Abstract
Outcome of unrelated donor marrow transplantation is influenced by donor-recipient matching for HLA. Prior studies assessing the effects of mismatches at specific HLA loci have yielded conflicting results. The importance of high-resolution matching for all HLA loci has also not been established. We therefore examined the effects of HLA matching (low or high resolution or both) on engraftment, graft-versus-host disease (GVHD), and mortality in 1874 donor-recipient pairs retrospectively typed at high resolution for HLA-A, -B, -C, -DRB1, -DQ, and -DP. Mismatches at HLA-A, -B, -C, and -DRB1 each had similar adverse effects on mortality. Only HLA-A mismatches demonstrated significant adverse effects on GVHD. These adverse effects on outcome were more evident in transplants with low-resolution versus only high-resolution mismatches. Mismatches for HLA-DQ or -DP did not significantly affect outcome. When high-resolution mismatches at HLA-A, -B, -C, and -DRB1 were considered together, adverse effects on survival and GVHD were observed. We therefore conclude that matching for HLA-C should be incorporated into algorithms for unrelated donor selection. High-resolution mismatches at HLA-A, -B, -C, and -DRB1 adversely affect outcome, but less so than low-resolution mismatches. When clinical circumstances allow, high-resolution class I typing may help optimize donor selection and improve outcome.
Introduction
Allogeneic hematopoietic stem cell transplantation can potentially cure a variety of lymphohematopoietic and congenital metabolic disorders.1-4 Whereas transplants between HLA-identical siblings produce the best outcomes,5 transplants from unrelated donors using marrow,6-11 peripheral blood stem cells,12 or umbilical cord blood,13-18 or using aggressively T cell–depleted mismatched related donors10,19,20 can also yield acceptable outcomes. Of these, the use of bone marrow from well-matched, unrelated donors has been, by far, the most commonly applied approach to date.
The use of unrelated donors introduces a number of questions and problems in donor selection, which do not occur in the context of transplantation from an HLA-identical sibling donor. Ideally, one would hope to identify unrelated donors who are genotypically identical to the patients at all HLA loci, analogous to transplantation between HLA-matched siblings. When a “perfectly” matched donor is not available, it remains uncertain whether some mismatches will be more forgiving than others. Controversies remain as to whether mismatches at some loci have more profound clinical consequences than those at other loci. Virtually every HLA locus has been reported to influence outcome of unrelated donor bone marrow transplantation (BMT),21-34 with conflicting results as to the relative importance of various class I and class II loci.21,24,25,27-29,31,33-37 Many of these studies were limited in size, making definitive conclusions difficult.
Additionally, many of the earlier studies relied on serologic typing approaches for HLA class I loci. Although serologically based HLA typing is accurate in most circumstances, some limitations are associated with this technique. In particular, discrimination among certain closely related HLA alleles is beyond the resolution of serologic typing. However, such polymorphisms can be detected by alloreactive T cells, either in the laboratory or clinically in association with graft-versus-host disease38 (GVHD) or graft rejection39 in transplant recipients. It remains uncertain whether mismatches that can only be detected using high-resolution (allele level) nucleic acid techniques are more permissive of clinical success than those mismatches that can be detected using serology or comparable low-resolution (antigen level) DNA-based typing approaches.
Resolving these questions has important implications with regard to the number of patients for whom acceptably matched unrelated donors can be identified and the search algorithms used for donor selection. To address these issues, we have performed retrospective high-resolution typing for HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1, and -DPB1 on more than 1800 donor-recipient transplant pairs. The effects of HLA mismatching at high and low resolution on transplant outcome are presented, and the implications of these findings for donor selection are discussed.
Patients, materials, and methods
Patient population
Between 1988 and 1996, among all BMTs performed under the auspices of the National Marrow Donor Program (NMDP), retrospective high-resolution typing for HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1, and -DPB1 was performed for 1874 donor-recipient pairs. This subset of transplantations was reflective of the overall case mix with the exceptions of the need for availability of pretransplant donor and recipient samples for retrospective high-resolution typing and the deliberate overrepresentation of chronic myelogenous leukemia (CML) cases (45.8% in this analysis versus 30.1% of controls) to allow a separate outcome study to be performed for that disease (E.P. et al, manuscript in preparation). Consequently, a reciprocal modest decrease was observed in patients with acute myelogenous leukemia (AML; 15.5% versus 21.0%) and acute lymphocytic leukemia (ALL; 16.0% versus 23.6%) among the patients typed by high resolution. This likely also accounts for their slightly higher median age (30 versus 26 years) and percent with Karnofsky scores 90 or higher (76% versus 70%). Approximately 9% had nonmalignant disorders, including severe aplastic anemia, Fanconi anemia, metabolic disorders, and immunodeficiency states. The high-resolution–typed group had a higher proportion of cases from the middle years of the study (1992-1994).
HLA typing
Retrospective high-resolution molecular typing of class II alleles (DRB1, DQA1, DQB1, DPA1, and DPB1)40 was performed mainly by sequence-specific oligonucleotide probe (SSOP) methods with approximately 64% of the samples tested in duplicate. Of these, 23% were tested by sequence-based typing (SBT) methods. All HLA-A, -B, and -C alleles were identified by SBT with duplicate typing performed independently using SSOP methodologies. Duplicate results were compared and discrepancies resolved41 (T.W., Tamara Winden, M.S. et al, manuscript in preparation).
HLA matching
Two levels of DNA-based HLA matching were considered in the analyses of clinical outcomes. Donors and recipients were considered high-resolution (allele level) matched for a given locus when their high-resolution typing was identical, indicating that they expressed the identical allele. Low-resolution (serology level or antigen level) DNA matching involved conversion of the DNA-based typing to its lower-level serologic equivalent, usually by collapsing the 4-digit typing result back to its first 2 digits, with the exception of a few HLA-B alleles that were mapped to their corresponding serologic specificities.42 For HLA-C, low-resolution HLA-C matching was performed by collapsing the 4-digit allele back to the first 2 digits, even though this is less rigorously supported by prior serology than is the case for HLA-A, -B, and -DR.
Although all typing was performed using DNA-based approaches in this study, donors and recipients were considered low-resolution or “serologically equivalent”-matched for a given locus when their low-resolution DNA-typing assignments for a given locus were identical. Low-resolution matching for a given locus indicates that a donor and recipient express similar gene products (and possibly identical ones) for the locus in question. Conversely, a low-resolution mismatch is one that can be detected using low-resolution typing, whereas a high-resolution mismatch could only be detected using high-resolution typing techniques. A high-resolution mismatch is used to indicate that the donor and recipient are matched at the serologic or low level of resolution, but differ with regard to the specific allele they express from within that serologic or low-resolution antigenic family.
In the case of HLA-DQ and HLA-DP, each of the 2 protein chains forming the heterodimeric cell surface molecule may contribute to donor-recipient disparities. If there were differences between one HLA-DQA1 type or one HLA-DQB1 type, these were assumed to result from a single haplotype and were scored as a single HLA-DQ disparity. Disparity for 2 HLA-DQA1 or 2 HLA-DQB1 high-resolution types was scored as 2 HLA-DQ disparities. Comparison of HLA-DP was analogous to HLA-DQ.
Clinical outcomes
Evaluation of clinical outcomes was performed using criteria standardized by the NMDP for all its studies. Diagnosis and grading of acute and chronic GVHD used standard criteria.43-45 Time to engraftment was defined as the first of 3 consecutive absolute neutrophil counts equal to or more than 5.0 × 108/L. Patients were considered evaluable for engraftment if they survived 21 days after transplant and evaluable for chronic GVHD if they survived at least 80 days.
Statistical methods
Comparisons between the subset of cases for which high-resolution typing was performed and the other NMDP-facilitated transplants used the Wilcoxon rank sum test for continuous variables (eg, age) and the likelihood ratio χ2 statistic for categorical variables (eg, gender).
Cumulative incidences were compared at 100 days for acute GVHD and engraftment, and at 2 years for chronic GVHD, treating death as a competing risk, and using a Taylor series linear approximation to estimate the variance.46,47 Survival rates were estimated up to the date when patient follow-up forms were due at the NMDP, calculated by the method of Kaplan and Meier,48 and compared using the log-rank statistic.49
Logistic regression was used for multivariate analysis of neutrophil engraftment and the proportional hazards model was used for the other outcomes.50 Each model included disease/stage as a covariate regardless of significance. For HLA-A, -B, -C, and -DRB1, donor-recipient matching was considered in 3 categories: high-resolution match, low-resolution match/high-resolution mismatch, and low-resolution mismatch. Two indicator variables (one for each type of mismatch) were included in the regression model for each of these 4 loci. A single indicator was included in the model for each of the other HLA loci (DQ and DP) without distinguishing between allele-level and low-resolution mismatches.
Other factors were included in the multivariate models if they demonstrated a statistically significant (Wald χ2P < .05) association with outcome. Factors considered were transplantation center; T-cell depletion; cell dose (T cell–replete cases only); recipient and donor age, sex, cytomegalovirus (CMV) serology, body mass index, and race; interval from diagnosis to transplantation; and year of transplantation. The interval from diagnosis to transplantation was modeled separately for each disease group. Due to nonlinear effects, the continuous variables of recipient age and interval from diagnosis to transplantation were divided into discrete categories.
Additional regression models were run replacing the HLA indicator variables with 2 continuous variables counting the total number of low-resolution and high-resolution mismatches, respectively. HLA-DQ and -DP were ignored in these models because they had demonstrated no statistically significant effect on transplantation outcome. Because of the large number of statistical comparisons performed in this study, only associations with a P value of less than .01 were considered statistically significant.
Results
Types of mismatches at HLA-A, -B, -C, and -DRB1 present in the study population
The number and types of mismatches detected by molecular typing in the 1874 cases analyzed are illustrated in Table 1. Table 1 summarizes the types of mismatches observed at each locus. As illustrated, approximately half the mismatches at HLA-A and HLA-B were detectable by low-resolution typing, and half required high-resolution typing for detection. Nearly twice as many HLA-C locus disparities were identified compared to HLA-A and -B. More than 80% of HLA-C mismatches were detectable with low-resolution typing. Most HLA-DR mismatches could only be detected using high-resolution typing for DRB1, whereas DQ mismatches were evenly split as to whether they required high-resolution typing for detection or were detectable using low-resolution techniques.
Number and types of mismatches by locus observed in 1874 donor-recipient pairs
. | Donor-recipient pairs, n = 1874 . | . | Alleles, n = 3748 . | . | ||
|---|---|---|---|---|---|---|
| Locus . | Total mismatched donor-recipient pairs at this locus, no. (%) . | Total mismatched alleles at this locus, no. (%) . | Mismatches detectable at low resolution, no. (%) . | Mismatches detected at high resolution only, no. (%) . | ||
| HLA-A | 374 (20) | 386 (10) | 219 (57) | 167 (43) | ||
| HLA-B | 477 (25) | 514 (14) | 209 (41) | 305 (59) | ||
| HLA-C | 749 (40) | 851 (23) | 734 (86) | 117 (14) | ||
| HLA-DRB1 | 311 (17) | 342 (9) | 52 (15) | 290 (85) | ||
| HLA-DQ | 415 (22) | 449 (12) | 219 (49) | 230 (51) | ||
| HLA-DP | 1648 (88) | 2255 (60) | NA | NA | ||
. | Donor-recipient pairs, n = 1874 . | . | Alleles, n = 3748 . | . | ||
|---|---|---|---|---|---|---|
| Locus . | Total mismatched donor-recipient pairs at this locus, no. (%) . | Total mismatched alleles at this locus, no. (%) . | Mismatches detectable at low resolution, no. (%) . | Mismatches detected at high resolution only, no. (%) . | ||
| HLA-A | 374 (20) | 386 (10) | 219 (57) | 167 (43) | ||
| HLA-B | 477 (25) | 514 (14) | 209 (41) | 305 (59) | ||
| HLA-C | 749 (40) | 851 (23) | 734 (86) | 117 (14) | ||
| HLA-DRB1 | 311 (17) | 342 (9) | 52 (15) | 290 (85) | ||
| HLA-DQ | 415 (22) | 449 (12) | 219 (49) | 230 (51) | ||
| HLA-DP | 1648 (88) | 2255 (60) | NA | NA | ||
Shown are the total number of mismatches detected in the 3748 alleles typed at each locus in the 1874 donor-recipient pairs. The absolute number and percentage of mismatches detectable at either low or high resolution only is shown.
The match/mismatch characteristics of the 1874 donor-recipient pairs are summarized in Table 2. Only 6% of the donor-recipient pairs were matched for all 8 of the loci studied. Thirty-six percent of the pairs were matched at high resolution for HLA-A, -B, -C, and -DRB1 but had a mismatch at HLA-DQ or -DP or both. Thus, between these 2 groups, 42% of the pairs were matched at high resolution for HLA-A, -B, -C, and -DRB1. Of the remaining 58% of the study pairs, 34% were matched for HLA-A, -B, -C, and -DRB1 at low resolution but had one or more mismatches at high resolution. Twenty-four percent had at least one HLA-A, -B, -C, or -DRB1 mismatch that could be detected at low resolution. Within the study population, 25% of pairs exhibited one HLA-A, -B, -C, or -DRB1 mismatch, 19% exhibited 2 mismatches, and 14% exhibited 3 or more mismatches at these loci (C.W., M.F.-V., W.H. et al, manuscript in preparation).
Mismatch characteristics in 1874 donor-recipient pairs
Mismatch characteristics . | Donor-recipient pairs, no. (%) . |
|---|---|
| Matched for all 8 loci | 108 (6) |
| High-resolution mismatch at HLA-A,B,C and/or DRB1 | 631 (34) |
| Low-resolution mismatch at HLA-A,B,C and/or DRB1 | 452 (24) |
| DQ and/or DP mismatch only | 683 (36) |
| Total | 1874 (100) |
| Total mismatched alleles at HLA-A, -B, -C, and/or -DRB1 | |
| 0 | 791 (42) |
| 1 | 469 (25) |
| 2 | 351 (19) |
| 3+ | 263 (14) |
| Total | 1874 (100) |
Mismatch characteristics . | Donor-recipient pairs, no. (%) . |
|---|---|
| Matched for all 8 loci | 108 (6) |
| High-resolution mismatch at HLA-A,B,C and/or DRB1 | 631 (34) |
| Low-resolution mismatch at HLA-A,B,C and/or DRB1 | 452 (24) |
| DQ and/or DP mismatch only | 683 (36) |
| Total | 1874 (100) |
| Total mismatched alleles at HLA-A, -B, -C, and/or -DRB1 | |
| 0 | 791 (42) |
| 1 | 469 (25) |
| 2 | 351 (19) |
| 3+ | 263 (14) |
| Total | 1874 (100) |
Shown are the types of mismatches detected between donor and recipient in the 1874 pairs. “High-resolution mismatch at HLA-A, -B, -C, or DRB1” indicates the presence of one or more mismatches at one of these loci detectable with high-resolution typing only. “Low-resolution mismatch at HLA-A,B,C or DRB1” indicates the presence of one or more mismatches at one of these loci detectable at low resolution with or without additional high-resolution mismatches. The 8 loci typed include HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1, and -DPB1.
Impact of mismatching at individual HLA loci on transplant-related outcomes
Table 3 shows the results of multivariate analyses of the impact of HLA mismatching on transplant-related outcomes. The cumulative incidence of engraftment in the overall study population was 95% ± 1%. Even among patients with HLA mismatches, engraftment rates remained high. HLA-C mismatching showed the strongest association with graft failure (OR of engraftment 0.54, P = .02), although this did not achieve the statistical significance threshold defined for this study. Multiple HLA-C mismatches were not associated with a significantly higher risk of nonengraftment (data not shown).
Impact of HLA mismatching at specific loci on transplant-related outcomes: associations between HLA mismatch (high- or low-resolution) at specific loci and outcome after unrelated donor BMT
. | . | Engraftment . | . | . | Grade III-IV acute GVHD . | . | . | Chronic GVHD . | . | . | Mortality . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mismatched locus . | No. . | OR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | ||||||||
| HLA-A | 374 | 0.68 | (0.39, 1.22) | .20 | 1.41 | (1.16, 1.71) | .0005 | 1.35 | (1.09, 1.66) | .006 | 1.33 | (1.15, 1.54) | .0002 | ||||||||
| HLA-B | 477 | 1.07 | (0.61, 1.88) | .82 | 1.24 | (1.03, 1.50) | .03 | 1.04 | (0.84, 1.30) | .71 | 1.22 | (1.06, 1.41) | .007 | ||||||||
| HLA-C | 749 | 0.54 | (0.33, 0.89) | .02 | 1.19 | (1.00, 1.41) | .05 | 1.01 | (0.84, 1.21) | .92 | 1.21 | (1.06, 1.38) | .005 | ||||||||
| HLA-DRB1 | 311 | 1.07 | (0.57, 2.02) | .83 | 1.26 | (1.01, 1.56) | .04 | 1.27 | (0.99, 1.64) | .07 | 1.23 | (1.04, 1.45) | .01 | ||||||||
| HLA-DQ | 415 | 0.67 | (0.39, 1.14) | .14 | 1.03 | (0.85, 1.26) | .76 | 0.93 | (0.74, 1.16) | .50 | 0.98 | (0.84, 1.14) | .80 | ||||||||
| HLA-DP | 1648 | 0.69 | (0.38, 1.25) | .22 | 1.19 | (0.99, 1.43) | .06 | 1.17 | (0.98, 1.39) | .08 | 1.07 | (0.89, 1.27) | .48 | ||||||||
. | . | Engraftment . | . | . | Grade III-IV acute GVHD . | . | . | Chronic GVHD . | . | . | Mortality . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mismatched locus . | No. . | OR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | ||||||||
| HLA-A | 374 | 0.68 | (0.39, 1.22) | .20 | 1.41 | (1.16, 1.71) | .0005 | 1.35 | (1.09, 1.66) | .006 | 1.33 | (1.15, 1.54) | .0002 | ||||||||
| HLA-B | 477 | 1.07 | (0.61, 1.88) | .82 | 1.24 | (1.03, 1.50) | .03 | 1.04 | (0.84, 1.30) | .71 | 1.22 | (1.06, 1.41) | .007 | ||||||||
| HLA-C | 749 | 0.54 | (0.33, 0.89) | .02 | 1.19 | (1.00, 1.41) | .05 | 1.01 | (0.84, 1.21) | .92 | 1.21 | (1.06, 1.38) | .005 | ||||||||
| HLA-DRB1 | 311 | 1.07 | (0.57, 2.02) | .83 | 1.26 | (1.01, 1.56) | .04 | 1.27 | (0.99, 1.64) | .07 | 1.23 | (1.04, 1.45) | .01 | ||||||||
| HLA-DQ | 415 | 0.67 | (0.39, 1.14) | .14 | 1.03 | (0.85, 1.26) | .76 | 0.93 | (0.74, 1.16) | .50 | 0.98 | (0.84, 1.14) | .80 | ||||||||
| HLA-DP | 1648 | 0.69 | (0.38, 1.25) | .22 | 1.19 | (0.99, 1.43) | .06 | 1.17 | (0.98, 1.39) | .08 | 1.07 | (0.89, 1.27) | .48 | ||||||||
Shown are results of multivariate analysis including odds ratio (OR) for engraftment and relative risk (RR) for GVHD and mortality, 95% confidence intervals (CI), and P values associated with mismatching at each locus. Statistically significant associations are shown in boldface. HLA-A mismatching showed significant associations (boldface) with acute GVHD, chronic GVHD, and mortality. HLA-B, -C, and -DR showed significant associations with mortality. Mismatch at HLA-C showed a trend to association with poorer engraftment, although not reaching the significance threshold defined for this study. Mismatched HLA-B, -C, -DR all showed trends to more frequent acute GVHD.
Mismatching for HLA-A was associated with a significantly increased risk of grades III/IV acute GVHD (RR = 1.41; P = .005). Mismatches for HLA-B, -C, -DR, and -DP were each associated with relative risks for grades III/IV acute GVHD of around 1.2, but these did not reach independent statistical significance (P = .03-.06). Recipients mismatched at HLA-DQ demonstrated no increased risk of developing grades III/IV acute GVHD (RR = 1.03; P = .76).
HLA-A mismatching was also associated with a significantly higher incidence of chronic GVHD (RR = 1.35; P = .006). HLA-DR and -DP mismatches were associated with somewhat higher relative risks of developing chronic GVHD, but these associations were not statistically significant. Mismatching for HLA-B, -C, and -DQ was not associated with increased risk of chronic GVHD.
The final and most important outcome variable analyzed was mortality. As illustrated in Table 3, mismatches for HLA-A, -B, -C, or -DR were each independently associated with significantly higher risks of mortality. In contrast, mismatching for HLA-DQ and -DP did not appear to exert any significant effect on survival.
Impact of high- and low-resolution mismatches at individual loci on transplant-related outcomes
Mismatches at HLA-A, -B, -C, and -DR were all associated with at least 1 significant effect on a major clinical outcome. Therefore, we next examined these associations after subdividing the mismatches into those detectable at low resolution versus those requiring high-resolution typing for detection. In this analysis, HLA-DQ and -DP mismatches were not included because these loci showed no significant independent association with any of the transplant outcomes studied.
Clinical outcomes were examined in the subsets of patients with either high- or low-resolution mismatches at each locus (Table 4) and compared with the results observed when these 2 subgroups were combined (Table 3). As shown in Table 3, mismatching for a particular HLA locus was associated with a statistically significant (P < .01) impact on transplant outcome in 6 cases. Specifically, mismatches at HLA-A, -B, -C, and -DR each demonstrated significant associations with mortality, whereas HLA-A mismatching also demonstrated significant associations with acute and chronic GVHD. As shown in Table 4, in all 6 of these instances a similar statistically significant adverse effect was observed in the subset of mismatches detectable with low-resolution typing. Within the limits of the current sample size, there was no instance, either in these 6 cases or the others analyzed, where high-resolution mismatching showed an independent statistically significant impact on outcome when analyzed for an individual HLA locus. However, if one compares the low- and high-resolution mismatched subsets, the relative risks are similar in 4 of the 6 cases noted above (HLA-A and -DR on mortality; HLA-A on acute and chronic GVHD).
Impact of HLA mismatching at specific loci on transplant-related outcomes: associations between high- versus low-resolution HLA mismatch at specific loci and outcome after unrelated donor BMT
. | . | Engraftment . | . | . | Grade III-IV acute GVHD . | . | . | Chronic GVHD . | . | . | Mortality . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | No. . | OR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | ||||||||
| HLA-A match | 1500 | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | ||||||||
| Hi-res mismatch | 157 | 0.95 | (0.40, 2.26) | .91 | 1.31 | (1.00, 1.70) | .05 | 1.25 | (0.91, 1.70) | .17 | 1.26 | (1.02, 1.55) | .03 | ||||||||
| Low-res mismatch | 217 | 0.51 | (0.24, 1.08) | .08 | 1.51 | (1.18, 1.95) | .001 | 1.42 | (1.09, 1.85) | .01 | 1.44 | (1.20, 1.74) | < .0001 | ||||||||
| HLA-B match | 1397 | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | ||||||||
| Hi-res mismatch | 270 | 1.66 | (0.75, 3.69) | .22 | 1.13 | (0.91, 1.42) | .27 | 1.07 | (0.83, 1.38) | .58 | 1.08 | (0.90, 1.28) | .41 | ||||||||
| Low-res mismatch | 207 | 0.72 | (0.34, 1.49) | .37 | 1.35 | (1.03, 1.76) | .03 | 0.95 | (0.68, 1.32) | .74 | 1.46 | (1.19, 1.80) | .0003 | ||||||||
| HLA-C match | 1125 | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | ||||||||
| Hi-res mismatch | 83 | 0.80 | (0.29, 2.24) | .68 | 0.83 | (0.58, 1.20) | .32 | 0.93 | (0.65, 1.32) | .66 | 0.96 | (0.72, 1.29) | .78 | ||||||||
| Low-res mismatch | 666 | 0.54 | (0.31, 0.93) | .03 | 1.24 | (1.03, 1.50) | .02 | 1.05 | (0.86, 1.28) | .61 | 1.21 | (1.06, 1.40) | .007 | ||||||||
| HLA-DRB1 match | 1563 | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | ||||||||
| Hi-res mismatch | 260 | 0.77 | (0.39, 1.50) | .44 | 1.29 | (1.02, 1.62) | .03 | 1.34 | (1.04, 1.74) | 0.03 | 1.21 | (1.02, 1.44) | |||||||||
| Low-res mismatch | 51 | 1.61 | (0.28, 9.26) | .59 | 1.42 | (0.88, 2.29) | .15 | 0.71 | (0.32, 1.57) | 0.40 | 1.64 | (1.13, 2.38) | .009 | ||||||||
. | . | Engraftment . | . | . | Grade III-IV acute GVHD . | . | . | Chronic GVHD . | . | . | Mortality . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus . | No. . | OR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | RR . | 95% CI . | P . | ||||||||
| HLA-A match | 1500 | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | ||||||||
| Hi-res mismatch | 157 | 0.95 | (0.40, 2.26) | .91 | 1.31 | (1.00, 1.70) | .05 | 1.25 | (0.91, 1.70) | .17 | 1.26 | (1.02, 1.55) | .03 | ||||||||
| Low-res mismatch | 217 | 0.51 | (0.24, 1.08) | .08 | 1.51 | (1.18, 1.95) | .001 | 1.42 | (1.09, 1.85) | .01 | 1.44 | (1.20, 1.74) | < .0001 | ||||||||
| HLA-B match | 1397 | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | ||||||||
| Hi-res mismatch | 270 | 1.66 | (0.75, 3.69) | .22 | 1.13 | (0.91, 1.42) | .27 | 1.07 | (0.83, 1.38) | .58 | 1.08 | (0.90, 1.28) | .41 | ||||||||
| Low-res mismatch | 207 | 0.72 | (0.34, 1.49) | .37 | 1.35 | (1.03, 1.76) | .03 | 0.95 | (0.68, 1.32) | .74 | 1.46 | (1.19, 1.80) | .0003 | ||||||||
| HLA-C match | 1125 | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | ||||||||
| Hi-res mismatch | 83 | 0.80 | (0.29, 2.24) | .68 | 0.83 | (0.58, 1.20) | .32 | 0.93 | (0.65, 1.32) | .66 | 0.96 | (0.72, 1.29) | .78 | ||||||||
| Low-res mismatch | 666 | 0.54 | (0.31, 0.93) | .03 | 1.24 | (1.03, 1.50) | .02 | 1.05 | (0.86, 1.28) | .61 | 1.21 | (1.06, 1.40) | .007 | ||||||||
| HLA-DRB1 match | 1563 | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | 1.00 | — | — | ||||||||
| Hi-res mismatch | 260 | 0.77 | (0.39, 1.50) | .44 | 1.29 | (1.02, 1.62) | .03 | 1.34 | (1.04, 1.74) | 0.03 | 1.21 | (1.02, 1.44) | |||||||||
| Low-res mismatch | 51 | 1.61 | (0.28, 9.26) | .59 | 1.42 | (0.88, 2.29) | .15 | 0.71 | (0.32, 1.57) | 0.40 | 1.64 | (1.13, 2.38) | .009 | ||||||||
Shown are results of multivariate analysis including odds ratio (OR) for engraftment and relative risk (RR) for GVHD and mortality, 95% confidence intervals (CI), and P values associated with mismatching at each locus. Statistically significant associations are shown in boldface. Low- but not high-resolution mismatching for HLA-A adversely affected acute GVHD, chronic GVHD, and mortality, whereas low-resolution mismatch for HLA-B, -C, and -DR showed significant effects on mortality. Similar adverse relative risks and trends (.01 < P < .05) were noted also with high-resolution mismatch at HLA-A for acute GVHD and mortality, HLA-DR for acute GVHD, chronic GVHD and mortality. Low-resolution but not high-resolution mismatching demonstrated trends toward adverse effects for HLA-B on acute GVHD and for HLA-C on engraftment and acute GVHD. Res indicates resolution.
If one compares the low-resolution mismatched subset (Table 4) to the composite group of mismatches (ie, low- and high-resolution mismatches combined [Table 3]), the relative risks are higher for the low-resolution mismatched subset in 5 of the 6 cases noted and the same in one (impact of HLA-C on mortality). Looking separately at the impact of HLA-A, -B, -C, or -DR mismatching on mortality, the statistical significance of the associations was actually stronger in the low-resolution subsets than in the composite (high and low) groups, despite the fact that the sample sizes were smaller and the degrees of freedom higher.
Thus, in each of these 6 instances where mismatching at one particular HLA locus was associated with an adverse effect on transplant outcome, the risks were most evident in those cases mismatched at low resolution. However, the number of patients within each of these groups is too small to formally prove whether high-resolution and low-resolution mismatches at any given locus truly differ from one another with regard to risk, despite the fact that low-resolution mismatches showed statistically significant associations with outcome events but high-resolution mismatches did not.
Impact of class I versus class II high-resolution mismatches on transplant outcome
Within the limits of the current study population, high-resolution HLA mismatches at any single locus did not demonstrate an independent statistically significant adverse effect on unrelated donor BMT outcome. However, when the high-resolution mismatches detectable from DNA-based typing for class I (HLA-A, -B, and -C) were pooled, additional effects on outcome could be demonstrated.
Using HLA-A, -B, and -DR low-resolution matched pairs as our starting group, we assessed the clinical impact of additional mismatches at either HLA class I loci versus mismatches at HLA-DRB1 in these patients. As illustrated in Table 5, the presence of a single mismatch at class I or a single mismatch for HLA-DRB1 had similar deleterious effects on the incidence of grades III/IV acute GVHD (8%-10% more frequent) and mortality (8%-12% worse at 5 years). Next, as illustrated in Table 6, we analyzed cases matched for HLA-A and -B at low resolution and HLA-DRB1 at high resolution to assess the impact of additional mismatches at HLA-A, -B, and -C versus mismatches for HLA-DQ and -DP on transplant outcome. In this group, often clinically described as “6 antigen matched,” additional mismatches for HLA-DQ and -DP had no impact on survival, whereas, in contrast, a single additional class I mismatch had a substantial adverse effect (7%-8% worse) on survival.
Impact of mismatching on transplant outcome: impact of DRB1 versus class I mismatching in HLA-A, B, DR serologically matched pairs
. | No. . | Grades III—IV acute GVHD, % . | 5-y survival, % . |
|---|---|---|---|
| No high-resolution mismatches | 791 | 30 ± 3 | 39 ± 3 |
| Single mismatch at class I | 317 | 40 ± 5 | 31 ± 5 |
| Single high-resolution mismatch at DRB | 77 | 38 ± 10 | 27 ± 10 |
. | No. . | Grades III—IV acute GVHD, % . | 5-y survival, % . |
|---|---|---|---|
| No high-resolution mismatches | 791 | 30 ± 3 | 39 ± 3 |
| Single mismatch at class I | 317 | 40 ± 5 | 31 ± 5 |
| Single high-resolution mismatch at DRB | 77 | 38 ± 10 | 27 ± 10 |
Shown are comparisons between mismatch at class I versus HLA-DRB1 on acute GVHD (P = .65) and 5-year survival (P = .37). Single mismatches at class I include high-resolution mismatches for HLA-A and -B and all mismatches for HLA-C.
Impact of mismatching on transplant outcome: impact of additional mismatching on survival among HLA-A, B low-resolution and DRB1 (“6 antigen”) matched pairs
. | No. . | 5-y survival, % . |
|---|---|---|
| High-resolution matched for 8 of 8 loci | 108 | 39 ± 9 |
| Mismatch at DQ/DP only | 683 | 39 ± 4* |
| Single class I mismatch | 317 | 31 ± 5† |
. | No. . | 5-y survival, % . |
|---|---|---|
| High-resolution matched for 8 of 8 loci | 108 | 39 ± 9 |
| Mismatch at DQ/DP only | 683 | 39 ± 4* |
| Single class I mismatch | 317 | 31 ± 5† |
Eight locus match includes high-resolution matching for HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1, and -DPB1. Class I mismatch refers to high-resolution mismatches for HLA-A and -B and all mismatches for HLA-C.
P = .93 compared to 8 of 8 matched pairs.
P = .08 compared to 8 of 8 matched pairs.
In prior reports, HLA-DQ mismatches have been shown to adversely affect transplant outcome.25,27 We therefore assessed whether the prior observation might have resulted from the ability of a mismatch for HLA-DQ to aggravate the impact of class I disparities that went undetected in those studies. In patients matched for HLA-DRB1 with a single disparity for HLA class I, additional disparity for HLA-DQ had no adverse effect on outcome (data not shown).
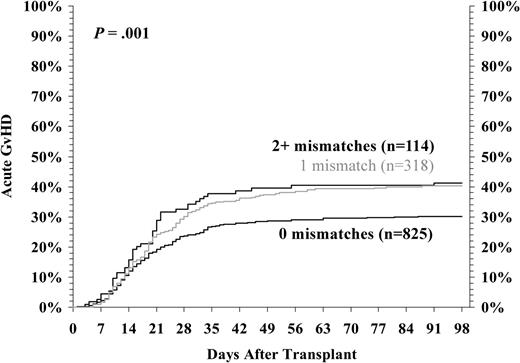
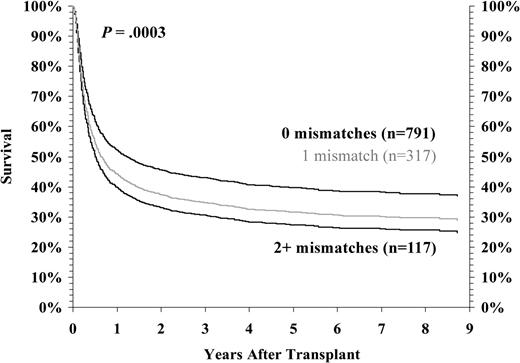
We then considered the cumulative impact of all allele-level mismatches for HLA-A, -B, -C, and -DRB1 on GVHD risk and survival in transplants performed between HLA-A, -B low-resolution matched pairs and DRB1 high-resolution matched pairs. As illustrated in Figure 1, there is a statistically significant increase in risk of developing grades III/IV acute GVHD with even a single class I mismatch. Similarly, survival progressively declined as the number of mismatches for HLA-A, -B, and -C increased (Figure 2). In multivariate analysis, the relative risk of grades III/IV acute GVHD increased from 1.0 to 1.53 to 1.78 as the number of HLA-A, -B, -C, and -DR mismatches increased from 0 to 1 to 2. Similarly, the risk of mortality increased from 1.0 to 1.32 to 1.53.
Grades III/IV acute GVHD among HLA-A, -B serologic, and -DRB1 allele-matched pairs by number of class I mismatched loci. The incidence of grades III/IV acute GVHD was analyzed as a function of the number of class I mismatches detected by high-resolution HLA typing in HLA-A, -B low-resolution, and -DRB1 high-resolution matched donor-recipient pairs. The data presented are adjusted for competing risk factors using the proportional hazards model, rather than univariate analysis. One or more additional mismatches led to more frequent GVHD (P = .001).
Grades III/IV acute GVHD among HLA-A, -B serologic, and -DRB1 allele-matched pairs by number of class I mismatched loci. The incidence of grades III/IV acute GVHD was analyzed as a function of the number of class I mismatches detected by high-resolution HLA typing in HLA-A, -B low-resolution, and -DRB1 high-resolution matched donor-recipient pairs. The data presented are adjusted for competing risk factors using the proportional hazards model, rather than univariate analysis. One or more additional mismatches led to more frequent GVHD (P = .001).
Risk-adjusted survival among HLA-A, -B serologic, and -DRB1 allele-matched pairs by number of mismatched class I loci. Survival after transplantation was analyzed as a function of the number of class I mismatches detected by high-resolution HLA typing in HLA-A, -B low-resolution, and -DRB1 high-resolution matched donor-recipient pairs. The data presented are adjusted for competing risk factors using the proportional hazards model, rather than univariate analysis. One or more additional mismatches led to poorer risk-adjusted survival (P = .0003).
Risk-adjusted survival among HLA-A, -B serologic, and -DRB1 allele-matched pairs by number of mismatched class I loci. Survival after transplantation was analyzed as a function of the number of class I mismatches detected by high-resolution HLA typing in HLA-A, -B low-resolution, and -DRB1 high-resolution matched donor-recipient pairs. The data presented are adjusted for competing risk factors using the proportional hazards model, rather than univariate analysis. One or more additional mismatches led to poorer risk-adjusted survival (P = .0003).
It should be noted that in the above analyses, both high- and low-resolution C locus disparities were included. If one restricts the analysis to pairs who are matched at low-resolution for HLA-C as well as the other loci, the trends observed are the same, although they do not reach statistical significance in these smaller groups (data not shown).
Because of the direct implications on donor selection, we constructed additional regression models to compare the impact of lowversus high-resolution mismatches at HLA-A, -B, -C, or -DR on GVHD or survival. Donor-recipient pairs with a single low-resolution mismatch were compared to pairs with a single high-resolution mismatch. Pairs with 2 high-resolution mismatches were compared to pairs with one high-resolution and one low-resolution mismatch (2 total mismatches). Patients with 3 and 4 mismatches were compared in similar fashion. In this model, mismatches detectable at low resolution were associated with similar risks of grades III/IV acute GVHD as high-resolution mismatches (RR = 1.16; P = .18). However, transplants with low-resolution mismatches were associated with significantly worse survival than those with only high-resolution mismatches (RR = 1.26; P = .006).
Frequency of high-resolution mismatches in low-resolution matched pairs
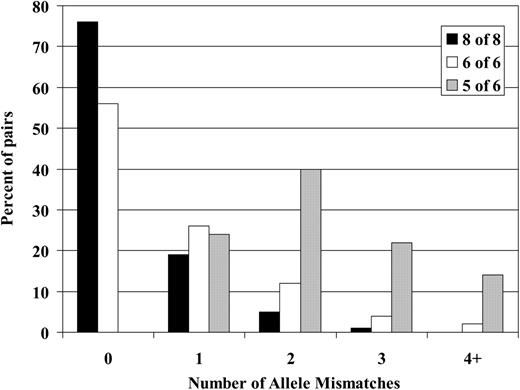
Because matches at HLA-A, -B, -C, and -DR were all shown to influence survival after unrelated donor BMT, we assessed the ability of low-resolution HLA-A, -B, -C, and -DR typing to predict the outcome of high-resolution typing for these same loci. These results are illustrated in Figure 3. Low-resolution matching for HLA-A, -B, and -DR (6 of 6 match) was associated only with a 56% chance of high-resolution matching for HLA-A, -B, -C, and DR. Among these HLA-A, -B, -DR low-resolution matched pairs, 26% had at least one high-resolution mismatch for HLA-A, -B, -C, or -DR, whereas 12% had 2, 4% had 3, and 2% had 4 or more mismatches, respectively. Among the donor-recipient pairs matched at low resolution for HLA-C as well as for HLA-A, -B, and -DR (8 of 8 match), the frequency of high-resolution matching for all 4 loci increased to 76%. Nineteen percent of these cases had a single high-resolution mismatch for HLA-A, -B, -C, or -DR and only 5% and 1% of cases exhibited 2 and 3 high-resolution mismatches, respectively. However, if a single low-resolution mismatch is detected for HLA-A, -B, or -DR (5 of 6 match), the likelihood of identifying additional occult mismatches after high-resolution typing increases substantially. After high-resolution typing of these 5 of 6 matched pairs, additional mismatches are identified in 76% of pairs. Forty percent had one additional mismatch beyond the original mismatch detected by low-resolution typing (2 mismatches in total), 22% had 3, and 14% had 4 HLA-A, -B, -C, or -DR mismatches.
Predictive value of HLA-A, -B, -C, and -DR low-resolution typing for subsequent matching after high-resolution typing. The frequency of unrecognized high-resolution mismatches in pairs selected using low-resolution typing for HLA-A, -B, and -DR ± HLA-C. “6of6” match refers to pairs matched at low resolution for HLA-A, -B, and -DR (n = 1422). “5 of 6” match refers to pairs with a single low-resolution mismatch for HLA-A, -B, or -DR (n = 429). “8 of 8” match refers to pairs matched at low resolution for HLA-A, -B, -C, and -DR (n = 1047). The data show the frequency of pairs with 0, 1, 2, 3, and 4 or more allele-level mismatches in each cohort.
Predictive value of HLA-A, -B, -C, and -DR low-resolution typing for subsequent matching after high-resolution typing. The frequency of unrecognized high-resolution mismatches in pairs selected using low-resolution typing for HLA-A, -B, and -DR ± HLA-C. “6of6” match refers to pairs matched at low resolution for HLA-A, -B, and -DR (n = 1422). “5 of 6” match refers to pairs with a single low-resolution mismatch for HLA-A, -B, or -DR (n = 429). “8 of 8” match refers to pairs matched at low resolution for HLA-A, -B, -C, and -DR (n = 1047). The data show the frequency of pairs with 0, 1, 2, 3, and 4 or more allele-level mismatches in each cohort.
Discussion
We have described a large and diverse analysis of unrelated donor transplant recipients assessing the impact of high-resolution HLA matching for all major class I and class II loci on transplant outcome. The results demonstrate strong negative effects of mismatching for either HLA-A, -B, -C, or -DRB1 on survival after unrelated donor BMT. Single mismatches at these loci were associated with significant decrements in survival, and the presence of multiple mismatches was even worse. Low-resolution mismatches appear to have a more severe impact on survival than mismatches detectable only with high-resolution typing techniques, but high-resolution mismatches were also associated with adverse outcomes.
Using rigorous statistical criteria, low- but not high-resolution HLA-DRB1 mismatching was associated with adverse effects on survival. Previous studies reported that HLA-DRB1 mismatching (low or high resolution) was associated with worse GVHD and survival.24,28 Since that time, HLA-DRB1 matching has been a priority in most transplant programs, driving the broad-based adoption of molecularly based DRB1 typing. During the last decade, relatively few patients proceeded to transplantation with HLA-DR low-resolution mismatched donors. When a high-resolution mismatch for HLA-DRB1 was unavoidable, some transplant centers have attempted to select donors whose DRB1 mismatches had fewer amino acid disparities, conservative amino acid substitutions, or substitutions in portions of the molecules thought to be less functionally significant. Data supporting these assumptions in donor selection have not been reported. However, it is conceivable that such practices have sufficiently skewed the types of DRB1 mismatches occurring in the present study so as to dilute the impact of mismatching at this locus on transplant outcome, partially accounting for the finding that high-resolution HLA-DRB1 mismatching was not clearly associated with any adverse effects on GVHD and survival using the study's rigorous statistical criteria. Additionally, the small subset of low-resolution DRB1 mismatches had the highest relative risk for mortality. Our data concerning HLA-DRB1 are consistent with earlier observations, despite the lack of an independently significant association, and we encourage continued use of high-resolution HLA-DRB1 matching as a criterion in donor selection.
Analysis of class I allele matching in Japan29 identified increased GVHD with HLA-A and HLA-C mismatching, and poorer survival with HLA-A mismatching. HLA-B associations with GVHD and survival were observed in univariate, but not multivariate analyses. HLA class II disparities did not affect outcome. Morishima et al37 subsequently reported that single high-resolution disparities at HLA-A, -B, and -C led to more acute GVHD and graft failure in Japan, whereas HLA-A and -B disparities worsened chronic GVHD and survival. In contrast to the earlier report of Sasazuki et al,29 HLA-DRB1 was also a risk factor for acute GVHD but not other transplant outcomes. These studies did not address the relative importance of low- versus high-resolution mismatches. Smaller studies of white populations have also raised the question as to whether HLA-C mismatching worsens GVHD and survival.22,23
In contrast, our study demonstrated significant adverse impact for HLA-A, -B, or -C mismatching on survival. Whereas only HLA-A demonstrated statistically significant effects on the incidence of grades III/IV acute GVHD, HLA-B, -C, and -DR mismatches all showed trends for more frequent acute GVHD (RR = 1.19-1.26; .01 < P < .05). Moreover, current techniques for scoring GVHD focus only on peak severity and do not capture information about resistance to therapy or the intensity and duration of treatment required. Thus, whether mismatches at HLA loci result in more resistant GVHD and worse survival, despite a similar overall incidence, requires further study. The reasons underlying differences in the consequences of HLA-DRB1 mismatching in Japan and North America are unclear. However, disparities at various HLA loci may differ between the Japanese population and the more heterogeneous NMDP population, which includes whites, African Americans, Hispanics, Asians, and individuals of mixed ancestry.
Prior North American and Japanese studies have suggested that HLA-C mismatches augment graft failure risks.26,37 Petersdorf et al33 reported increased risks of graft failure with low-resolution or multiple high-resolution mismatches for HLA class I loci, though this was primarily in patients with CML. We observed only a statistically insignificant trend to greater graft failure risks with HLA-C mismatching, but not with other loci.
Our analysis shows strong adverse effects on survival from mismatches at HLA-A, -B, -C, or -DRB1. The observation concerning HLA-C is particularly important because this locus is omitted in most matching algorithms. The present study suggests that HLA-C exerts significant effects on survival, comparable in magnitude to HLA-A, -B, and -DRB1. Strong linkage disequilibrium between HLA-B and -C results in their frequent coordinate matching, but it is important that the favorable, independent impact of HLA-C matching on survival be recognized and included in algorithms used for unrelated donor selection.51
The impact of HLA-DQ and -DP has not been clearly established from prior reports. Consistent with the report of Sasazuki et al,29 in our analysis mismatches at HLA-DQ and -DP had little impact on transplant outcome. This is in contradistinction to other reports which found HLA-DQ and -DP mismatching to be risk factors for GVHD and other posttransplant outcomes.25,27,31,33 In the present study, HLA-DQ showed an RR of 1.03 for grades III/IV acute GVHD and an RR of 0.98 for mortality with P = 0.76 and .80, respectively. To reconcile the present findings concerning HLA-DQ with prior observations,25,27,31,33 we considered the possibility that HLA-DQ disparities may have worsened the clinical consequences of high-resolution class I mismatches,30,52 which went undetected because they were not consistently typed for in the earlier studies.25,27,31,33 We therefore assessed outcomes of patients with HLA-DQ disparities with or without associated class I disparities. These 2 groups of patients behaved identically, and thus matching at HLA-DQ was not confirmed as critical to successful outcome of unrelated donor BMT. Linkage disequilibrium between the DR and DQ loci may have limited the spectrum of DQ mismatches in the study population to more limited mismatches such as DQB1*0301 versus DQB1*0302. It is also conceivable that, analogous to HLA-DRB1, donor selection practices may have similarly skewed the HLA-DQ mismatches in the study to those perceived to be more clinically permissible. However, in contrast to HLA-DRB1, the statistical analysis does not suggest any deleterious effects independently associated with HLA-DQ mismatch.
Prior matching algorithms have often favored class II matching over class I matching when a complete match could not be identified. HLA-DP is particularly problematic because this locus shows little positive genetic linkage disequilibrium with the rest of the major histocompatibility complex. The strong impact of HLA-A, -B, -C, and -DRB1 on survival and the lack of any significant impact of HLA-DQ and -DP suggest that when mismatching is unavoidable, it may be appropriate to accept a mismatch for HLA-DQ if this will facilitate better matching for HLA-class I loci and DR. The current data do not support expending the resources nor incurring the delays required for HLA-DP typing.
The present study is the first to analyze the relative clinical importance of high- versus low-resolution mismatching for HLA-class I loci for mortality and GVHD. In all 6 instances where mismatching for HLA-A, -B, -C, or -DR demonstrated a significant effect on clinical outcome (Table 3, boldface data), a statistically significant effect was also noted for the corresponding subset of low-resolution mismatched cases (Table 4, boldface data). Moreover, in the stratified regression analyses performed to compare the relative effects of high- versus low-resolution mismatches, a low-resolution mismatch was associated with significantly worse posttransplant survival. In contrast, we observed similar adverse impact of low- or high-resolution mismatching on acute GVHD. Petersdorf52 has reported that HLA mismatches detectable at low resolution carry a higher risk of graft rejection than mismatches only detectable using high-resolution typing. This is consistent with the concept that low-resolution mismatched donor-recipient pairs are likely to differ at a larger number of immunogenic epitopes than would usually occur in the setting of a high-resolution mismatch. Although these data argue that certain high-resolution mismatches are perhaps more permissive than their low-resolution counterparts, high-resolution mismatches do, when pooled together, still exhibit adverse effects on transplant outcome. In HLA-A, -B, -DR low-resolution matched pairs, the adverse effects revealed by additional typing information for HLA-A, -B, and -C versus HLA-DRB1 are similar in the 2 subgroups. Thus the current data would indicate that there is demonstrable clinical benefit to using high-resolution class I typing to guide unrelated donor selection.
It is noteworthy that low-resolution HLA-A, -B, and -DR matching, a commonly used starting point for donor selection, provides little more than a 50% likelihood that a donor will be matched for HLA-A, -B, -C, and -DRB1 after high-resolution typing is performed.53-56 The addition of HLA-C typing to the algorithm increases the predictive value, in part by eliminating the HLA-C mismatched cases and in part by helping to match for haplotypes rather than alleles.
The current study underscores the importance of recognizing the added information gained from high-resolution typing for HLA-A, -B, -C, and -DRB1. Adding more typing information to the process of donor selection should not be used to exclude more patients from the potentially curative benefits of this therapy. Rather it should be used as a means of stratifying patients' risk and allowing risk-adapted treatment strategies to be based on more complete and precise matching information. Ultimately, new approaches to immune suppression, which selectively or preferentially eliminate or permanently inactivate alloreactive T cells while sparing other T-cell populations, will be required to obviate the adverse effects of HLA mismatching on posttransplant survival.
In summary, the present analysis demonstrates significant adverse clinical effects for HLA-A, -B, -C, and -DR mismatch on survival after unrelated donor BMT. Most important, HLA-C should clearly be included in search algorithms. High-resolution class I typing for HLA-A, -B, and -C provides important additional prognostic information regarding transplant outcome. The ability to resolve the DNA sequences of the HLA alleles in even larger numbers of donors and recipients will allow us to address questions such as whether multiple amino acid substitutions will have more significance than fewer substitutions, whether conservative amino acid substitutions will be more permissive clinically, and whether substitutions in some portions of HLA molecules will be better tolerated than others. Answering these questions will allow us to more precisely understand the structural basis of alloreactivity and to hopefully define a subset of HLA disparities with little clinical consequence, which can be clinically tolerated when a perfectly matched donor is unavailable.
Prepublished online as Blood First Edition Paper, June 10, 2004; DOI 10.1182/blood-2004-03-0803.
Supported by funding from the National Marrow Donor Program, the Health Resources and Services Administration no. 240-97-0036 and the Office of Naval Research no. N00014-93-1-0658, N00014-95-1-0055, and N00014-96-2-0016. The views expressed in this article do not reflect the official policy or position of the Department of the Navy, the Department of Defense, or the US government.
Presented in abstract form at the 43rd annual meeting of the American Society of Hematology, Orlando, FL, December 10, 2001.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal