoviruses are prototype strains from the American Type Culture Collection (Rockville, MD).
Retroviral constructs
Murine stem cell virus (MSCV)–Hexon–internal ribosomal entry site (IRES)–green fluorescent protein (GFP). A 2.9-kb fragment containing hexon was isolated by digesting the Adeasy1 vector29 with BssHII and BglII. This fragment was cloned into the BssHII and BglII sites of the shuttle vector pSL1190 (Amersham Biosciences, Piscataway, NJ). From the pSL1190 plasmid, the hexon open reading frame (ORF) was isolated using a ClaI-SalI digest and cloned into the retroviral expression vector MSCV-IRES-GFP digested with the restriction enzymes XhoI and BstBI.
MSCV-Penton-I-GFP. A 1.9-kb fragment containing penton was isolated by digesting the Adeasy1 vector with DraI and NotI and cloned into the EcoRV and NotI sites of pSL1190. From pSL1190, the penton ORF, isolated using an EcoRI-NruI digest, was cloned into the EcoRI and NruI sites of the retroviral expression vector MSCV-IRES-GFP.
Generation of retroviral producer lines. Transient supernatant was made by transfecting 293T cells with a total of 10 μg DNA from 3 plasmids: (1) PSRalpha-G (vesicular stomatitis virus-G [VSV-g] envelope), (2) PeqPam3-E (Moloney murine leukemia virus [MoMLV]–gagpol), and (3) MSCV-Hexon-IRES-GFP or MSCV-Penton-IRES-GFP, at a ratio of 2:3:3. A total of 1 × 104 FLYRD18 cells30 were plated into each well of a 6-well plate and transduced daily (8-10 days) with 1.5 mL of the transient supernatant and 1 μL polybrene (10 mg/mL). High GFP-expressing cells were single cell sorted using a FACScan flow cytometer (BD Biosciences, San Jose, CA). Eight clones for each construct were expanded and the supernatants titered on HeLa cells. Clones with the highest titer were expanded and used for subsequent transductions.31
Transduction of skin fibroblasts with retroviral constructs. A total of 1 × 105 fibroblast cells were plated into each well of a 6-well plate, transduced with 1.5 mL of the retroviral supernatant and 1 μL polybrene (10 mg/mL), incubated at 37° C for 2 hours, and then supplemented with fibroblast medium. The transduction process was repeated until all cells were GFP positive.
Predictive algorithms
The T-cell epitope prediction programs BIMAS32 and SYFPEITHI33,34 were used to predict peptides that were capable of binding into HLA-A*1, A*2, A*24, and B*7 molecules. Predicted peptides were synthesized by Genemed Synthesis (San Francisco, CA). The overlapping hexon peptide library was synthesized by Alta Bioscience (Birmingham, United Kingdom).
Flow cytometry
For all flow cytometric analyses, a FACSCalibur instrument (BD) and CellQuest software (BD) were used. Antibodies were purchased from BD or Immunotech (Marseille, France). Isotype controls for antibody staining were immunoglobulin G1–phycoerythrin (IgG1-PE), IgG1–peridinin chlorophyll protein (IgG1-perCP), and IgG1–fluorescein isothiocyanate (IgG1-FITC). Phosphate-buffered saline (PBS; Sigma, St Louis, MO) with 2% FBS and 0.1% sodium azide (Sigma) was used as wash buffer. PBS with 0.5% paraformaldehyde (Sigma) was used as fixative solution. Cells were washed once, pelleted, and antibodies were added in saturating amounts (5 μL). After 15 minutes of incubation at 4° C in the dark, cells were washed twice, fixed, and analyzed. For surface staining of DCs, a lineage cocktail of antibodies was used.15 For Vβ staining of T cells, the antibodies were Vβ5.3-PE, Vβ7.1-PE+FITC, Vβ3-FITC, Vβ9-PE, Vβ17-PE+FITC, Vβ16-FITC, Vβ18-PE, Vβ5.1-PE+FITC, Vβ20-FITC, Vβ13.1-PE, Vβ13.6-PE+FITC, Vβ8-FITC, Vβ5.2-PE, Vβ2-PE+FITC, Vβ12-FITC, Vβ23-PE, Vβ1-PE+FITC, Vβ21.3-FITC, Vβ11-PE, Vβ22-PE+FITC, Vβ14-FITC, Vβ13.2-PE, Vβ4-PE+FITC, and Vβ7.2-FITC (Immunotech).
Tetramers were constructed in the Tetramer Core Facility of St Jude Children's Research Hospital, Memphis, TN (HLA-A*1–TDLGQNLLY = TDL, HLA-A*24–TYFSLNNKF = TYF) and the MHC Tetramer Core Facility, Houston, TX (HLA-B*7–KPYSGTAYNSL). For staining, cells were resuspended at 5 × 107/mL in 20 μL wash buffer. Tetramers were used at 1:100 final dilution together with antibodies against CD3 and CD8. After incubation for 30 minutes at 4° C in the dark, cells were washed twice, fixed, and analyzed immediately; 1 × 105 live events were acquired.
DC generation
DCs were prepared as previously described,15 matured, and used as stimulators following transduction with Ad vectors for 2 hours at the indicated MOIs.
CTL generation
For activation, 2 protocols were used.
Cryopreserved nonadherent PBMCs were used as responders and Ad5f35GFP-transduced mature DCs as stimulators.15,35 For the first stimulation responders were plated at 2 × 106 per well in a 24-well plate in 2 mL CTL medium (RPMI 1640 supplemented with 45% Click medium, Irvine Scientific, Santa Ana, CA, 2 mM GlutaMAX-I, and 10% FBS), and the responder to stimulator ([R/S] PBMC to DC) ratio was 10:1. On day 9 or 10, responders were harvested, plated as 2 × 106 per well, and stimulated at an R/S (CTL to DC) of 10:1. Responders were fed every 2 to 3 days with a half-medium change, harvested on day 17 or 18, and stimulated for a third time using an R/S (CTL to Ad5f35-transduced LCL) of 4:1.
Fresh PBMCs were transduced with the Ad5f35null vector at an MOI of 200 and used as both stimulators (monocytes) and responders (T cells), as previously described.15 For the first stimulation, cells were plated at 2 × 106 per well in a 24-well plate in 2 mL CTL medium; the precise R/S ratio was unknown, because the monocyte fraction varied from donor to donor. On day 9 or 10, responder T cells were harvested, plated at 2 × 106 per well, and restimulated with autologous, irradiated Ad5f35-transduced PBMCs, again at an unknown R/S. Responders were fed every 2 to 3 days with a half-medium change, harvested on day 17 or 18, and stimulated for a third time with adenovirus-transduced LCLs using an R/S (CTL to Ad5f35-transduced LCL) of 4:1.15
Single cell cloning
On day 24, CD8+ T cells were isolated using miniMACS (MACS, Miltenyi Biotec, Bergisch Gladbach, Germany) positive selection columns, according to the manufacturer's instructions. The isolated CD8+ T cells were seeded at 1 cell per round-bottomed microtest plate well using irradiated (40 Gy [4000 rad]) autologous Ad5f35GFP-transduced LCLs (MOI 500) as stimulators. The cultures were maintained in interleukin-2 (IL-2)–supplemented medium as previously described.36,37
Enzyme-linked immunospot (ELISPOT) assay
The previously described enzyme-linked immunospot (ELISPOT) assay was used to determine the HLA restriction of adenovirus-specific interferon-γ (IFN-γ)–secreting T-cell clones.37,38 Autologous and allogeneic LCLs, sharing 1 or 2 HLA class I alleles, were transduced with the Ad5f35GFP vector at an MOI of 500 and used as stimulator cells in an ELISPOT assay at 1 × 105 per well. Responder cells were T-cell clones plated at 100 cells per well.
To determine the epitope specificity of the adenovirus-specific CD8 clones restricted through HLA-A*1, A*2, A*24, and B*7, autologous LCLs were pulsed with 1 μg/mL of the predicted peptides for 1 hour, excess peptide was washed off, and then these peptide-loaded LCLs were stimulator cells in the ELISPOT assay at 1 × 105 per well, with LCLs alone as a negative control. Responder cells were T-cell clones plated at 1000 to 5000 cells per well.37 Alternatively, CTL lines were directly loaded with 1 μg/mL of peptide and plated out at 1 × 105 to 1 × 104 per well. After 20 hours of incubation, plates were developed as previously described.15 After overnight drying at room temperature in the dark, plates were sent for evaluation to Zellnet Consulting, New York, NY. Spot-forming cells (SFCs) and input cell numbers were plotted, and a linear regression was calculated after exclusion of plateau data points. IFN-γ production was expressed as specific SFCs after subtraction of the background (ie, the frequency of unstimulated responding cells).
Cytotoxicity assay
A chromium-51 release assay was performed to assess the cytolytic activity of responders.25 Fibroblasts were infected with adenovirus at an MOI of 1 to 5 at 48 hours and then treated with 100 U/mL IFN-γ (R&D Systems, Minneapolis, MN). Mock-infected and allogeneic targets were used as controls. Fibroblasts were infected with retroviral constructs expressing hexon and penton, treated with 100 U/mL IFN-γ (R&D Systems), and used for a minimum of 48 hours after infection. The percent specific lysis was calculated as ([experimental release – spontaneous release]/[maximum release – spontaneous release]) × 100.
Results
Specificity and cross-reactivity of an adenovirus-specific CTL line
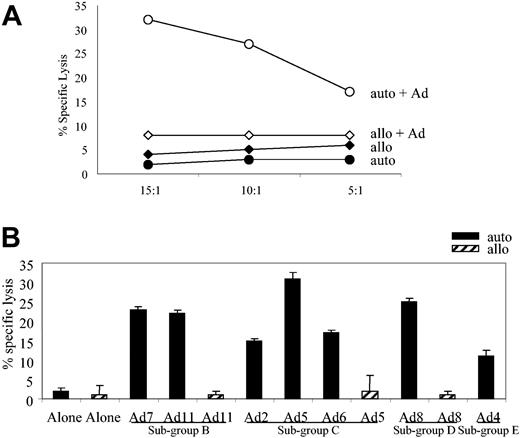
Using CTL-generation protocol no. 1 described in “Materials and methods,” we generated an adenovirus-specific CTL line from the PBMCs of seropositive donor 1. To analyze the specificity of the line, we used a cytotoxicity assay and autologous Ad5f35GFP-transduced fibroblasts as targets; controls were autologous fibroblasts alone and allogeneic HLA-mismatched fibroblasts either alone or infected with the Ad5f35GFP vector. The CTLs specifically recognized and killed autologous virus-infected targets but not the allogeneic targets or autologous fibroblasts alone (Figure 1A).
Adenovirus-specific, serotype cross-reactive CTL line from donor AL. (A) At an E/T ratio of 20:1, the CTLs showed specific cytolytic activity against autologous adenovirus-infected targets in a standard chromium release assay with only slight or negligible killing of allogeneic, infected or uninfected targets or autologous fibroblasts alone. (B) Adenovirus-specific CTLs lysed autologous fibroblast targets transduced with Ad2, Ad4, Ad5, Ad6, Ad7, Ad8, and Ad11, but not allogeneic fibroblast targets, either alone or transduced with Ad5, Ad8, and Ad11.
Adenovirus-specific, serotype cross-reactive CTL line from donor AL. (A) At an E/T ratio of 20:1, the CTLs showed specific cytolytic activity against autologous adenovirus-infected targets in a standard chromium release assay with only slight or negligible killing of allogeneic, infected or uninfected targets or autologous fibroblasts alone. (B) Adenovirus-specific CTLs lysed autologous fibroblast targets transduced with Ad2, Ad4, Ad5, Ad6, Ad7, Ad8, and Ad11, but not allogeneic fibroblast targets, either alone or transduced with Ad5, Ad8, and Ad11.
Human adenoviruses comprise 51 different serotypes divided into 6 subgroups. To test whether our CTL line could recognize Ad serotypes from different subgroups we assessed the cytolytic activity toward autologous fibroblasts infected with a panel of viruses: Ad7 and Ad11 (subgroup B); Ad2, Ad5, and Ad6 (sub-group C); Ad8 (subgroup D); and Ad4 (subgroup E). Controls were allogeneic HLA-mismatched fibroblasts either alone or infected with Ad5, Ad8, and Ad11. As shown in Figure 1B, at an effector-target (E/T) ratio of 20:1 the CTLs lysed targets infected with each of the test viruses but failed to recognize the allogeneic targets or the autologous fibroblasts alone. Thus, as previously established, CTLs generated from an adenovirus-seropositive donor are both subgroup and serotype cross-reactive.15
Adenovirus-specific CTL clones recognize peptides presented through 2 different HLA alleles
To analyze the specificity of individual clones within the CTL line of donor 1, we isolated and cloned CD8+ T cells from the CTL line shown in Figure 1. Altogether, 5 adenovirus-specific clones were established. To confirm their clonality we analyzed Vβ usage by fluorescence-activated cell sorting (FACS) using a panel of antibodies covering more than 70% of the normal human T-cell receptor (TCR) Vβ repertoire. All T-cell clones were uniformly CD8+ and monoclonal. Vβ usage was characterized as Vβ12 (clones 9 and 10), Vβ17 (clones 11 and 20), and Vβ20 (clone 16) (data not shown). To determine the HLA restriction of these clones, we tested their recognition of partially HLA class I–matched allogeneic Ad5f35GFP-transduced LCLs in an ELISPOT assay. Detailed results for 2 clones, c11 and c16, are shown in Figure 2A, which indicates only shared alleles. Clone 11 reacted strongly against the autologous Ad5f35GFP-transduced LCLs as well as Ad5f35GFP-transduced LCLs matched on the A*1 and B*8 alleles. However, no significant reactivity was detected against a line matched exclusively at the B*8 allele, indicating that this clone recognized an epitope presented in the context of HLA-A*1. When clone 16 was tested in a similar manner, it showed reactivity only when HLA-A*24 was present, suggesting that this response was A*24 restricted. Neither clone reacted with nontransduced LCLs (data not shown). Of the 5 adenovirus-specific clones we generated, 4 were A*1 restricted, while clone 16 was A*24 restricted.
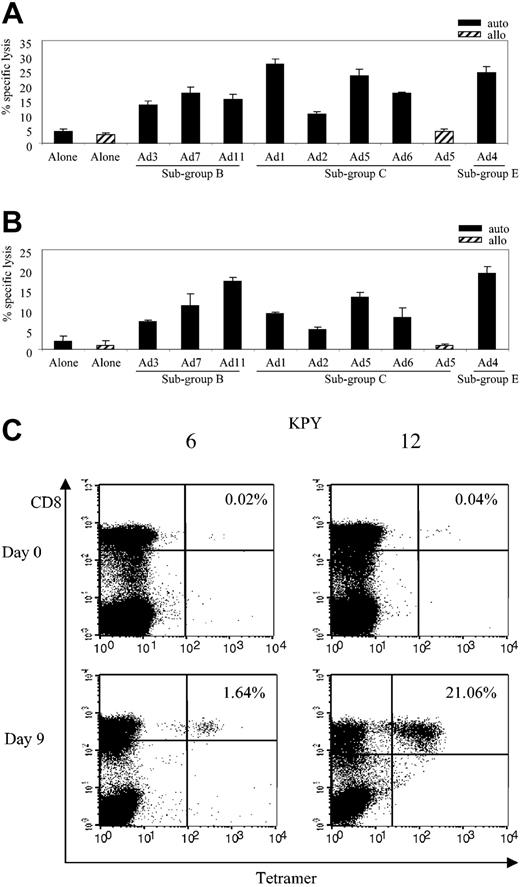
Clone HLA restriction and antigen specificity. (A) Adenovirus-specific CD8+ T-cell clones from donor AL are A*1 and A*24 restricted. Adenovirus-specific CD8 T-cell clones c11 and c16 were incubated with autologous and partially HLA class I–matched LCL targets infected with Ad5f35GFP; only shared alleles are indicated. Results are expressed as SFCs per 100 T cells. (B) In vitro–reactivated adenovirus-specific T-cell clones recognize a capsid protein, hexon. (i) A1-restricted clone 11 tested at E/T ratios of 5:1 and 2.5:1. (ii) A*24-restricted clone 16 tested at E/T ratios of 30:1, 20:1, and 10:1. Killing was assessed against autologous fibroblast targets alone or fibroblasts transduced with the MSCV–Hexon–internal ribosome entry site [IRES]–GFP viral supernatant (retro-Hexon) or with MSCV-Penton-IRES-GFP viral supernatant (retro-Penton). Error bars reflect standard deviation of triplicates.
Clone HLA restriction and antigen specificity. (A) Adenovirus-specific CD8+ T-cell clones from donor AL are A*1 and A*24 restricted. Adenovirus-specific CD8 T-cell clones c11 and c16 were incubated with autologous and partially HLA class I–matched LCL targets infected with Ad5f35GFP; only shared alleles are indicated. Results are expressed as SFCs per 100 T cells. (B) In vitro–reactivated adenovirus-specific T-cell clones recognize a capsid protein, hexon. (i) A1-restricted clone 11 tested at E/T ratios of 5:1 and 2.5:1. (ii) A*24-restricted clone 16 tested at E/T ratios of 30:1, 20:1, and 10:1. Killing was assessed against autologous fibroblast targets alone or fibroblasts transduced with the MSCV–Hexon–internal ribosome entry site [IRES]–GFP viral supernatant (retro-Hexon) or with MSCV-Penton-IRES-GFP viral supernatant (retro-Penton). Error bars reflect standard deviation of triplicates.
The virion protein hexon is the antigen recognized by T-cell clones
The clones described above were isolated from an adenovirus-specific CTL line generated with a replication-incompetent Ad vector. Because these vectors express their own genome only when infecting target cells at a high MOI, we inferred that the clones were likely specific for a component of the virion. To confirm this prediction and to determine the antigen specificity of the response, we made retroviral constructs expressing 2 of the most abundant capsid proteins, hexon and penton. Protein expression was confirmed by Western blot (data not shown). Retroviral supernatants from the resultant producer lines were used to transduce autologous fibroblasts, which were used as targets in a cytotoxicity assay. The results from 2 clones, c11 (A*1 restricted) and c16 (A*24 restricted), are shown in Figure 2B. In both cases, cytolytic activity was seen only when the fibroblasts expressed hexon. Subsequently, the remaining 3 clones were tested and found to be uniformly hexon specific (data not shown).
Cross-reactivity and epitope mapping of A*1-restricted adenovirus-specific clones
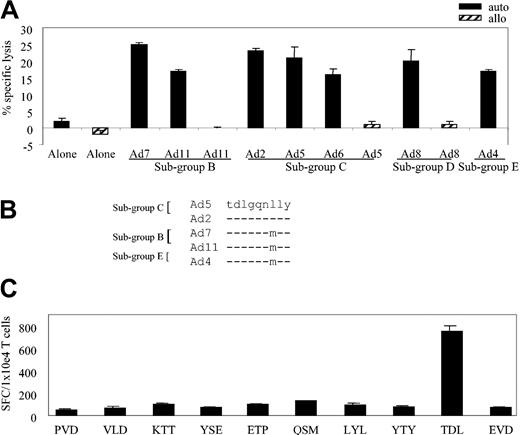
We next asked whether the cross-reactivity noted for the bulk CTL line (Figure 1B) extended to the various A*1-restricted clones. Figure 3A assesses the ability of clone 11 to kill autologous fibroblast targets infected with a cross-section of Ad serotypes representing 4 subgroups: Ad7 and Ad11 (subgroup B); Ad2, Ad5, and Ad6 (subgroup C); Ad8 (subgroup D); and Ad4 (subgroup E). Controls were allogeneic HLA-mismatched fibroblasts either uninfected or infected with Ad5, Ad8, and Ad11. At an E/T of 20:1, clone 11 showed cross-reactive properties, recognizing autologous targets infected with each of the Ad serotypes (Figure 3A) while failing to react appreciably with allogeneic targets or autologous fibroblasts alone.
HLA-A*1–restricted adenovirus-specific T-cell clones are both sub-group and serotype cross-reactive. (A) Cytolytic activity of adenovirus-specific, A*1-restricted clone 11 (E/T 20:1) against autologous and allogeneic fibroblast targets either alone or transduced with the indicated Ad serotypes. (B) Epitope alignment of sequences from the indicated Ad serotypes. (C) Hexon-specific, A*1-restricted T-cell response (clone 11 from donor AL) by ELISPOT assay. Stimulators are autologous LCLs pulsed with 1 μg/mL of the synthesized peptides, designated by the first 3 amino acids of the peptide sequence. Results are expressed as SFCs per 1 × 104 T cells.
HLA-A*1–restricted adenovirus-specific T-cell clones are both sub-group and serotype cross-reactive. (A) Cytolytic activity of adenovirus-specific, A*1-restricted clone 11 (E/T 20:1) against autologous and allogeneic fibroblast targets either alone or transduced with the indicated Ad serotypes. (B) Epitope alignment of sequences from the indicated Ad serotypes. (C) Hexon-specific, A*1-restricted T-cell response (clone 11 from donor AL) by ELISPOT assay. Stimulators are autologous LCLs pulsed with 1 μg/mL of the synthesized peptides, designated by the first 3 amino acids of the peptide sequence. Results are expressed as SFCs per 1 × 104 T cells.
To identify the HLA-A*1–restricted, cross-reactive epitope recognized by this A*1-restricted clone, we used 2 T-cell epitope prediction algorithms. Because the clone was (1) hexon specific, (2) HLA-A*1 restricted, and (3) subgroup cross-reactive, we expected that the epitope sequence would be highly conserved among Ad serotypes. The hexon sequence from Ad5 was chosen as the reference sequence and was applied to the prediction programs BIMAS and SYFPEITHI. Hexon sequences from 4 other serotypes (Ad2 [subgroup C], Ad7 and Ad11 [subgroup B], and Ad4 [subgroup E]) were aligned with the reference sequence, and the conserved predicted peptides were chosen for further study. For example, one peptide shown in Figure 3B contained the preferred A*1 binding motif even though the sequence was not absolutely conserved among the serotypes. On the basis of serotype alignment combined with the epitope prediction programs, a total of 15 peptides were synthesized (Table 1) and tested by ELISPOT assay.
Amino acid position . | Peptide sequence . |
|---|---|
| A*1 peptides* | |
| aa 70-78 | PVDREDTAY |
| aa 72-80 | DREDTAYSY |
| aa 93-101 | VLDMASTSF |
| aa 226-234 | KTTPMKPCY |
| aa 287-295 | YSEDVDIET |
| aa 294-302 | ETPDTHISY |
| aa 318-326 | QSMPNRPNY |
| aa 405-413 | GTEDELPNY |
| aa 479-487 | LYLPDKLKY |
| aa 573-581 | LLLPGSYTY |
| aa 638-646 | TNDQSFNDY |
| aa 699-707 | YTYSGSIPY |
| aa 886-894 | TDLGQNLLY |
| aa 906-914 | EVDPMDEPT |
| aa 909-917 | PMDEPTLLY |
| A*24 peptides† | |
| aa 37-45 | TYFSLNNKF |
| aa 369-377 | SYQLLLDSI |
| aa 393-401 | SYDPDVRII |
| aa 473-481 | LYSNIALYL |
| aa 471-479 | NFLYSNIAL |
| aa 527-535 | DYMDNVNPF |
Amino acid position . | Peptide sequence . |
|---|---|
| A*1 peptides* | |
| aa 70-78 | PVDREDTAY |
| aa 72-80 | DREDTAYSY |
| aa 93-101 | VLDMASTSF |
| aa 226-234 | KTTPMKPCY |
| aa 287-295 | YSEDVDIET |
| aa 294-302 | ETPDTHISY |
| aa 318-326 | QSMPNRPNY |
| aa 405-413 | GTEDELPNY |
| aa 479-487 | LYLPDKLKY |
| aa 573-581 | LLLPGSYTY |
| aa 638-646 | TNDQSFNDY |
| aa 699-707 | YTYSGSIPY |
| aa 886-894 | TDLGQNLLY |
| aa 906-914 | EVDPMDEPT |
| aa 909-917 | PMDEPTLLY |
| A*24 peptides† | |
| aa 37-45 | TYFSLNNKF |
| aa 369-377 | SYQLLLDSI |
| aa 393-401 | SYDPDVRII |
| aa 473-481 | LYSNIALYL |
| aa 471-479 | NFLYSNIAL |
| aa 527-535 | DYMDNVNPF |
Fifteen potential hexon-specific peptides were synthesized to identify the peptide epitope presented in the context of the A*1 allele.
Six potential hexon-specific peptides were synthesized to identify the peptide epitope presented in the context of the A*24 allele.
We pulsed autologous LCLs with these peptides and used them as stimulators in an ELISPOT assay with individual clones as responders. The results for clone 11, which was first screened against 10 of the 15 synthesized peptides, are shown in Figure 3C. Responses were detected against the TDLGQNLLY (TDL) peptide, amino acids (aa) 886 to 894, which was predicted only by the SYFPEITHI program. Subsequently, all other A*1-restricted clones were tested in the ELISPOT assay and all responded to the TDL peptide.
Epitope mapping of HLA A*24-restricted adenovirus-specific clone 16
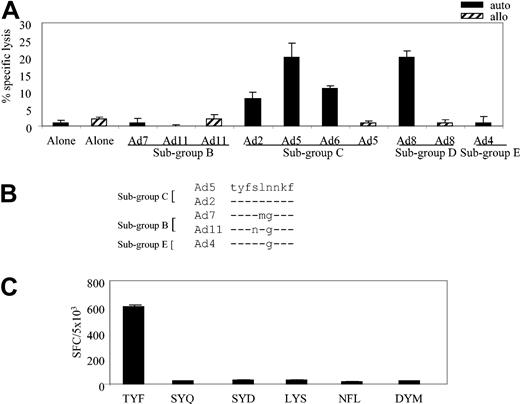
To identify the hexon-specific epitope recognized in the context of the A*24 allele, we first determined whether clone 16 could recognize autologous fibroblasts infected with our panel of Ad serotypes: Ad7 and Ad11 (subgroup B); Ad2, Ad5, and Ad6 (subgroup C); Ad8 (subgroup D); and Ad4 (subgroup E). Controls were allogeneic HLA-mismatched fibroblasts, either alone or infected with Ad5, Ad8, and Ad11. The results obtained with clone 16, at an E/T of 20:1, are shown in Figure 4A, which demonstrates recognition and killing of subgroup C (Ad2, Ad5, and Ad6) and subgroup D (Ad8) serotypes. By contrast, neither the allogeneic targets nor the autologous fibroblasts alone were recognized by clone 16; nor was there any recognition of autologous targets infected with Ad7 or Ad11 (subgroup B) or Ad4 (subgroup E).
HLA-A*24–restricted adenovirus-specific T-cell clone 16 is subgroup C and D specific. (A) Cytolytic activity of adenovirus-specific, A*24-restricted clone 16 (E/T 20:1), incubated with autologous and allogeneic fibroblast targets, either alone or transduced with the indicated Ad serotypes. (B) Alignment of peptide sequences from the indicated Ad serotypes. (C) Hexon-specific, A*24-restricted T-cell response (clone 16 from donor 1) by ELISPOT assay. Autologous LCL targets were pulsed with 1 μg/mL of the indicated peptides and then used as stimulators in an ELISPOT assay, with clone 16 T cells serving as responder cells. Results are expressed as SFCs per 5 × 103 T cells.
HLA-A*24–restricted adenovirus-specific T-cell clone 16 is subgroup C and D specific. (A) Cytolytic activity of adenovirus-specific, A*24-restricted clone 16 (E/T 20:1), incubated with autologous and allogeneic fibroblast targets, either alone or transduced with the indicated Ad serotypes. (B) Alignment of peptide sequences from the indicated Ad serotypes. (C) Hexon-specific, A*24-restricted T-cell response (clone 16 from donor 1) by ELISPOT assay. Autologous LCL targets were pulsed with 1 μg/mL of the indicated peptides and then used as stimulators in an ELISPOT assay, with clone 16 T cells serving as responder cells. Results are expressed as SFCs per 5 × 103 T cells.
To identify the epitope sequence, we concentrated on predicted peptides that were conserved within subgroup C viruses but were mutated in the other subgroups; one such peptide is shown in Figure 4B. Altogether, 6 peptides (Table 1) met these criteria and were synthesized for analysis by ELISPOT assay. Autologous LCLs were pulsed with 1 μg/mL of each peptide and used as stimulators. As shown in Figure 4C, specific IFN-γ release was detected only when the TYFSLNNKF (TYF) peptide, aa 37 to 45, was present. Thus, Ad cellular immunity is made of subgroup cross-reactive and subgroup C– and D-specific T cells.
Reactivity against the TDL and TYF epitopes in other HLA-A*1 and A*24 donors
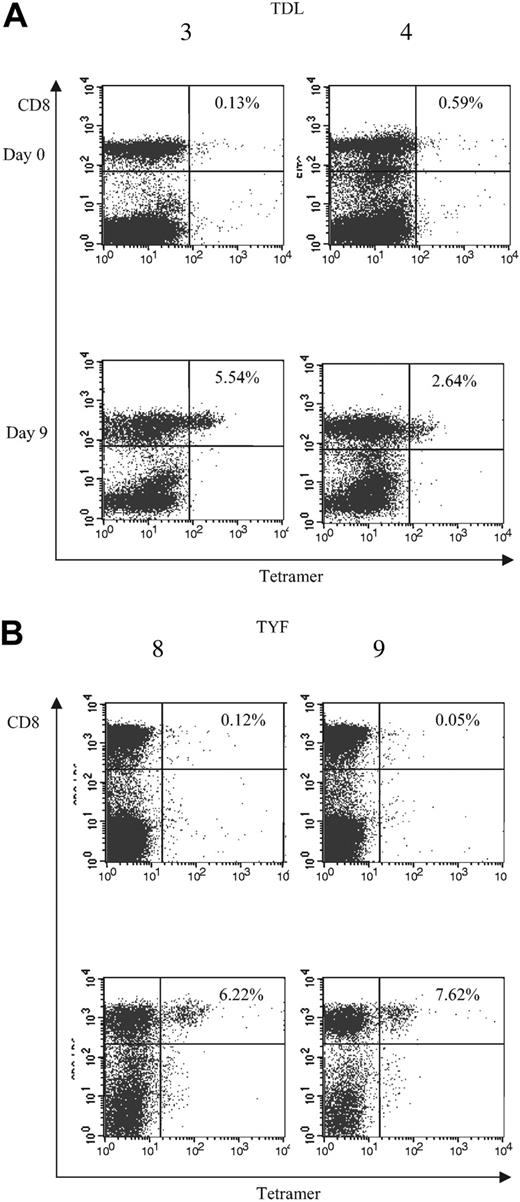
To determine whether the subgroup cross-reactive A*1-restricted and subgroup C– and D-specific A*24-restricted responses were frequently detected among adenovirus-seropositive donors, we generated TDL and TYF tetramers. The function of the tetramers was confirmed using the clones (Figure 5). We then analyzed reactivity of the tetramers with PBMCs and adenovirus-stimulated responder cells from 6 A*1- and 4 A*24-seropositive donors. An HLA-mismatched donor served as a negative control (data not shown). Figure 6A shows the tetramer analysis of 2 representative HLA-A*1–positive donors, 3 and 4. The frequency of tetramer-positive cells in PBMCs from 6 A*1-positive donors ranged from less than 0.1% to 0.6% of CD8+ cells, but after one stimulation with the Ad5f35 vector it increased to a range of 0.63% to 5.54%. ELISPOT assays done in parallel detected 0 to 86 SFCs per 106 PBMCs, which after one stimulation increased to 1325 to 9825 SFCs (Table 2). The TYF response was also detectable in all 4 A*24 donors screened by ELISPOT and tetramer analysis. Results for donors 8 and 9 are shown in Figure 6B. Tetramer-positive cells in PBMCs ranged from less than 0.1% to 0.23% of the total CD8+ cells but increased following one stimulation with Ad vector to a range of 2.64% to 7.62%. A corresponding increase in frequency was detectable by ELISPOT assay, with 7.5 to 32.5 measurable SFCs per 106 PBMCs, which after one stimulation increased to 1310 to 2630 IFN-γ spots per 106 cells (Table 2). Thus, in A*1 and A*24 donors, the TDL and TYF epitopes appear to be common targets.
Tetramer analysis of adenovirus-specific CD8+ T-cell clones, clone 11 and clone 16. TDL-specific CD8+ T-cell clone 15 and TYF-specific T-cell clone 1 from donor 1 were screened for the percentage of tetramer-positive T cells.
Tetramer analysis of adenovirus-specific CD8+ T-cell clones, clone 11 and clone 16. TDL-specific CD8+ T-cell clone 15 and TYF-specific T-cell clone 1 from donor 1 were screened for the percentage of tetramer-positive T cells.
Analysis of T-cell frequencies by tetramer. Frequencies of (A) TDL- and (B) TYF-specific T cells in PBMCs and CTL lines following one Ad stimulation. PBMCs (day 0) and CTL lines (day 9) from A*1-positive (3 and 4) and A*24-positive donors (8 and 9) were screened for the percentage of tetramer-positive T cells.
Analysis of T-cell frequencies by tetramer. Frequencies of (A) TDL- and (B) TYF-specific T cells in PBMCs and CTL lines following one Ad stimulation. PBMCs (day 0) and CTL lines (day 9) from A*1-positive (3 and 4) and A*24-positive donors (8 and 9) were screened for the percentage of tetramer-positive T cells.
. | SFCs per 106 cells . | . | |
|---|---|---|---|
. | PBMCs . | CTLs* . | |
| A*1 donor | |||
| 1 | 33 | 2760 | |
| 2 | 67 | 4400 | |
| 3 | 6 | 9825 | |
| 4 | 86 | 4237 | |
| 5 | 25 | 1862 | |
| 6 | 0 | 1325 | |
| A*24 donor | |||
| 1 | 7.5 | 2630 | |
| 7 | 32.5 | 1640 | |
| 8 | 7.5 | 1810 | |
| 9 | 10 | 1310 | |
. | SFCs per 106 cells . | . | |
|---|---|---|---|
. | PBMCs . | CTLs* . | |
| A*1 donor | |||
| 1 | 33 | 2760 | |
| 2 | 67 | 4400 | |
| 3 | 6 | 9825 | |
| 4 | 86 | 4237 | |
| 5 | 25 | 1862 | |
| 6 | 0 | 1325 | |
| A*24 donor | |||
| 1 | 7.5 | 2630 | |
| 7 | 32.5 | 1640 | |
| 8 | 7.5 | 1810 | |
| 9 | 10 | 1310 | |
Responses detected after a single stimulation with the Ad5f35 vector.
Identification of additional hexon-specific CD8+ T-cell epitopes
Because hexon appears to be a strong target for adenovirus-specific CD8+ T cells, we synthesized a panel of peptides (20 mers overlapping by 15 amino acids [aa]) covering the 952 aa unique sequence of the hexon serotype 5 protein. These peptides were then used to screen CTL lines and T-cell clones generated from 3 seropositive donors (donor 11, HLA-A*2, A*3, B*7, B*44; donor 7, HLA-A*24, B*7, B*40; and donor 10, HLA-A*2, B*8, B*57) in an effort to identify additional epitopes. To minimize the size of the initial screening we used 63 pools, each containing 3 adjacent peptides from the hexon sequence, an approach already established for identifying both CD8 and CD4 T-cell epitopes from a variety of different proteins.37,39-41 Once reactivity was detected within a peptide pool, each of the lines was rescreened against the individual 20 mers from the relevant pools and against shorter peptides from the same region. This method revealed an A*2-restricted peptide, TFYLNHTFKKV (TFY), aa 711 to 721, identified in donor 10 and two B*7-restricted peptides, KPYSGTAYNSL(KPY), aa 114 to 124, and MPNRPNYIAF (MPN), aa 320 to 329, identified in donors 11 and 7 (Table 3). To determine whether these epitopes were cross-reactive, we assessed the ability of MPN-specific c32 (Figure 7A) and KPY-specific c12 (Figure 7B) to kill autologous fibroblast targets infected with a cross-section of Ad serotypes representing 3 subgroups: Ad3, Ad7, and Ad11 (subgroup B); Ad1, Ad2, Ad5, and Ad6 (subgroup C); and Ad4 (subgroup E). Controls were allogeneic HLA-mismatched fibroblasts either uninfected or infected with Ad5. At an E/T of 20:1 the clones showed cross-reactive properties, recognizing autologous targets infected with each of the Ad serotypes (Figure 7A-B) while failing to react appreciably with allogeneic targets or autologous fibroblasts alone.
CD8+ hexon epitope
Amino acids . | HLA restriction . | Epitope sequence . |
|---|---|---|
| 37-45 | A*24 | TYFSLNNKF |
| 114-124 | B*7 | KPYSGTAYNSL |
| 320-329 | B*7 | MPNRPNYIAF |
| 711-721 | A*2 | TFYLNHTFKK |
| 886-894 | A*1 | TDLGQNLLY |
Amino acids . | HLA restriction . | Epitope sequence . |
|---|---|---|
| 37-45 | A*24 | TYFSLNNKF |
| 114-124 | B*7 | KPYSGTAYNSL |
| 320-329 | B*7 | MPNRPNYIAF |
| 711-721 | A*2 | TFYLNHTFKK |
| 886-894 | A*1 | TDLGQNLLY |
Cross-reactivity and frequency of HLA-B*7–restricted adenovirus-specific T-cell responses. (A) Cytolytic activity of adenovirus-specific, MPN-specific B*7-restricted clone 32 (E/T 20:1) against autologous and allogeneic fibroblast targets either alone or transduced with the indicated Ad serotypes. (B) Cytolytic activity of adenovirus-specific, KPY-specific B*7-restricted clone 12 (E/T 20:1) against autologous and allogeneic fibroblast targets either alone or transduced with the indicated Ad serotypes. (C) PBMCs (day 0) and CTL lines (day 9) from B*7-positive donors (6 and 12) were screened for the percentage of KPY-specific tetramer-positive T cells.
Cross-reactivity and frequency of HLA-B*7–restricted adenovirus-specific T-cell responses. (A) Cytolytic activity of adenovirus-specific, MPN-specific B*7-restricted clone 32 (E/T 20:1) against autologous and allogeneic fibroblast targets either alone or transduced with the indicated Ad serotypes. (B) Cytolytic activity of adenovirus-specific, KPY-specific B*7-restricted clone 12 (E/T 20:1) against autologous and allogeneic fibroblast targets either alone or transduced with the indicated Ad serotypes. (C) PBMCs (day 0) and CTL lines (day 9) from B*7-positive donors (6 and 12) were screened for the percentage of KPY-specific tetramer-positive T cells.
To analyze the frequency and the magnitude of the KPY-specific response in other B*7 donors, we generated a KPY-specific tetramer and confirmed its function using a clone from donor 11 (data not shown). We then analyzed reactivity of the KPY tetramer with PBMCs and adenovirus-stimulated responder cells from 5 B*7-seropositive donors. An HLA-mismatched donor served as a negative control (data not shown). The frequency of tetramer-positive cells in PBMCs from 5 B*7-positive donors screened ranged from 0% to 0.04% of CD8+ cells, but after one stimulation with Ad5f35 vector it increased to a range of 1.64% to 21.06%. Figure 7C shows results using 2 representative HLA-B*7–positive donors, 6 and 12.
Discussion
Although it has been suggested that T cells specific for adenoviruses can cross-react with different Ad serotypes, cross-reactive CD8+ epitopes have not been identified.14-16 Because the molecular basis for this putative cross-reactivity was unknown, it has not been clear whether T-cell immunity is due to recognition of multiple cross-reactive epitopes or to a combination of type-specific and type cross-reactive responses. We addressed this question by isolating single-cell CD8+ T-cell clones from adenovirus-specific T-cell lines and identifying their antigen and epitope specificity. This strategy revealed 5 hexon-specific, CD8+ T-cell epitopes, the first HLA class I–restricted epitopes to be reported for adenoviruses. Four epitopes restricted by HLA-A*1, HLA-A*2, and HLA-B*7 were processed and presented in a subgroup cross-reactive manner, while the remaining epitope, recognized in the context of HLA-A*24, was specific for viruses within subgroups C and D. Importantly, peptide-specific responses were detectable by tetramer and ELISPOT assay in 6 of 6 A*1-, 4 of 4 A*24-, and 5 of 5 B*7-seropositive donors screened for TDL, TYF, and KPY responses, respectively, suggesting that (1) these epitopes are dominant components of adenovirus-specific immunity, and (2) cellular immunity to adenoviruses is characterized by subgroup cross-reactive responses that may be therapeutically exploitable.
The ability of T cells to cross-react among different viral strains has previously been described for influenza virus, where repeated in vitro stimulations with naturally occurring viral variants resulted in the expansion of influenza A nucleoprotein (NP)–specific T-cell lines that were cross-reactive among different viral strains.42 More recently, a report of T-cell epitope conservation among vaccinia and variola viruses suggested an analogous phenomenon among poxviruses.43 However, this report is the first to describe the coexistence of 2 populations of cross-reactive CD8+ T cells within a single viral system—ie, those that recognize 2 subgroups in parallel with those that recognize at least 4 subgroups. The adenovirus-specific memory T-cell pool likely reflects the frequency of exposure to a particular subgroup relative to others and may offer broad protection against all possible infections.42,44 Whether the pattern of differential cross-reactivity seen in the adenovirus system also exists in other viruses is unknown but may well be a unique feature of adenoviruses, which stably maintain a large variety of different serotypes and have a temporal infection pattern allowing for the maintenance of subgroup cross-reactive immunity.4,5
To identify the epitopes recognized by the hexon-specific A*1-, A*2-, B*7-, and A*24-resticted clones generated in this study, we used predictive algorithms and hexon sequence alignments of Ad serotypes from subgroups B, C, and E. Analysis of the resultant A*1 peptides identified TDLGQNLLY as the epitope recognized by our A*1-restricted clones. Alignment of the TDL peptide sequences showed a leucine (l) to methionine (m) change at position 7 in subgroup B and E viruses, but this peptide still conformed to the A*1 binding motif,45 while the TCR-binding elements remained identical. Further sequence analysis indicated that this also held true for subgroups A and F. Although the hexon sequence was not available for every adenovirus serotype—for example, Ad8 (subgroup D)—cells infected by this wild-type virus were killed in a cytotoxicity assay, implying that the A*1 motif is also conserved within Ad8. Similar methodology was utilized to identify the HLA-A*2–restricted TFY epitope and the HLA-B*7–restricted KPY and MPN epitopes. The TFY peptide is completely conserved between all serotypes and subgroups. The KPY peptide is highly conserved between viruses from subgroups A to F except for a serine (s) at position 10 in all subgroups excluding C. In subgroup C viruses the MPN epitope has an alanine (a) at position 9, which is mutated to a glycine (g) at this position in all other subgroups, and at position 1 representative adenoviruses from subgroups A and F have an alanine (a) rather than a methionine (m). Nevertheless, the KPY- and MPN-specific clones were capable of recognizing serotypes from subgroups B, C, and E in a cytotoxicity assay (Figure 7). The A*24 TYF epitope was highly conserved (100%) among the subgroup C viruses but mutated in all serotypes within representative adenoviruses from subgroups A to F, excluding D. Although the hexon sequence was not available for Ad8, cells infected with this wild-type virus were killed in a cytotoxicity assay, implying that the TYF sequence is conserved between C and D adenoviruses.
A major question raised by our findings was whether broadly cross-reactive T cells are more common than those directed against only 2 subgroups. We therefore analyzed the frequency of T-cell responses to the TDL and TYF epitopes in A*1- and A*24-seropositive donors, respectively. The broadly cross-reactive TDL and subgroup C– and D-specific TYF responses were detected by ELISPOT and tetramer analysis in 6 of 6 and 4 of 4 A*1 and A*24 donors, respectively, with similar frequencies (Figure 6). These T cells can be expanded rapidly following antigen stimulation, confirming that these are true memory responses. Interestingly, the magnitude of these responses, detected after one stimulation with antigen, is comparable to that seen against immunodominant T-cell epitopes recognized by donors chronically infected with cytomegalovirus (CMV), suggesting either that T-cell immunity against acutely infecting viruses can be as strong as T-cell memory to herpesviruses that persist life-long, and that provide constant stimulation to circulating T cells, or may reflect the long-term persistence of adenoviruses.35
The sequence conservation of the cross-reactive epitopes suggests that they are located in areas of the viral genome crucial to survival and/or function. Indeed, the TYF peptide resides within the N-terminal region of the hexon protein, an area important for structural stability. The TDL and TFY peptides map to the V2 domain, and the KPY and MPN peptides map to the V1 domain; the V1 and V2 domains are 2 of the components making up the hexon base (Supplementary Figure 1, available at the Blood website; see the Supplemental Figure link at the top of the online article). These regions are highly conserved among adenoviruses from different species because they are in areas of the molecule likely important for capsid assembly, structure, and infectivity.46 By contrast, the serotype-specific B-cell epitopes are probably located in hypervariable regions (HVRs) of the hexon molecule, occurring in loops that lie on the exterior of the virion.47 It is at these HVRs that the hexon structure is amenable to specific alteration, but any changes in the core N-terminal V1 and V2 domains may have profound detrimental effects on capsid assembly.48 Thus, our results have implications for the use of Ad vectors in gene therapy. One of the limitations of these vectors is that they induce potent humoral and cellular immune responses, which reduce the initial effectiveness of gene transfer and the survival of the transduced cells. Studies in mice, whose cellular immune response to human adenoviruses is dominated by the immediate early proteins E1A and E1B,23,49 have indicated that one way of evading this response is to delete these genes, while humoral immunity detected mainly against the capsid proteins may be overcome by repeat gene delivery using vectors derived from several different subgroups of viruses.50 Our study suggests that such an option may not translate to humans, in whom a significant portion of the cellular immune response is directed against the conserved hexon-derived viral domain likely critical for viral assembly.
The cross-reactivity of CTL responses to adenoviruses has promising implications for immunotherapy of adenovirus-related complications in immunocompromised patients. Thus, CTLs raised against one serotype may be effective in vivo against infections caused by most other serotypes. Whether these cross-reactive CD8+ T cells will be protective against adenovirus in vivo remains to be addressed, particularly because a number of in vitro studies have found that CD4+ T cells form the bulk of the cellular immune response. Nevertheless, tetramer studies indicate that the newly identified adenovirus-specific CD8+ T-cell epitopes may form a substantial portion of the immune response to adenoviruses and should therefore also be useful for monitoring adenovirus-specific responses in vivo, in immunocompromised patients at risk for disease, and following adoptive transfer of adenovirus-specific CTL lines.
In summary, we have shown that adenovirus-specific T-cell immunity can be cross-reactive and that the abundant capsid protein, hexon, is a preferred target for specific T cells. Thus, human T-cell immunity to adenoviruses may predominantly target a small number of cross-reactive epitopes, whose sequences are critical for virus assembly and function and are therefore precluded from mutation as a viral immune escape mechanism. If this suggestion is correct, establishing adoptive immunotherapy for adenovirus infections may be less laborious than the range of subgroups suggests; conversely, development of effective and safe gene transfer with Ad vectors may be more challenging than originally hoped.
Prepublished online as Blood First Edition Paper, July 20, 2004; DOI 10.1182/blood-2004-02-0646.
Supported by grants from the Methodist Foundation and the Southwest Texas Fund (A.M.L.), grants from the National Cancer Institute (NIH1RO1 CA61384) and the Dana Foundation (C.M.R.).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Helen Huls for excellent technical assistance and Tatiana Goltsova for flow cytometric analysis. PAdeasy1 vector was kindly provided by Tong-Chuan He and Bert Vogelstein, and MoMLV-gagpol was provided by Patrick Kelly.

![Figure 2. Clone HLA restriction and antigen specificity. (A) Adenovirus-specific CD8+ T-cell clones from donor AL are A*1 and A*24 restricted. Adenovirus-specific CD8 T-cell clones c11 and c16 were incubated with autologous and partially HLA class I–matched LCL targets infected with Ad5f35GFP; only shared alleles are indicated. Results are expressed as SFCs per 100 T cells. (B) In vitro–reactivated adenovirus-specific T-cell clones recognize a capsid protein, hexon. (i) A1-restricted clone 11 tested at E/T ratios of 5:1 and 2.5:1. (ii) A*24-restricted clone 16 tested at E/T ratios of 30:1, 20:1, and 10:1. Killing was assessed against autologous fibroblast targets alone or fibroblasts transduced with the MSCV–Hexon–internal ribosome entry site [IRES]–GFP viral supernatant (retro-Hexon) or with MSCV-Penton-IRES-GFP viral supernatant (retro-Penton). Error bars reflect standard deviation of triplicates.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/8/10.1182_blood-2004-02-0646/6/m_zh80200468200002.jpeg?Expires=1769135692&Signature=lO265DU3up5K9a3xwY-1k85uTG6dcnkiyBDGBCMCKdFRt0fRM1n34crpiYxiOFes~NLlvXIAqYFnX4dmG3nPAYZa5K7Cc7t9beYNXrfdNR69J7ckh6FTcpKSE~1Ybw14n6GtHffAbeHlkHH0S7GYzAsSJOhw4xkHdMcM4uGGdqmWjjyJ6HlU7hAAElwZ5kbrB8MgyVKdw0ic-wCxCp4lbve1GdYdKvJi8Ct2LvwskimKExuTFvpc8X5DHpGOYJIfJE9wRO8GiMW38RWtpIte4f6S1Sfl~iNLjPgkTboivNmykHpSJITsYL-fJHxnYiGrGw-~CG3NDgwK4N4zw6YX2A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal