Abstract
Complete interleukin-12/interleukin-23 receptor β1 (IL-12Rβ1) deficiency is the most frequent known genetic etiology of the syndrome of Mendelian susceptibility to mycobacterial disease. The patients described to date lack IL-12Rβ1 at the surface of their natural killer (NK) and T cells due to IL12RB1 mutations, which either interrupt the open reading frame or disrupt protein folding. We describe a patient with a large in-frame deletion of 12165 nucleotides (nt) in IL12RB1, encompassing exons 8 to 13 and resulting in the surface expression of nonfunctional IL-12Rβ1. These 6 exons encode the proximal NH2-terminal half of the extracellular domain downstream from the cytokine-binding domain. Five of 6 monoclonal anti-IL-12Rβ1 antibodies tested recognized the internally truncated chain on the cell surface. However, IL-12 and IL-23 did not bind normally to the patient's IL-12Rβ1-containing respective heterodimeric receptors. As a result, signal transducer and activator of transcription-4 (STAT4) was not phosphorylated and interferon-γ (IFN-γ) production was not induced in the patient's cells upon stimulation with even high doses of IL-12 or IL-23. The functional defect was completely rescued by retrovirus-mediated IL-12Rβ1 gene transfer. Thus, the detection of IL-12Rβ1 on the cell surface does not exclude the possibility of complete IL-12Rβ1 deficiency in patients with mycobacteriosis or salmonellosis. Paradoxically, the largest IL12RB1 mutation detected is associated with the cell surface expression of nonfunctional IL-12Rβ1, defining a novel genetic form of IL-12Rβ1 deficiency. (Blood. 2004;104:2095-2101)
Introduction
Mendelian susceptibility to mycobacterial disease (MSMD) (Mendelian Inheritance in Man, MIM209950; Online Mendelian Inheritance in Man [OMIM]: http://www.ncbi.nlm.nih.gov/Omim/)1 is a rare syndrome predisposing affected individuals to infectious diseases caused by poorly virulent mycobacteria, such as bacille Calmette-Guérin (BCG) vaccines and environmental mycobacteria (EM), and poorly virulent Salmonella strains, such as nontyphoidal “minor” serovars. Patients are also susceptible to infections caused by the more virulent Mycobacterium tuberculosis and typhoidal “major” Salmonella serotypes.1,2 Unlike patients with “classic” immunodeficiencies, these patients are otherwise quite healthy and only rarely suffer from other unusually severe bacterial, viral, fungal, or parasitic diseases.2,3 The spectrum of infections is narrow, but the spectrum of severity is broad—from disseminated BCG disease in infancy to localized environmental mycobacterial disease in the elderly. Moreover, whereas some sporadic and most familial cases seem to involve autosomal recessive heredity, the syndrome has been found to segregate in an autosomal dominant4,5 or X-linked recessive6 pattern in other families, further suggesting genetic heterogeneity.
Five disease-causing autosomal genes have been identified since 1996,7,8 and allelic heterogeneity accounts for the existence of 9 defined disorders, all of which result in impaired interferon-γ (IFN-γ)-mediated immunity.1,2 Null recessive mutations in the IFN-γ receptor ligand-binding chain (IFN-γR1)-encoding gene (IFNGR1) abolish either receptor expression7,8 or the binding of surface-expressed receptors to IFN-γ.9,10 Partial recessive11 and dominant4 IFN-γR1 deficiencies have also been described. Different recessive mutations in the gene encoding the IFN-γ signaling chain (IFN-γR2), IFNGR2, are responsible for complete12 or partial13 IFN-γR2 deficiency. A dominant mutation in STAT1 is responsible for partial signal transducer and activator of transcription-1 (STAT-1) deficiency and defines the remaining disease in which cellular responses to IFN-γ are impaired.5 Complete recessive STAT-1 deficiency is a related but distinct disorder involving susceptibility to both mycobacteria and viruses, due to the impairment of IFN-γ- and IFN-α/β-mediated immunity.14
In about half of all patients with MSMD and a well-defined genetic disorder, cellular responses to IFN-γ are normal, but the interleukin-12 (IL-12)- and interleukin-23 (IL-23)-dependent production of IFN-γ is severely impaired. Nineteen children homozygous for null mutations in IL12B, encoding the p40 subunit of IL-12 and IL-23, have been identified.15-17 Null recessive IL12RB1 mutations have been identified in 54 other patients with IL-12 and IL-23 receptor β1 chain deficiency.17-28 Patients with IL-12 p40 and IL-12Rβ1deficiency share a number of common clinical characteristics: low penetrance of genetic susceptibility to mycobacteriosis and salmonellosis; high proportion of extraintestinal salmonellosis among symptomatic patients; broad resistance to other microorganisms; and a favorable clinical outcome.29,30 From a molecular point of view, 53 of 54 known patients with complete IL-12Rβ1 deficiency17-26,28 have no detectable IL-12Rβ1 on the cell surface, due to mutations that either interrupt the open reading frame (ORF) (nonsense and frameshift mutations) or disrupt folding of the protein (missense mutations). We report here the molecular investigation of a patient with complete IL-12Rβ1 deficiency despite the presence of IL-12Rβ1 at the cell surface.
Patients, materials, and methods
The patient
The patient (P) is a 6-year-old boy born to first-cousin parents of Bedouin origin living in Israel (Figure 1). He was not inoculated with BCG and was first seen at the age of 12 months with disseminated Salmonella enteritidis disease (septicemia and multiple adenitis). Between the ages of 1 and 3 years, the patient suffered 8 recurrences of systemic Salmonella infection, with the same serovar implicated on each occasion. The detailed clinical and bacteriologic features of these infections have been reported elsewhere,27 and this patient was patient 10.II.3 in a previous study26 in which his genotype was described. The negative control (C-) used in this study was also previously described (patient 20.II.126 ) and is homozygous for a nonsense mutation resulting in a premature stop in the ORF (Q32X).
A large in-frame deletion in IL12RB1. (A) Schematic representation of the wild-type IL-12Rβ1 chain containing 17 coding exons (Arabic numerals) encoding 662 amino acids, with a peptide leader sequence (L), extracellular domain (exons 2 to 13, EC), transmembrane domain (exon 14, TM), and an intracellular, cytoplasmic domain (exons 15 to 17, IC). The mutation found in P is also indicated (700 + 362_1619-944 del). The mature IL-12Rβ1 chain contains 5 fibronectin III (FNIII) domains shown in the bottom row in light gray. (B) Schematic representation of the mutant protein, lacking the sequences encoded by 6 of the exons (8 to 13) in the wild-type gene. The mutant protein contains 356 amino acids and only the first 2 FNIII domains of the extracellular domain but has intact transmembrane and intracellular domains. In the family tree, the patient is indicated by an arrow.
A large in-frame deletion in IL12RB1. (A) Schematic representation of the wild-type IL-12Rβ1 chain containing 17 coding exons (Arabic numerals) encoding 662 amino acids, with a peptide leader sequence (L), extracellular domain (exons 2 to 13, EC), transmembrane domain (exon 14, TM), and an intracellular, cytoplasmic domain (exons 15 to 17, IC). The mutation found in P is also indicated (700 + 362_1619-944 del). The mature IL-12Rβ1 chain contains 5 fibronectin III (FNIII) domains shown in the bottom row in light gray. (B) Schematic representation of the mutant protein, lacking the sequences encoded by 6 of the exons (8 to 13) in the wild-type gene. The mutant protein contains 356 amino acids and only the first 2 FNIII domains of the extracellular domain but has intact transmembrane and intracellular domains. In the family tree, the patient is indicated by an arrow.
Our study was approved by the Institutional Review Board of the Université de Paris René Descartes. Informed consent was obtained from the patient's family according to the Declaration of Helsinki.
Cell culture and stimulation
Epstein-Barr virus-transformed lymphoblastoid cell lines (EBV-B cell lines) were cultured as previously described.16 Peripheral blood mononuclear cells (PBMCs) were cultured in RPMI 1640 supplemented with 10% heat-inactivated pooled human AB serum and activated by incubation with phytohemagglutinin-P (PHA) (Bacto, Becton Dickinson, Heidelberg, Germany) for 72 hours to generate PHA-activated T cells. PHA-T-cell blasts were restimulated every 48 hours with IL-2 (40 IU/mL) (Chiron, Amsterdam, The Netherlands) and cultured in Panserine 401 (Pan Biotech, Aidenbach, Germany) with 10% heat-inactivated pooled human AB serum and 2 mM l-glutamine. For cytokine stimulation, we plated 0.5 × 106 PHA-T-cell blasts in complete medium in each well of a 48-well plate on day 6 and added IL-23 and IL-12p70 (both from R&D Systems, Minneapolis, MN) at various concentrations to a final volume of 500 μL. As a positive control for activation, PHA-T-cell blasts were stimulated with 10-7 M phorbol myristate acetate (PMA) (Sigma-Aldrich, St Louis, MO) and 10-5 M ionomycin. Supernatants were harvested after 48 hours.
ELISA and cell surface flow cytometry
Cell culture supernatants were assayed for IFN-γ by enzyme-linked immunosorbent assay (ELISA), according to the kit manufacturer's recommendations (Pelikin Compact, CLB, Amsterdam, The Netherlands). IFN-γ concentration was calculated per 1 million PHA-T-cell blasts. For flow cytometry, PHA-T-cell blasts and/or EBV-transformed B cells were first incubated with an IL-12Rβ1-specific mouse immunoglobulin G1 (IgG1) monoclonal antibody (mAb) (24E6), an IL-12Rβ1-specific rat IgG2a mAb (2B10), or matched isotypic control mAbs; then with a biotinylated rat anti-mouse Ab or a biotinylated mouse anti-rat Ab; and finally with streptavidin-phycoerythrin (streptavidin-PE) (all reagents were from Pharmingen, San Diego, CA). Mouse antibodies B101, B103, and 12RB44 were all generously provided by the Genetics Institute (Andover, MA). One additional commercial mAb—an anti-human IL-12Rβ1 mAb (clone 69310 coupled to R-phycoerythrin from R&D Systems)—and a matched isotype control were tested for IL-12Rβ1 staining. The cells were fixed by incubation in 4% paraformaldehyde for 30 minutes and were then stained. All washing and incubation steps were performed in the presence of 0.1% saponin (Sigma-Aldrich). Signals were analyzed with a FACScan and the Cellquest software (Becton Dickinson Immunocytometry Systems, San Jose, CA).
Fluorescent IL-12/IL-23 binding and phospho-STAT4 detection
IL-12p70 or IL-23 fluorescence binding experiments were performed as follows: 400 000 day 6 PHA-T-cell blasts were incubated in 20 μL phosphate-buffered saline (PBS) with (or without) 50 ng IL-12p70 or 100 ng recombinant human IL-23 (rhIL-23) (R&D Systems) for 30 minutes at 4°C and then with mouse anti-IL-12p40-p70 IgG1, biotinylated rat anti-mouse IgG1, and finally with streptavidin-PE (all reagents and antibodies were from Pharmingen). Phospho-STAT4 detection by flow cytometry was adapted from Uzel et al31 : PHA-T-cell blasts were either left unstimulated or stimulated by IFN-α (105 U/mL during 30 minutes) or IL-12 (100 ng/mL during 15 minutes) at 37°C. Cells were then fixed with 4% paraformaldehyde (PFA) in PBS, followed by 100% methanol fixation while vortexing, permeabilized with saponin, and stained with rabbit polyclonal anti-STAT4 Ab or rabbit polyclonal antiphospho-STAT4 Ab (both from Zymed, South San Francisco, CA) (or matched isotype control), followed by goat anti-rabbit Alexa Fluor 488 (Molecular Probes, Eugene, OR). Signals were analyzed with a FACScan using Cellquest software (Becton Dickinson).
Retroviral-mediated gene transfer
The retroviral vector, MND-IL-12Rβ1 (myeloproliferative sarcoma virus enhancer, negative control region deleted, dl587 rev primer-binding site substituted), was constructed using the MND-X-IRES-EGFP vector (a gift from Dr D. B. Kohn, Children's Hospital, Los Angeles, CA) and by replacing the internal ribosome entry site-enhanced green fluorescent protein (IRES-EGFP) fragment with human IL-12Rβ1 cDNA (gift from Dr J. J. O'Shea, National Institute of Arthritis and Musculoskeletal and Skin Diseases [NIAMS], National Institutes of Health [NIH], Bethesda, MD). Infectious retroviral particles were generated using the PG13 cell line32 as previously described.33 Retroviral supernatant stocks were produced by incubating producer cells in Dulbecco modified Eagle medium (DMEM) (Life Technologies, Bethesda, MD), 10% fetal bovine serum (FBS) for 72 hours at 32°C. PHA-T-cell blasts (1 × 106/mL) were incubated for 24 hours in fibronectin-coated plates (20 μg/mL Retronectin, Takara Bio, Shiga, Japan) preloaded with retroviral supernatant. The transduction procedure was repeated the following day. After 48 to 72 hours, cells were stained with anti-human IL-12Rβ1 (24E6 or 2B10), stimulated with IFN-α and IL-12, followed by intracellular flow cytometry phospho-STAT4 detection or stimulated with increasing doses of IL-12. In this last case, supernatants were harvested after 48 hours, and IFN-γ was measured by ELISA (hIFN-γ Quantikine kit; R&D Systems).
DNA and RNA extraction, cDNA synthesis, and PCR amplification
Genomic DNA and total RNA were extracted from EBV-transformed B cells or T-cell blasts as previously described.16 RNA was reverse transcribed in the presence of oligo(dT) with Superscript II reverse transcriptase (Invitrogen Life Technologies, Paisley, United Kingdom).16 The IL12RB1 cDNA, coding exons, and flanking intron regions were amplified using pairs of primers and polymerase chain reaction (PCR) conditions available in Table S1 of the supplementary material (at the Blood website, see the Supplemental Table link at the top of the online article).
Sequencing
PCR was carried out with pairs of intron primers flanking each IL12RB1 exon, under conditions available upon request. PCR products were sequenced by dideoxynucleotide termination with nested primers (Table S1) and the ABI PRISM dGTP BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Courtaboeuf, France). PCR products were sequenced on an ABI Prism 3100 apparatus and analyzed with Sequencing Analysis software (Applied Biosystems).
Results
A large in-frame deletion in IL12RB1
We previously reported a patient for whom we were unable to amplify exons 8 to 13 of IL12RB1 (10.II.2 26 ). The sequencing of introns 7 and 13 made it possible to identify the genomic breakpoints of a large deletion of 12165 nucleotides (12165 nt), and the mutation was designated 700 + 362_1619-944 del 34 (the deletion occurred beginning 362 nucleotides [nt] 3′ of exon 7, which ends at nt 700, and ending 944 nucleotides 5′ of nt 1619, which is the first nucleotide of exon 14). The patient was homozygous for this mutation, inherited from his 2 heterozygous parents. No other mutations of IL12RB1 were found. Amplification of the IL12RB1 ORF from cDNAs produced from both an EBV-transformed B-cell line and PHA-T-cell blasts yielded a fragment of lower molecular weight than was obtained for the control (not shown). Sequencing showed that exon 7 was directly spliced to exon 14 (Figure 1). Exon 1 encodes the signal peptide, and exon 14 encodes the IL-12Rβ1 transmembrane domain. The aberrant mRNA detected is in frame, contains no novel codon, and is predicted to result in the production of a 356-amino acid protein with an internally truncated extracellular domain but intact transmembrane and intracellular domains. In comparison, the wild-type (WT) protein contains 662 amino acids (Figure 1). The putative mature mutant protein (following cleavage of the signal peptide) would thus lack 306 (59%) of the 521 extracellular amino acids and sequences corresponding to 6 of the 12 exons encoding the mature extracellular domain.
Detection of an IL-12Rβ1 chain by intracellular staining
By Northern blot analysis, we detected a transcript with a lower molecular weight than the WT transcript, although their molecular amounts as determined by PhosphorImager (Molecular Dynamics, Sunnyvale, CA) quantification were equal (not shown). Because the deletion was in frame, we first tried to detect a mutant receptor chain by intracellular flow cytometry (fluorescence-activated cell sorter [FACS]) analysis. Staining of day 5 PHA-T-cell blasts with the mouse IgG1 anti-IL-12Rβ1 mAb 24E6 resulted in the detection of an intracellular chain in P, although staining was less intense than in the positive control (C+), with this chain not detected in cells from the negative control (C-) (not shown). Staining with rat IgG2a anti-IL-12Rβ1 mAb 2B10 was negative in C+, C-, and P (not shown). We also assessed the intracellular staining of IL-12Rβ1 with 4 other mAbs: clearly positive results were obtained for C+ and P with clones B101, 12RB44, and 69310, whereas the signal obtained with clone B103 was weak in C+, and no signal was detected in P (not shown). These results were confirmed by the intracellular staining of EBV-transformed B-cell lines, although the signal was less intense for both C+ and P in these cell lines (not shown). The mutant receptor encoded by the IL12RB1 allele in P, who carries the large 700 + 362_1619-944 deletion, can therefore be detected by flow cytometry in 2 types of cell, EBV-transformed B cells and PHA-T-cell blasts, with 4 of the 5 mAbs that stained the wild-type IL-12Rβ1 chain.
A detectable IL-12Rβ1 chain on the cell surface
Because intracellular IL-12Rβ1 was detectable in P, we used FACS analysis to investigate IL-12Rβ1 expression on the cell surface. IL-12Rβ1 was present in large amounts on the surface of PHA-T-cell blasts on day 5 in C+, as shown by staining with all 6 mAbs tested, including 3 commercially available (clones 24E6, 2B10, 69310) mAbs. PHA-T-cell blasts from P also tested positive with 5 of the 6 mAb tested (clones 24E6, B101, B103, 12RB44, 69310). No signal was obtained with the 2B10 mAb in P (Figure 2). Similar results were found when staining EBV-B cells of C+, P, and C- (not shown). The 700 + 362_1619-944 del IL12RB1 allele therefore encodes a detectable surface-expressed IL-12Rβ1 chain in our patient. Remarkably, none of the other 53 patients with IL-12Rβ1 deficiency described to date17-26,28 were found to express detectable levels of these receptors at the cell surface. In contrast, IL-12Rβ1 was present in large amounts at the cell surface in P and was detected with 5 of the 6 mAbs tested, including 2 of the 3 commercially available mAbs. With the 5 mAbs, the level of surface expression of the mutant IL-12Rβ1 chain detected is reduced. This may be due to an impaired surface expression of the protein or to an abnormal conformation of the molecule, as suggested by the various levels of expression found using different mAbs.
IL-12Rβ1 chain detected at the cell surface by FACS analysis. PHA-T-cell blasts from a positive control (C+), the patient (P), and a negative control (C-) were stained with various purified mouse monoclonal antibodies (24E6, B101, B103, 12RB44), rat mAb (2B10), or matched isotype control, followed by biotinylated matched Ab and phycoerythrin-conjugated streptavidin. IL-12Rβ1 clone 69310, directly conjugated to R-PE, was compared with a matched conjugated isotype control. Specific signals are represented as plain lines; matched isotype controls are represented as dotted lines.
IL-12Rβ1 chain detected at the cell surface by FACS analysis. PHA-T-cell blasts from a positive control (C+), the patient (P), and a negative control (C-) were stained with various purified mouse monoclonal antibodies (24E6, B101, B103, 12RB44), rat mAb (2B10), or matched isotype control, followed by biotinylated matched Ab and phycoerythrin-conjugated streptavidin. IL-12Rβ1 clone 69310, directly conjugated to R-PE, was compared with a matched conjugated isotype control. Specific signals are represented as plain lines; matched isotype controls are represented as dotted lines.
A lack of phosphorylated STAT4 upon IL-12 stimulation
STAT4 is a major transducer of the signals mediated by IL-12R.35,36 It is phosphorylated by the activated Janus kinases TYK2 and JAK2 upon the binding of IL-12 to its heterodimeric receptor (IL-12Rβ1 and IL-12Rβ2). Homodimers of phosphorylated STAT4 (P-STAT4) are formed and translocate to the nucleus, where they induce IFNG and other target genes. We thus tried to detect STAT4 phosphorylation following the stimulation of PHA-T-cell blasts, by means of intracellular FACS, as previously described.31 In unstimulated PHA-T-cell blasts from C+, C-, and P, no P-STAT4 was detected, whereas unphosphorylated STAT4 was clearly present (not shown). Following 30 minutes of stimulation with IFN-α, P-STAT4 was detected in PHA-T-cell blasts from C+, C-, and P, with no change in the total amount of STAT4 present (not shown). Following stimulation with IL-12, P-STAT4 was detected in PHA-T-cell blasts from C+ but not in those from P and C- (Figure 3). Thus, despite the presence of IL-12Rβ1 at the cell surface, P cells did not respond to IL-12, as detected by STAT4 phosphorylation, a critical early activation event.
Lack of phosphorylated STAT4 upon IL-12 stimulation, as shown by FACS analysis. PHA-T-cell blasts from a positive control (C+), a negative control (C-), and the patient (P) were left unstimulated (plain line) or were stimulated (dotted line) with IFN-α (105 U/mL) (left) for 30 minutes or with IL-12 (100 ng/mL) (right) for 15 minutes. Cells were fixed by PFA and methanol, permeabilized with saponin, and stained with a phospho-STAT4 rabbit polyclonal Ab (Zymed) (or matched isotype control), followed by goat anti-rabbit Alexa Fluor 488 (Molecular Probes).
Lack of phosphorylated STAT4 upon IL-12 stimulation, as shown by FACS analysis. PHA-T-cell blasts from a positive control (C+), a negative control (C-), and the patient (P) were left unstimulated (plain line) or were stimulated (dotted line) with IFN-α (105 U/mL) (left) for 30 minutes or with IL-12 (100 ng/mL) (right) for 15 minutes. Cells were fixed by PFA and methanol, permeabilized with saponin, and stained with a phospho-STAT4 rabbit polyclonal Ab (Zymed) (or matched isotype control), followed by goat anti-rabbit Alexa Fluor 488 (Molecular Probes).
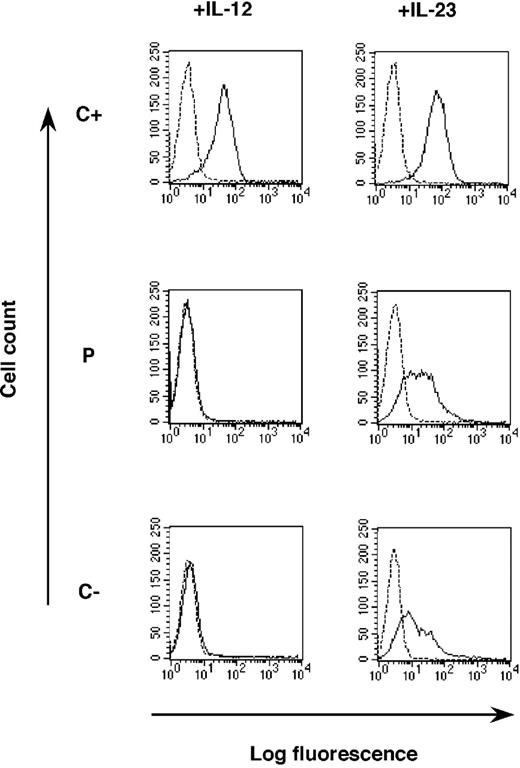
A lack of IFN-γ secretion in response to IL-12
We then investigated the impact of the IL12RB1 mutation on a more distal and equally crucial event—the induction of IFN-γ—by stimulating whole blood with BCG alone or BCG plus IL-12.26 By ELISA, no IFN-γ was induced by IL-12 in P cells, in contrast to what was observed in C+ (not shown), implying that peripheral T and natural killer (NK) cells do not respond to IL-12. Indeed, we have shown in another study that IFN-γ is secreted by both NK and T cells in this assay (J.F., in preparation). We then stimulated PHA-T-cell blasts from P, C+, and C- with various doses of IL-12 (1 pg/mL to 100 ng/mL) (Figure 4A). PHA-T-cell blasts from C+ produced large amounts of IFN-γ in response to IL-12, with a dose-dependent response up to 10 ng/mL, where a plateau was reached. In contrast, cells from P, like PHA-T-cell blasts from C-, did not respond to even high doses of IL-12, ruling out a partial defect with residual signaling in P. The cells from the patient's mother, who is heterozygous for the large deletion and expresses both wild-type and mutant receptors, as detected by flow cytometry, showed a normal response to IL-12, ruling out a dominant negative effect of the mutant allele for IL-12 responsiveness (not shown). Homozygosity for 700 + 362_1619-944 del is thus associated with a cellular phenotype of complete IL-12Rβ1deficiency, as shown by early (STAT4 phosphorylation) and late (IFN-γ induction) events in both NK and T cells ex vivo and in PHA-T-cell blasts in vitro.
Lack of IFN-γ production in response to IL-12 and IL-23. PHA-T-cell blasts from a positive control (C+), a negative control (C-), and the patient (P) were plated in 24-well plates and were left unstimulated (NS) or were stimulated with increasing concentrations of IL-12 for 48 hours (A) or IL-23 for 72 hours (B). Supernatants were harvested, and IFN-γ was quantified by ELISA.
Lack of IFN-γ production in response to IL-12 and IL-23. PHA-T-cell blasts from a positive control (C+), a negative control (C-), and the patient (P) were plated in 24-well plates and were left unstimulated (NS) or were stimulated with increasing concentrations of IL-12 for 48 hours (A) or IL-23 for 72 hours (B). Supernatants were harvested, and IFN-γ was quantified by ELISA.
A cellular phenotype of complete IL-23R deficiency
IL-12Rβ1 binds to the IL-23R chain to generate the heterodimeric receptor for a recently described cytokine, IL-23, that is composed of the p19 specific subunit and the p40 subunit that also forms part of IL-12.37-39 Several patients with IL-12Rβ1 deficiency due to a lack of surface receptor expression were previously shown not to respond to IL-23.25,40 We therefore stimulated PHA-T-cell blasts with various doses of IL-23 (5 pg/mL to 500 ng/mL). Cells from C+ produced IFN-γ in a dose-dependent manner in response to IL-23 (although less than in response to IL-12), whereas cells from P and C- did not respond to even high concentrations of IL-23 (Figure 4B). Cells from P therefore respond neither to IL-12 nor to IL-23 (Figure 4). P thus displays complete IL-12 and IL-23 receptor deficiency despite the presence of detectable IL-12Rβ1 at the cell surface.
The absence of cytokine binding to IL-12Rβ1 molecules at the cell surface
We investigated the reasons for the lack of response to IL-12 and IL-23 in P despite the presence of the IL-12Rβ1 chain at the cell surface. We performed fluorescence binding assays to assess the binding of IL-12 and IL-23 to PHA-T-cell blasts. A mAb specific for IL-12p40 was added to PHA-T-cell blasts after their incubation with large doses of recombinant IL-12 or IL-23. IL-12 and IL-23 binding was detectable by FACS analysis in most cells from C+, indicating that this antibody recognizes both IL-12 (p40-p35) and IL-23 (p40-p19). As expected, in the absence of cell surface IL-12Rβ1inC-, there was no detectable binding of IL-12. Similarly, no IL-12 binding was detected in P either (Figure 5). Thus, although our assay does not exclude the possibility that IL-12 binds to Pcells with a low affinity, the cell surface IL-12R heterodimers comprising mutant IL-12Rβ1 molecules in P did not bind normally their natural ligand, IL-12. C- displayed residual binding of IL-23, indicating that IL-23 binds to the IL-23R chain in the absence of IL-12Rβ1 (Figure 5). Moreover, Pshowed similar levels of IL-23 binding to C- whereas C+ displayed much higher levels of binding, indicating that the heterodimeric IL-23 receptors comprising mutant IL-12Rβ1 chains in P were impaired in their ability to normally recognize IL-23 and that the weak binding to IL-23R was not sufficient to trigger stimulation. Despite residual binding of IL-23 (and possibly of IL-12 to an even lower extent), complete IL-12 and IL-23 receptor defects in P therefore result at least in part from the impairment of IL-12 and IL-23 binding to heterodimers comprising the mutant IL-12Rβ1 chains at the cell surface. Our data do not exclude the possibility that the mutant IL-12Rβ1 chains do not interact normally with other signaling components, further contributing to the functional cytokine receptor defect.
Lack of cytokine binding to the surface of PHA-T-cell blasts. PHA-T-cell blasts from a positive control (C+), a negative control (C-), and the patient (P) were incubated without (dotted line) or with (plain line) IL-12p70 or IL-23 for 30 minutes at 4°C. The PHA-T-cell blasts were then incubated with a purified mouse anti-IL-12 p40 antibody, followed by a biotinylated anti-mouse antibody, and antibody binding was detected by incubation with PE-conjugated streptavidin.
Lack of cytokine binding to the surface of PHA-T-cell blasts. PHA-T-cell blasts from a positive control (C+), a negative control (C-), and the patient (P) were incubated without (dotted line) or with (plain line) IL-12p70 or IL-23 for 30 minutes at 4°C. The PHA-T-cell blasts were then incubated with a purified mouse anti-IL-12 p40 antibody, followed by a biotinylated anti-mouse antibody, and antibody binding was detected by incubation with PE-conjugated streptavidin.
IL-12Rβ1 expression and function are restored by retroviral transduction
We checked that the lack of response to IL-12 was truly caused by the IL12RB1 genotype, and not by a defect in another receptor chain or signaling molecule, by complementing the cellular defect by means of retrovirus-mediated transfer. We demonstrated that STAT4 was phosphorylated and activated in response to IFN-α but not to IL-12 in P. The transduction of T-cell blasts from P with a retrovirus encoding WT IL-12Rβ1 restored normal IL-12Rβ1 expression, as detected by the 2B10 mAb, which did not recognize the mutant chain (Figure 6A). The expression of a WT IL-12Rβ1 chain was accompanied by the restoration of STAT4 phosphorylation upon the IL-12 stimulation of transduced T cells, as shown by intracellular FACS analysis (not shown). WT IL-12Rβ1 expression not only restored STAT4 phosphorylation but also the ability of T-cell blasts to respond to IL-12 in terms of IFN-γ production (Figure 6B). The complete lack of response to IL-12 documented in P despite the presence of IL-12Rβ1 molecules at the cell surface is therefore due to the absence of surface IL-12Rβ1 molecules able to bind IL-12 normally.
Correction of the patient's IL-12Rβ1 defect by retroviral-mediated gene tranfer. (A) PHA-T-cell blasts from the patient (P), a negative control (C-), MND-IL-12Rβ1-transduced PHA-T-cell blasts from the patient (Ptd), or the negative control (C-td) were stained with anti-IL-12Rβ1 mAb (24E6 or 2B10, plain line) or matched isotype control (dotted line). (B) IFN-γ production in response to IL-12. PHA-T-cell blasts from the patient (P), a negative control (C-), and MND-IL-12Rβ1-transduced PHA-T-cell blasts from the patient (Ptd) or the negative control (C-td) were plated in 24-well plates and were either left unstimulated (NS) or were stimulated with increasing concentrations of IL-12 for 48 hours. Supernatants were harvested, and IFN-γ was quantified by ELISA.
Correction of the patient's IL-12Rβ1 defect by retroviral-mediated gene tranfer. (A) PHA-T-cell blasts from the patient (P), a negative control (C-), MND-IL-12Rβ1-transduced PHA-T-cell blasts from the patient (Ptd), or the negative control (C-td) were stained with anti-IL-12Rβ1 mAb (24E6 or 2B10, plain line) or matched isotype control (dotted line). (B) IFN-γ production in response to IL-12. PHA-T-cell blasts from the patient (P), a negative control (C-), and MND-IL-12Rβ1-transduced PHA-T-cell blasts from the patient (Ptd) or the negative control (C-td) were plated in 24-well plates and were either left unstimulated (NS) or were stimulated with increasing concentrations of IL-12 for 48 hours. Supernatants were harvested, and IFN-γ was quantified by ELISA.
Discussion
IL-12Rβ1 deficiency is the most frequent genetic defect responsible for the syndrome of MSMD, with 54 patients from 16 countries reported to date.17-28 All patients except the patient reported here lack IL-12Rβ1 at the surface of all cells examined, due to mutations creating a premature stop codon in the coding region or to disrupting protein folding and stability. We describe here a patient with a large in-frame deletion of 12165 nt, which results in the surface expression of internally truncated IL-12Rβ1 chains, as documented by flow cytometry with 5 of the 6 mAbs tested. Impaired binding of IL-12 and IL-23 to their surface-expressed heterodimeric receptors, including IL-12Rβ1, accounts at least in part for the complete absence of response to both cytokines. The abnormal conformation of the receptor may also affect its interaction with other signaling molecules. This patient thus defines a novel genetic form of complete IL-12Rβ1deficiency. A similar situation had been found for complete IFN-γR1 deficiency, caused by the lack of surface receptors,1,2,7,8 or surface-expressed nonfunctional receptors.9,10 However, the mutations in the latter patients were much smaller, consisting of short in-frame deletions or missense mutations.9,10
What molecular lessons can we learn from this experiment of nature? The mutant IL-12Rβ1 protein is stable, expressed on the cell surface, and lacks the proximal half of the extracellular domain. The wild-type IL-12Rβ1 chain is a member of the glycoprotein (gp) 130 family of receptors (type I cytokine receptor), the extracellular domain of which contains 5 fibronectin type III (FNIII) domains, each about 100 amino acids long.41 An FNIII domain contains 7-stranded β-sandwich motifs organized in an antiparallel manner.42 In IL-12Rβ1, the first 2 FNIII domains consist essentially of the translation products of exons 2 to 7 (Figure 1). In our patient, who lacks exons 8 to 13, the 3 C-terminal FNIII domains are removed by the large deletion. The truncated protein is stable, probably because the first 2 FNIII domains are intact, and the remaining 3 are completely lacking. Consistent with this view, another IL-12Rβ1-deficient patient (19.II.226 ) lacking IL-12Rβ1 surface expression bears another in-frame deletion, encompassing only exon 13 (1483 + 182-1619-1073 del). The protein generated from the 1483 + 182-1619-1073 del allele is not stable, probably due to the disruption of only half of the fifth FNIII domain—normally encoded by exons 12 and 13.
The cytokine-binding domain of receptors of the gp 130 family is located in the 200 N-terminal amino acids of the mature chain and consists, more precisely, of the first 2 FNIII domains, which are linked by a short proline-rich hinge, allowing an 80-degree elbow for the binding of the ligand.42,43 These 2 FNIII domains are also called “hematopoietin receptor domains”,41 or the “cytokine-binding homology region” (CHR).43 The first N-terminal FNIII domain (D1) contains 3 amino acids forming the CXW motif (CSW in IL-12Rβ1). The second N-terminal FNIII domain (D2) contains the SWXSW motif (SWKSW in IL-12Rβ1). Both motifs are signatures of the gp 130 family of cytokine receptors.41 Horsten et al44 have demonstrated that D1 and D2 are necessary and sufficient for the binding of IL-6 to gp130. In our patient, however, despite the integrity of these 2 FNIII domains, the recognition and binding of IL-12 is profoundly impaired. The difference between the 2 situations may be due to the different receptors involved (IL-6R and IL-12R) and possibly to IL-12 being itself a “truncated” receptor that may need to interact not only with the CHR of its receptor but also with other IL-12Rβ1 FNIII domains. Alternatively, IL-12Rβ1 CHR folding may be influenced by extracellular residues outside the CHR itself.
The impact of the (700 + 362_1619-944 del) mutation on IL-12Rβ1-specific Ab recognition was much less pronounced than that on cytokine binding. Indeed, the deletion of 6 exons from IL12RB1 is consistent not only with receptor expression at the cell surface but also with receptor recognition by 5 of the 6 available IL-12Rβ1-specific antibodies. This study therefore makes it possible to map the epitopes of some anti-human IL-12Rβ1 antibodies. The 5 mAbs that bound (clones 24E6, B101, B103, 12RB44, 69310) were probably generated against the first 2 FNIII domains (the CHR). The 5 epitopes located in the IL-12 recognition site are not significantly altered by the large (700 + 362_1619-944 del) deletion, which respects the first 2 FNIII domains. However, the low levels of receptor expression detected with these mAbs may result from an abnormal conformation of the receptor. Moreover, one epitope (recognized by 2B10) either maps outside the CHR or, if it is located within the CHR, is conformational and strictly depends on residues located in the other 3 FNIII domains, which are lacking in our patient.
Our report highlights the lack of correlation between the IL12RB1 genotype and IL-12Rβ1 expression: Paradoxically, the 700 + 362_1619-944 del mutation is the largest deletion described in IL12RB1 and the only known mutation allowing cell surface expression. Whereas a small IL12RB1 genomic lesion, such as a missense mutation, may be responsible for the lack of protein at the cell surface due to misfolding and degradation,21-24,26 a very large deletion of 12165 nt, encompassing half the exons encoding the extracellular domain, can lead to the presence of detectable receptors at the cell surface. This report also demonstrates that a diagnosis of IL-12Rβ1deficiency should not be excluded solely on the basis of a conserved surface expression on flow cytometry. The clinical implications of these findings are important for individual patients. Therapeutic options can best be tailored to the patient, on a rational basis, if accurate molecular diagnosis is achieved. Indeed, recombinant IFN-γ administration can save the lives of IL-12Rβ1-deficient patients. Our report thus stresses the importance of in-depth molecular diagnostic investigation in patients with MSMD.
Prepublished online as Blood First Edition Paper, June 3, 2004; DOI 10.1182/blood-2004-02-0584.
Supported by the Fondation pour la Recherche Médicale (FRM) (C.F.), the Fondation Banque National de Paris (BNP)-Paribas, the Fondation Schlumberger, and European Commission grant QLK2-CT-2002-00846.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the patients and their families for their trust, the members of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions, Dr Dominique Recan for the EBV transformation of B cells, and Betty Cazeau and Marianne O'Donnell from the Genetics Institute (Andover, MA) for providing 3 mAbs (B101, B103, 12RB44).






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal