Abstract
The role of the tumor necrosis factor (TNF) superfamily member receptor activator of nuclear factor kappa B ligand (RANKL) in promoting the differentiation of osteoclasts has been extensively characterized. In this study, we have investigated the effect of TNF-related apoptosis-inducing ligand (TRAIL), a member of the TNF superfamily of cytokines, in osteoclastogenesis, by using human peripheral blood mononuclear cells and the RAW264.7 murine monocytic cell line. Both cell models differentiate into osteoclast-like cells in presence of RANKL plus macrophage-colony-stimulating factor (M-CSF), as evaluated in terms of tartrate-resistant acid phosphatase (TRAP)-positive multinucleated cells and bone resorption activity. Unexpectedly, when added in culture in combination with RANKL plus M-CSF, TRAIL inhibited osteoclastic differentiation in both cell models. To investigate the molecular mechanism underlining such inhibitory activity, we analyzed the effect of TRAIL on the mitogen-activated protein kinases (MAPKs) pathways, which play a key role in osteoclastogenesis. Treatment with RANKL plus M-CSF activated both the ERK1/2 and p38/MAPK pathways, which are essential for proliferation and differentiation of preosteoclasts, respectively. Of note, the addition of TRAIL to RANKL plus M-CSF did not affect ERK1/2 but it profoundly inhibited p38/MAPK phosphorylation. Thus, our data demonstrate that TRAIL blocks osteoclastic differentiation and suggest that inhibition of the p38/MAPK pathway by TRAIL likely plays an important role in this process. (Blood. 2004;104:2044-2050)
Introduction
Osteoclasts are multinucleated cells, belonging to the monocytic/macrophagic lineage, that form by fusion of mononuclear precursors. This multistep differentiation process is under the control of the bone microenvironment, which includes stromal cells, osteoblasts, and local factors.1 One of the key factors mediating osteoclastogenesis is the tumor necrosis factor (TNF) family member receptor activator of nuclear factor kappa B ligand (RANKL),2 also referred to as osteoclast differentiation factor,3 TNF-related activation-induced cytokine,4 and osteoprotegerin ligand.5 RANKL, which is expressed on the surface of osteoblastic/stromal cells, was found to be essential for osteoclastic differentiation.3,6-8 Its cognate receptor RANK, one of the TNF receptor family members, is expressed in osteoclasts and their circulating precursor cells.6,8,9 Although RANKL in conjunction with macrophage-colony-stimulating factor (M-CSF) has been recognized as one of the key osteoclastogenic signals expressed by osteoblasts and stromal cells, the downstream signaling pathways activated by this cytokine have not been fully characterized. However, a series of data points to the mitogen-activated protein kinases (MAPKs) as key pathways in osteoclastogenesis.10-13 In particular, it has been shown that activation of p38/MAPK is essential in mediating osteoclastic differentiation, whereas activation of ERK1/ERK2 is implicated in preosteoclastic proliferation but counteracts osteoclastic differentiation.10-13
One important member of the TNF family of cytokines is TNF-related apoptosis-inducing ligand (TRAIL)/Apo-2 ligand (L), which exists as either a type II membrane protein or as a soluble form.14-16 TRAIL interacts with 4 high-affinity membrane receptors belonging to the apoptosis-inducing TNF receptor (R) family. TRAIL-R1 (DR4) and TRAIL-R2 (DR5) transduce apoptotic signals upon binding of TRAIL, whereas TRAIL-R3 (DcR1) and TRAIL-R4 (DcR2) are homologous to TRAIL-R1 and TRAIL-R2 in their cysteine-rich extracellular domain, but lack apoptosis-inducing capability. TRAIL shows the unique property to induce apoptosis in a variety of neoplastic cells,16 displaying minimal or absent toxicity on most normal cells. In spite of its potential as an anticancer therapeutic agent both in vitro and in vivo,17,18 the wide expression of TRAIL and TRAIL-Rs in many normal tissues14-16 suggests that the physiologic role of TRAIL is more complex than induction of apoptosis in cancer cells.
An interesting link between the RANKL and TRAIL systems is represented by evidence that both RANKL and TRAIL bind to osteoprotegerin (OPG),19,20 a secreted TNF-R family member, which increases bone density when overexpressed in transgenic mice. Moreover, OPG inhibits osteoclastogenesis in vitro, suggesting that increased bone density results from decreased numbers of mature osteoclasts. It has been shown that OPG binds to TRAIL in vitro,20 but with much lower affinity at physiologic temperature (37°C) with respect to RANKL.21
Materials and methods
Cell cultures and treatments
As model systems of osteoclastogenesis, we used human peripheral blood mononuclear cells (PBMCs) and the RAW264.7 murine monocytic/macrophagic cell line. Both cell types differentiate into osteoclast-like cells in the presence of RANKL plus M-CSF.13,22-24 Human PBMCs from healthy donors were separated by gradient centrifugation with Lymphocyte Cell Separation medium (Cedarlane Laboratories, Hornby, ON, Canada) and seeded in 6-well plates at a density of 10 × 106 cells/well. After incubation for 18 hours, nonadherent PBMCs were removed, and remaining adherent cells were referred to as “adherent PBMCs.” Cultures were maintained in RPMI medium containing 10% fetal bovine serum (FBS; Gibco BRL, Gaithersburg, MD). RAW264.7 cells, which are derived from a tumor induced by Abelson murine leukemia virus, were purchased from American Type Culture Collection (ATCC; Rockville, MD). Cells were cultured in 6-well plates at a density of 6 × 105 cells/well in ATCC medium: RPMI 1640 medium with 2 mM l-glutamine adjusted to contain 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, 10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], and 1.0 mM sodium pyruvate and 5% FBS.
For osteoclastic differentiation, cells were cultured for 10 to 12 days in the presence of 50 ng/mL human M-CSF (for PBMCs; PeproTech, London, United Kingdom) or murine M-CSF (for RAW264.7; R&D Systems, Minneapolis, MN) and 50 ng/mL human RANKL (for both PBMCs and RAW264.7; Alexis Biochemicals, Lausen, Switzerland). When indicated, cultures were treated with recombinant histidine 6-tagged TRAIL(114-281) purified by chromatography as previously described.25 The absence of endotoxin contamination in the recombinant TRAIL preparation (< 0.1 endotoxin units/mL) was assessed by Limulus Amoebocyte Lysate (LAL) assay (BioWhittaker, Walkersville, MD). The optimal TRAIL concentration (10 ng/mL) used in most experiments was determined based on preliminary assays in which scalar TRAIL doses (ranging from 0.1 ng/mL to 1000 ng/mL) were tested. TRAIL was added to the cultures simultaneously with RANKL plus M-CSF (day 0), or, when indicated, after 3 to 6 days. In selected experiments, cells were preincubated for 40 minutes with pharmacologic inhibitors of the ERK1/2 (PD98059; 20 μM) and p38/MAPK (SB203580; 10 μM) pathways (both from Calbiochem, La Jolla, CA), prior to the addition of cytokines. Medium and treatments were replaced every 3 days. At 10 to 12 days of culture, the overall number of viable cells was scored under a microscope (× 10 magnification).
Flow cytometric analysis
For flow cytometric analysis, adherent cells were harvested from culture plates by gentle scraping on ice. Surface cell staining was performed at 4°C for 40 minutes by incubating 3 × 105 cells in 200 μL phosphate-buffered saline (PBS; containing 1% bovine serum albumin [BSA] and 5% human plasma) with the indicated antibodies (Abs). Expression of monocytic/macrophagic markers was documented by using phycoerythrin (PE)-conjugated anti-CD14 (Immunotech, Marseille, France) and anti-CD36 monoclonal antibodies (MoAbs; BD Pharmingen, San Diego, CA), and fluorescein isothiocyanate (FITC)-conjugated anti-CD64 MoAb (Immunotech). Nonspecific fluorescence was assessed by incubation with irrelevant isotype-matched conjugated MoAbs. Surface expression of TRAIL-Rs was evaluated by indirect staining with primary MoAbs antihuman TRAIL-R1, TRAIL-R2, TRAIL-R3, and TRAIL-R4 (all from Alexis Biochemicals), followed by PE-conjugated antimouse secondary Abs (Immunotech). Nonspecific fluorescence was assessed using normal mouse immunoglobulin G (IgG) followed by secondary Abs. Flow cytometric analyses were performed by FACScan (Becton Dickinson, San Jose, CA).
Cytochemical staining
For DAPI (4′,6-diamidino-2-phenylindole) staining of nuclei, cells were washed with PBS, fixed in paraformaldehyde 4% for 10 minutes, permeabilized in Triton X-100 0.1% for 10 minutes, washed again with PBS, and incubated with 500 ng/mL DAPI (Sigma Chemical, St Louis, MO) in PBS for 15 minutes at 37°C in a dark, humidified chamber. After several washes in PBS, the coverslips were mounted on PBS/glycerin and the intercalation of DAPI was visualized by means of an Axiophot fluorescence microscope (Zeiss, Oberlochen, Germany).
Staining for tartrate-resistant acid phosphatase (TRAP) was carried out using a leukocyte acid phosphatase kit (387-A; Sigma Chemical), according to the manufacturer's instructions. After the staining, cells were rinsed with PBS, photographed under a light microscope, and large TRAP-positive cells that contained more than 3 nuclei were counted.
For apoptosis detection, cells were labeled with the CaspaTag Pancaspase in situ assay kit, fluorescein (Chemicon, Temecula, CA), according to the manufacturer's instruction.
Bone resorption assay
PBMCs and RAW264.7 cells were plated on 24-well plates coated with artificial bone slides (OAAS; Osteogenic Core Technologies, Choongnam, Korea) and allowed to attach overnight. Cytokines were added starting from the next day. After 10 to 12 days plates were processed according to the manufacturer's instructions, and resorption lacunae were visualized using a light microscope (Eclipse TE200 Inverted microscope; Nikon, Tokyo, Japan).
Western blot analyses
TRAIL-Rs expression was analyzed in adherent PBMCs and RAW264.7 cells by Western blot on enriched cell membrane lysates, obtained as previously described25 and using the following rabbit Abs: anti-TRAIL-R1 (DR4; Santa Cruz Biotechnology, Santa Cruz, CA) anti-TRAIL-R2 (DR5; R&D Systems), anti-TRAIL-R3 (Axxora, San Diego, CA), and anti-TRAIL-R4 (Stressgen Biotechnologies, Victoria, BC, Canada), which detect human and mouse TRAIL-Rs in Western blot.
For the analysis of OPG expression, at different days of cultures cells were harvested in lysis buffer containing 1% Triton X-100, Pefablock (1 mM), aprotinin (10 μg/mL), pepstatin (1 μg/mL), leupeptin (10 μg/mL), NaF (10 mM), and Na3VO4 (1 mM). For the analysis of MAPK pathways, cells were subjected to partial FCS reduction (to 0.5%) for 18 hours prior to the addition of TRAIL or RANKL plus M-CSF, used alone or in combination.
Protein determination was performed by Bradford assay (Bio-Rad, Richmond, CA). Equal amounts of protein (50 μg) for each sample were migrated in acrylamide gels and blotted onto nitrocellulose filters. The following Abs were used: anti-OPG (R&D Systems), antitubulin (Sigma), anti-ERK1/2, and antiphospho-ERK1/2 (both from Promega, Madison, WI), anti-p38/MAPK and antiphospho-p38/MAPK (both from Cell Signaling Technology, Beverly, MA). Blotted filters were probed with Abs for the phosphorylated forms of ERK1/2 and p38/MAPK and for the respective total protein kinase content for verifying loading evenness. After incubation with peroxidase-conjugated anti-rabbit IgG, specific reactions were revealed with the enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). Densitometry values were estimated by the ImageQuant TL software (Amersham). Multiple film exposures were used to verify the linearity of the samples analyzed and to avoid saturation of the film.
RNA extraction and analysis by RT-PCR
For measurement of OPG RNAm, RNA purification was performed using the SV total RNA isolation system (Promega) following the manufacturer's protocol. Synthesis of first-strand cDNA and amplification were performed by using the Access reverse transcriptase-polymerase chain reaction (RT-PCR) system (Promega). OPG RNAm amplification was performed using the human/mouse OPG PCR primer pair (R&D Systems), specifically designed for RT-PCR (product: 468 bp). Amplification of β-actin (product: 661 bp) was performed with primers purchased from Stratagene (San Diego, CA). Moreover, aliquots of supernates of the same cultures were collected and stored at -80°C until measurement by enzyme-linked immunosorbent assay (ELISA; detection limit: 3 pM; Alexis Biochemicals), according to the manufacturer's directions. Experiments were always performed in duplicate.
Statistical analysis
The results were evaluated by using analysis of variance with subsequent comparisons by Student t test for paired or nonpaired data, as appropriate. Statistical significance was defined as P < .05. Values are reported as means plus or minus standard deviation (SD).
Results
Expression of TRAIL-Rs in adherent PBMCs and RAW264.7
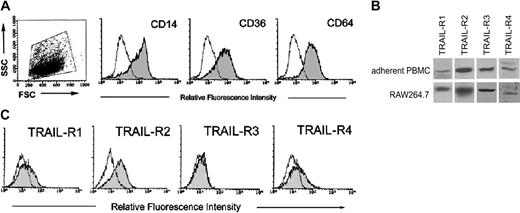
The first group of experiments was designed to investigate the expression of TRAIL-Rs in human and murine (RAW264.7) preosteoclastic precursors. For this purpose, PBMCs were let to adhere for 3 to 5 days before performing analysis of TRAIL receptors. At this time point, adherent PBMCs were positive for the monocytic/macrophagic markers CD14, CD36, and CD64 (Figure 1A). Western blot analysis revealed that both human adherent PBMCs and the RAW264.7 murine cell line expressed all TRAIL-Rs (Figure 1B). Additional analysis, for surface expression of TRAIL-Rs, was performed by flow cytometry, but only in human adherent PBMCs since no Abs for flow cytometry are currently available for mouse cells. As shown in Figure 1C, adherent PBMCs expressed surface TRAIL-R1 (mean fluorescence intensity [MFI]: 18 ± 15; means ± SD of 7 independent cultures), TRAIL-R2 (MFI: 36 ± 9; means ± SD of 7 independent cultures), and TRAIL-R4 (MFI: 20 ± 6; means ± SD of 7 independent cultures), at variable levels among the different (n = 7) donors analyzed, whereas surface TRAIL-R3 was never detected (Figure 1C). The highest variability was observed for surface TRAIL-R1, which was barely detectable in 4 of 7 samples and clearly detectable in the remaining 3 samples, including the one shown in Figure 1C. Thus, in line with previous findings of other authors,26 some TRAIL-Rs, and, in particular TRAIL-R3, were expressed at the cytoplasm level but not at the cell surface, in human preosteoclastic cells.
Expression of TRAIL-Rs in adherent human PBMCs and in RAW264.7. PBMCs were let to adhere for 3 to 5 days before performing Western blot and phenotypic analysis. At this time point, adherent PBMCs were either stained for monocytic/macrophagic markers CD14, CD36, and CD64 (A) or analyzed for TRAIL-R expression (B, C). (A) Dot plot shows the forward and side scatter (FSC/SSC) profile and the gate, on viable cells, considered for the phenotypic analysis. In (B) TRAIL-R expression was evaluated in adherent PBMCs and in RAW264.7 by Western blot analysis. A representative of 3 separate experiments is shown. (C) Surface TRAIL-R expression was evaluated in adherent PBMCs by flow cytometry. A representative of 7 separate experiments is shown. In panels A and C, shadowed histograms represent cells stained with MoAbs specific for the indicated surface antigens (CD14, CD36, CD64, and TRAIL-Rs) whereas unshadowed histograms represent the background fluorescence obtained from the staining of the same cultures with isotype-matched control MoAbs.
Expression of TRAIL-Rs in adherent human PBMCs and in RAW264.7. PBMCs were let to adhere for 3 to 5 days before performing Western blot and phenotypic analysis. At this time point, adherent PBMCs were either stained for monocytic/macrophagic markers CD14, CD36, and CD64 (A) or analyzed for TRAIL-R expression (B, C). (A) Dot plot shows the forward and side scatter (FSC/SSC) profile and the gate, on viable cells, considered for the phenotypic analysis. In (B) TRAIL-R expression was evaluated in adherent PBMCs and in RAW264.7 by Western blot analysis. A representative of 3 separate experiments is shown. (C) Surface TRAIL-R expression was evaluated in adherent PBMCs by flow cytometry. A representative of 7 separate experiments is shown. In panels A and C, shadowed histograms represent cells stained with MoAbs specific for the indicated surface antigens (CD14, CD36, CD64, and TRAIL-Rs) whereas unshadowed histograms represent the background fluorescence obtained from the staining of the same cultures with isotype-matched control MoAbs.
TRAIL blocks maturation of preosteoclasts and bone resorption activity induced by RANKL plus M-CSF
In the next group of experiments, adherent PBMCs were cultured in the absence or presence of predetermined optimal concentrations of RANKL plus M-CSF for 10 to 12 days. RANKL plus M-CSF induced dramatic morphologic changes, characterized by the appearance of TRAP-positive giant multinucleated cells with more than 3 nuclei (Figure 2A), as better evidenced by DAPI staining (Figure 2B). Moreover, several resorption pits were selectively observed in cultures treated with RANKL plus M-CSF in plates coated with carbonated calcium phosphate (Figure 2C), clearly demonstrating that the multinucleated TRAP-positive cells possessed major characteristics of functional osteoclasts.
Effect of TRAIL on differentiation of adherent PBMCs into functional osteoclasts. Adherent PBMCs were left untreated (without cytokines) or cultured in the presence of TRAIL and with RANKL plus M-CSF with or without TRAIL. After 10 days cells were analyzed for osteoclastic differentiation. (A) Representative fields of the cultures, treated as indicated, after TRAP staining (magnification, × 10; numerical aperture of the objective lens [NA], 0.25). Similar results were observed in 7 independent experiments performed in duplicate. (B) Magnification of a representative TRAP-positive multinucleated cell (i), and a representative field of RANKL plus M-CSF-treated cultures observed by light microscopy (ii) and by fluorescence microscopy after DAPI staining (iii). *Polynucleated cells characteristic of RANKL plus M-CSF cultures. Original magnification and NA: i, × 40 0.75 NA; ii-iii, × 20, 0.25 NA. In panel C, adherent PBMCs were plated on an artificial bone matrix slide and were cultured with RANKL plus M-CSF, in the absence or presence of TRAIL (10 ng/mL), as indicated. After 12 days, the slides were fixed and stained, and resorption was determined by examining pit formation under a light microscope (magnification, × 20; NA, 0.40). Representative fields are shown. (D-E) Cultures were treated as indicated and the number of TRAP-positive multinucleated cells containing 3 or more nuclei was scored. Data represent the means ± SD of 3 to 7 different experiments (*P < .05, compared with RANKL plus M-CSF).
Effect of TRAIL on differentiation of adherent PBMCs into functional osteoclasts. Adherent PBMCs were left untreated (without cytokines) or cultured in the presence of TRAIL and with RANKL plus M-CSF with or without TRAIL. After 10 days cells were analyzed for osteoclastic differentiation. (A) Representative fields of the cultures, treated as indicated, after TRAP staining (magnification, × 10; numerical aperture of the objective lens [NA], 0.25). Similar results were observed in 7 independent experiments performed in duplicate. (B) Magnification of a representative TRAP-positive multinucleated cell (i), and a representative field of RANKL plus M-CSF-treated cultures observed by light microscopy (ii) and by fluorescence microscopy after DAPI staining (iii). *Polynucleated cells characteristic of RANKL plus M-CSF cultures. Original magnification and NA: i, × 40 0.75 NA; ii-iii, × 20, 0.25 NA. In panel C, adherent PBMCs were plated on an artificial bone matrix slide and were cultured with RANKL plus M-CSF, in the absence or presence of TRAIL (10 ng/mL), as indicated. After 12 days, the slides were fixed and stained, and resorption was determined by examining pit formation under a light microscope (magnification, × 20; NA, 0.40). Representative fields are shown. (D-E) Cultures were treated as indicated and the number of TRAP-positive multinucleated cells containing 3 or more nuclei was scored. Data represent the means ± SD of 3 to 7 different experiments (*P < .05, compared with RANKL plus M-CSF).
Recombinant soluble human TRAIL (10 ng/mL) did not induce osteoclastic differentiation of adherent PBMCs (Figure 2A,D) also when added in combination with M-CSF (data not shown). However, when added to the RANKL plus M-CSF combination, unexpectedly TRAIL significantly (P < .01) inhibited the differentiative phenotype promoted by RANKL plus M-CSF in a dose-dependent manner, from 10 ng/mL onwards (Figure 2A,D). Moreover, the TRAIL-mediated inhibition of RANKL plus M-CSF-primed osteoclastogenesis was time-sensitive (Figure 2D). In fact, inhibition of osteoclast formation was maximal when TRAIL was added simultaneously to RANKL plus M-CSF (day 0) and it progressively declined when TRAIL was added 3 or 6 days after RANKL plus M-CSF. Consistent with its ability to abrogate the appearance in culture of giant multinucleated cells, TRAIL completely blocked the bone resorption activity (Figure 2C). To further characterize the interplay between TRAIL, RANKL, and M-CSF, adherent PBMCs were pretreated with M-CSF for 3 days, to foster the proliferation of preosteoclastic cells, and then treated with RANKL plus M-CSF or RANKL alone, in the absence or presence of TRAIL. As expected on the basis of previous studies,1 maximal osteoclastic differentiation was achieved when pretreatment with M-CSF was followed by the combination of RANKL plus M-CSF with respect to RANKL alone (Figure 2E). Of note, TRAIL significantly (P < .01) inhibited osteoclastic differentiation induced by either RANKL plus M-CSF or RANKL alone, suggesting that TRAIL mainly affected the differentiation rather than the proliferative phase of osteoclastogenesis.
In a parallel group of experiments, we could demonstrate that the inhibitory activity of TRAIL on osteoclastogenesis was not confined to human adherent PBMCs as recombinant TRAIL also blocked osteoclastic differentiation of the RAW264.7 murine cell line cultured with RANKL plus M-CSF (Figure 3A-B).
Effect of TRAIL on differentiation of RAW264.7 cells into functional osteoclasts. RAW264.7 cells were left untreated (without cytokines) or cultured in the presence of TRAIL and with RANKL plus M-CSF with or without TRAIL for 10 days. (A) Representative fields of the cultures, treated as indicated, after TRAP staining (magnification, × 10; NA, 0.25). (B) The number of TRAP-positive multinucleated cells containing 3 or more nuclei was scored. Data represent the means ± SD of 7 different experiments.
Effect of TRAIL on differentiation of RAW264.7 cells into functional osteoclasts. RAW264.7 cells were left untreated (without cytokines) or cultured in the presence of TRAIL and with RANKL plus M-CSF with or without TRAIL for 10 days. (A) Representative fields of the cultures, treated as indicated, after TRAP staining (magnification, × 10; NA, 0.25). (B) The number of TRAP-positive multinucleated cells containing 3 or more nuclei was scored. Data represent the means ± SD of 7 different experiments.
The antidifferentiative activity of TRAIL on osteoclastogenesis is not due to cytotoxic effects or to induction of OPG
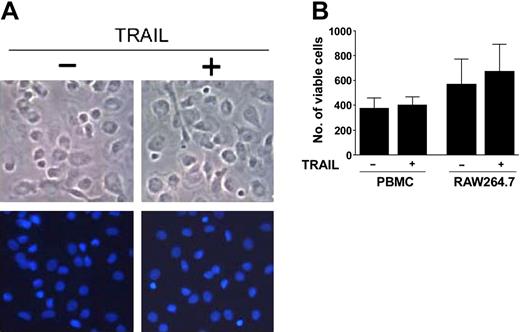
Taking into account that human adherent PBMCs and RAW264.7 cells express both “death receptors” TRAIL-R1 and TRAIL-R2, we have analyzed the possibility that the inhibitory activity of TRAIL on osteoclastogenesis might be due to a cytotoxic effect on osteoclastic precursors. This hypothesis was excluded since no apoptotic cells were noticed after 24 to 48 hours from the addition of recombinant TRAIL to human adherent PBMCs, as observed by light microscopy and fluorescence microscopy after DAPI staining (Figure 4A) or CaspaTag Pan-caspase in situ assay (data not shown). Moreover, the overall number of viable cells of both PBMCs and RAW264.7 were not significantly different among cultures left untreated or exposed to TRAIL for up to 10 to 12 days (Figure 4B).
Lack of cytotoxic effects of TRAIL on osteoclastic precursors. Adherent PBMCs and RAW264.7 cells were cultured in the absence or presence of TRAIL. (A) PBMCs were let to adhere for 3 days and then were exposed to TRAIL for 48 hours. Cultures were examined for apoptosis and cell viability by light microscopy (top panels) and by DAPI staining and fluorescence microscopy (bottom panels). Shown are single fields that are representative of untreated and TRAIL-treated cultures. Original magnification, × 20. (B) The number of viable cells was scored in at least 6 random fields at × 10 magnification of PBMCs and RAW264.7 cells after 12 days of culture. Data represent the means ± SD of 7 different experiments for each cell system.
Lack of cytotoxic effects of TRAIL on osteoclastic precursors. Adherent PBMCs and RAW264.7 cells were cultured in the absence or presence of TRAIL. (A) PBMCs were let to adhere for 3 days and then were exposed to TRAIL for 48 hours. Cultures were examined for apoptosis and cell viability by light microscopy (top panels) and by DAPI staining and fluorescence microscopy (bottom panels). Shown are single fields that are representative of untreated and TRAIL-treated cultures. Original magnification, × 20. (B) The number of viable cells was scored in at least 6 random fields at × 10 magnification of PBMCs and RAW264.7 cells after 12 days of culture. Data represent the means ± SD of 7 different experiments for each cell system.
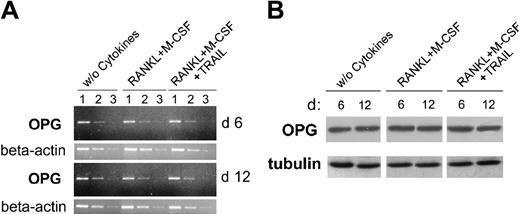
It has been clearly shown that the soluble decoy receptor OPG plays a key role in inhibiting osteoclastogenesis in vitro and in vivo by binding at high affinity to RANKL19 and that myeloid cells produce and secrete OPG.27 Therefore, in order to exclude the possibility that the antiosteoclastic activity of TRAIL might be due to OPG induction, the cellular levels of OPG were measured by RT-PCR and Western blot. As shown in Figure 5, OPG was readily detectable at both mRNA and protein levels in adherent PBMCs. However, it did not show any significant differences in cultures left untreated or treated with RANKL plus M-CSF in the absence or presence of TRAIL. Moreover, OPG was undetectable (below the detection limit of 3 pM) in all the culture supernatants collected at various time points from cultures treated with RANKL plus M-CSF with or without TRAIL. Taken together, these findings indicate that TRAIL does not affect osteoclastogenesis by up-regulating OPG expression.
Analysis of OPG expression during osteoclastic differentiation in the presence of TRAIL. At 6 and 12 days of cultures, cells were harvested and OPG expression was analyzed by RT-PCR (A) and Western blotting (B). (A) Semiquantitative OPG RT-PCR was performed on serial dilutions (1-3) of RNA extracted from cultures treated as indicated. β-actin amplification was used to confirm comparability of the samples. The ethidium bromide-stained agarose gels of PCR products are shown. Data are representative of 2 independent experiments. (B) Equivalent amounts of protein lysates were analyzed by Western blot with anti-OPG MoAb. A representative of 2 separate experiments is shown. Equal loading of protein in each lane was confirmed by staining with the antibody to tubulin.
Analysis of OPG expression during osteoclastic differentiation in the presence of TRAIL. At 6 and 12 days of cultures, cells were harvested and OPG expression was analyzed by RT-PCR (A) and Western blotting (B). (A) Semiquantitative OPG RT-PCR was performed on serial dilutions (1-3) of RNA extracted from cultures treated as indicated. β-actin amplification was used to confirm comparability of the samples. The ethidium bromide-stained agarose gels of PCR products are shown. Data are representative of 2 independent experiments. (B) Equivalent amounts of protein lysates were analyzed by Western blot with anti-OPG MoAb. A representative of 2 separate experiments is shown. Equal loading of protein in each lane was confirmed by staining with the antibody to tubulin.
TRAIL selectively inhibits the activation of the p38/MAPK pathway induced by RANKL plus M-CSF
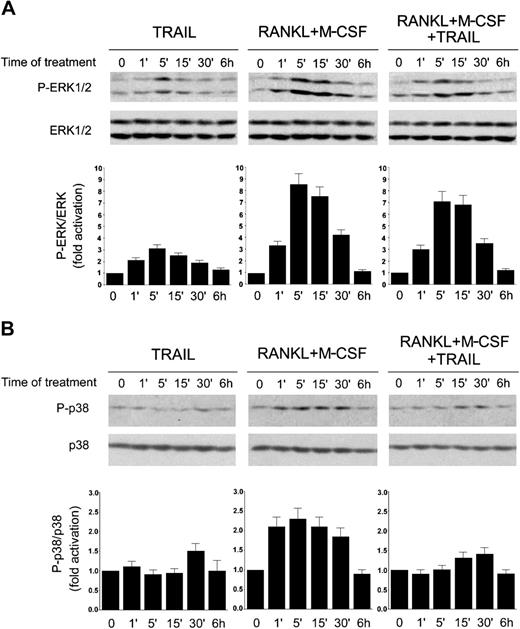
In the effort to elucidate the intracellular mechanism or mechanisms underlying the inhibitory activity of TRAIL, we next focused our attention of the MAPK family members ERK1/2 and p38/MAPK, which are known to play a major role in promoting proliferation and differentiation of preosteoclastic precursors, respectively.10-13 TRAIL alone induced phosphorylation of ERK1/ERK2, peaking at 5 minutes and declining thereafter (Figure 6A), whereas it did not significantly affect p38/MAPK phosphorylation (Figure 6B). As expected on the basis of previous studies,10-13 the combination of RANKL plus M-CSF strongly activated both the ERK1/2 (Figure 6A) and the p38/MAPK (Figure 6B) pathways in a time frame between one minute and 30 minutes. Of note, the addition of TRAIL to RANKL plus M-CSF did not significantly modulate ERK1/2 phosphorylation (Figure 6A), whereas it almost completely suppressed the p38/MAPK phosphorylation elicited by RANKL plus M-CSF (Figure 6B).
Time-course analysis of ERK1/2 and p38/MAPK phosphorylation in response to TRAIL. RAW264.7 cells were made quiescent by reduction of serum to 0.5% in culture medium overnight, before stimulation with TRAIL and RANKL plus M-CSF, used alone or in combination. Equal amounts of cell lysates, harvested at the indicated time points, were analyzed for ERK1/2 (A) and p38/MAPK (B) phosphorylation by Western blot analyses using Abs specific for the native form of the kinases and for residues that are phosphorylated (P-) in each kinase upon activation. Protein bands were quantified by densitometry and levels of P-ERK1/2 (A) and P-p38 (B) were calculated for each time point, after normalization to ERK1/2 or p38, respectively, in the same sample. Unstimulated basal expression was set as unity. Error bars indicate SD. Results are representative of 3 separate experiments.
Time-course analysis of ERK1/2 and p38/MAPK phosphorylation in response to TRAIL. RAW264.7 cells were made quiescent by reduction of serum to 0.5% in culture medium overnight, before stimulation with TRAIL and RANKL plus M-CSF, used alone or in combination. Equal amounts of cell lysates, harvested at the indicated time points, were analyzed for ERK1/2 (A) and p38/MAPK (B) phosphorylation by Western blot analyses using Abs specific for the native form of the kinases and for residues that are phosphorylated (P-) in each kinase upon activation. Protein bands were quantified by densitometry and levels of P-ERK1/2 (A) and P-p38 (B) were calculated for each time point, after normalization to ERK1/2 or p38, respectively, in the same sample. Unstimulated basal expression was set as unity. Error bars indicate SD. Results are representative of 3 separate experiments.
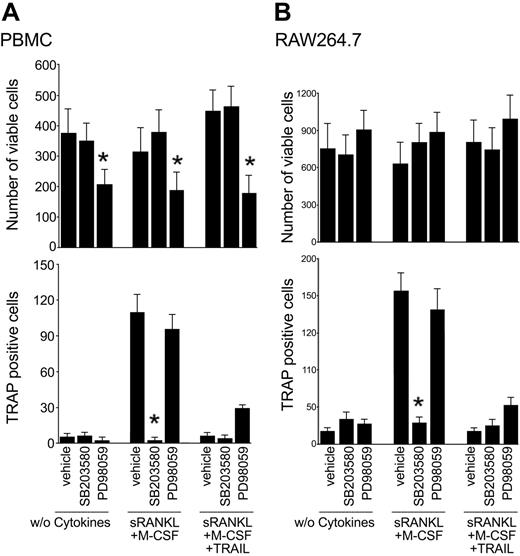
In the next experiments, adherent PBMCs were cultured with RANKL plus M-CSF with or without TRAIL in the presence or absence of either SB203580 or PD98059 inhibitors of the p38/MAPK and ERK1/2 pathways, respectively. Of note, SB203580 abrogated osteoclastic differentiation in cultures supplemented with RANKL plus M-CSF, without inducing cytotoxicity (Figure 7A), thus confirming the key role of the p38/MAPK in promoting osteoclastic differentiation.10-13 On the other hand, PD98059 induced a significant decrease in the total number of cells, but it did not affect the percentage of TRAP-positive, multinucleated cells (Figure 7A), confirming the predominant role of ERK1/2 in preosteoclastic proliferation. Similar results were obtained when SB203580 or PD98059 were added to the RAW264.7 cell line (Figure 7B).
Effect of pharmacologic inhibitors on osteoclastic differentiation. Adherent PBMCs (A) and RAW264.7 cells (B) were cultured in the absence of cytokines or with RANKL plus M-CSF with or without TRAIL. Cultures were preincubated with vehicle (0.1% DMSO), PD98059 (20 μM), SB203580 (10 μM) prior to the addition of cytokines. After 10 days, the number of viable cells and the number of TRAP-positive multinucleated cells were scored. Data represent the means ± SD of 4 different experiments (*P < .05, inhibitor vs vehicle).
Effect of pharmacologic inhibitors on osteoclastic differentiation. Adherent PBMCs (A) and RAW264.7 cells (B) were cultured in the absence of cytokines or with RANKL plus M-CSF with or without TRAIL. Cultures were preincubated with vehicle (0.1% DMSO), PD98059 (20 μM), SB203580 (10 μM) prior to the addition of cytokines. After 10 days, the number of viable cells and the number of TRAP-positive multinucleated cells were scored. Data represent the means ± SD of 4 different experiments (*P < .05, inhibitor vs vehicle).
Discussion
Bone is continuously remodeled by bone formation and resorption processes, and the cooperative bone metabolism is tightly regulated to maintain homeostasis. Deviation from the normal conditions of bone resorption would result in bone diseases such as osteoporosis and osteopetrosis. Osteoclasts, which are responsible for bone resorption in bone metabolism, are multinucleated cells formed by the fusion of circulating hematopoietic precursor cells of the monocyte/macrophage lineage.1 In the bone microenvironment, osteoclast formation requires cell-to-cell interactions of osteoclast precursor cells with osteoblasts and can be achieved by coculturing bone marrow precursor cells with osteoblasts/stromal cells.24,27
A series of hormones (vitamin D3, parathormon), prostaglandin E2, and cytokines (TNF-α, IL-1, transforming growth factor β 1 [TGF-β1]) have been involved in promoting osteoclastogenesis28-32 ; however, a key role in this process has recently been attributed to the RANKL/RANK/OPG system (reviewed in Boyle et al1 ). In vitro, RANKL and TNF-α, the prototype member of the TNF family of cytokines, have a similar osteoclast-inductive capacity and the concomitant presence of RANKL and TNF-α shows a cooperative impact of these cytokines on osteoclastogenesis.31 However, the special characteristic of RANKL is that, at variance to TNF-α, it is not proinflammatory.32 In line with a predominant role of the TNF family members of cytokines in inducing osteoclastogenesis, it has recently been demonstrated that TNF-like weak inducer of apoptosis (TWEAK), an additional member of this family, also induces osteoclastogenesis in RAW264.7 cells.33 On the contrary, we have here documented for the first time that TRAIL is the only member of this family of cytokines with antiosteoclastic activity. In fact, TRAIL significantly inhibited osteoclastic maturation induced by RANKL plus M-CSF, blocking the formation of giant multinucleated cells and totally abolishing their bone resorption activity.
The complexity of TRAIL biologic effects on osteoclastogenesis is not unprecedented. For instance, TGF-β1 and IFN-γ direct responsive M-CSF-dependent precursors toward osteoclastic or cytocidal activities, respectively.32 Just as TGF-β1 deactivates cytocidal macrophages, IFN-γ deactivates the osteoclast pathway. In this respect, the ability of TRAIL, like INF-γ, to deactivate the osteoclastic differentiation of monocytes is particularly noteworthy in light of the study of Kumar-Sinha et al,34 who demonstrated by microarray technology the existence of a cross-talk between the pathways triggered by TRAIL and IFN. In a recent study, it has been reported that TRAIL gene-deficient mice did not show evidence of altered gross bone density, and no alterations in frequency or in vitro differentiation of bone marrow precursor osteoclasts.35 Therefore, the data of Sedger et al,35 together with our present findings, suggest that TRAIL is likely a redundant negative regulator of physiologic osteoclastogenesis. The lack of TRAIL gene expression/function is likely vicariate by INF-γ and/or other cytokines, at least in the mouse model.
In the attempt to elucidate the molecular mechanisms underlining the inhibitory activity of TRAIL on osteoclastogenesis, we have analyzed the phosphorylation levels of ERK/MAPK and p38/MAPK pathways, which play a key role in preosteoclastic proliferation and differentiation, respectively.10-13 Consistent with a key role of p38/MAPK in mediating osteoclastic differentiation, the inhibitory activity of TRAIL was paralleled by its ability to suppress the RANKL plus M-CSF-induced p38 phosphorylation. Moreover, SB203580, a pharmacologic inhibitor of the p38/MAPK pathway, mimicked the biologic activity of TRAIL both in terms of inhibition of p38/MAPK and block of osteoclastic differentiation.
The implications of our study are particularly noteworthy for the potential clinical application of recombinant TRAIL in the treatment of malignancies with osteolitic lesions, and, in particular, multiple myeloma (MM). In fact, several studies have shown that TRAIL, either used alone or in combination with chemotherapy or antiangiogenic therapy, is effective in inducing cytotoxicity of malignant plasma cells.36-40 In MM, bone resorption is increased and associated with the presence of increased numbers of osteoclasts, whereas bone formation is reduced. This uncoupling of resorption and formation, in association with an increase in the frequency of bone remodeling units, leads to rapid bone loss and the development of osteolytic bone lesions.41 Recent studies have shown that human MM cells up-regulated RANKL expression in stromal cells of primary human bone marrow. Moreover, immunohistochemical staining performed on bone marrow biopsy specimens showed an increase of RANKL and a reduction of OPG expression in patients with MM as compared with healthy subjects, suggesting that the RANKL/OPG system is involved in the pathogenesis of MM-induced bone disease.42-44 Thus, our present findings suggest that the therapeutic potential of TRAIL on MM is 2-fold: induction of cytotoxicity of malignant plasma cells and block of osteoclastogenesis, which represent a major pathogenetic aspect of MM.
Prepublished online as Blood First Edition Paper, June 22, 2004; DOI 10.1182/blood-2004-03-1196.
Supported by grants from the Fondo per gli Investimenti della Ricerca di Base (FIRB) (P.S. and G.Z) and the Associazione Italiana per la Ricerca sul Cancro (AIRC) (G.Z.). E.R. is a recipient of a fellowship from Fondazione Rose della Salute, the Organizzationi Non Lucrative di Attivita Sociale (ONLUS).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 2. Effect of TRAIL on differentiation of adherent PBMCs into functional osteoclasts. Adherent PBMCs were left untreated (without cytokines) or cultured in the presence of TRAIL and with RANKL plus M-CSF with or without TRAIL. After 10 days cells were analyzed for osteoclastic differentiation. (A) Representative fields of the cultures, treated as indicated, after TRAP staining (magnification, × 10; numerical aperture of the objective lens [NA], 0.25). Similar results were observed in 7 independent experiments performed in duplicate. (B) Magnification of a representative TRAP-positive multinucleated cell (i), and a representative field of RANKL plus M-CSF-treated cultures observed by light microscopy (ii) and by fluorescence microscopy after DAPI staining (iii). *Polynucleated cells characteristic of RANKL plus M-CSF cultures. Original magnification and NA: i, × 40 0.75 NA; ii-iii, × 20, 0.25 NA. In panel C, adherent PBMCs were plated on an artificial bone matrix slide and were cultured with RANKL plus M-CSF, in the absence or presence of TRAIL (10 ng/mL), as indicated. After 12 days, the slides were fixed and stained, and resorption was determined by examining pit formation under a light microscope (magnification, × 20; NA, 0.40). Representative fields are shown. (D-E) Cultures were treated as indicated and the number of TRAP-positive multinucleated cells containing 3 or more nuclei was scored. Data represent the means ± SD of 3 to 7 different experiments (*P < .05, compared with RANKL plus M-CSF).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/7/10.1182_blood-2004-03-1196/6/m_zh80190467380002.jpeg?Expires=1767700368&Signature=k6VHlFCPnkfC6ty2WXPbU8HI14U9MZMKET1EyBLYO-60819QKLGAmxU37z13iDI3QthXs-rJRyE8FotT~f6IhmhwvYWnOJpeffPtuzXXi7W~rhEFvoy~YAXvhAvcRwwyAB21bfSlDHh~ZGOO~ktwRAIN~IQvactizXVP0ZkzPS7rPn7gBqvPgPKJZvECEkYEXWscBPeFPE31xBUwOV5dU42uYZ-IoWzKCTbaLxoPN0ecPOBGdXvEriMM596b-o1fc1dLdI6rKA-IUFGimarJfqir~LxNwiPZwV4~ivrK8-w-ribodhB6NjuUmUf~aII0txP1rjAUFk6JJ2lMoxHyYA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal