Abstract
A region of the von Willebrand factor (VWF) promoter has been identified that is necessary to confer endothelial cell-specific activation to the VWF promoter. This region spans sequences +155 to +247 and contains binding sites for GATA6 and NFY transcription factors. To identify potential DNA binding transcription factors that directly interact with these sequences in an endothelial-specific manner, we have performed extensive gel mobility assays with use of 7 overlapping DNA probes that collectively span this entire region. An endothelial-specific protein DNA complex was formed with an oligonucleotide that corresponded to sequences +155 to +184 of the VWF gene. Mutation analysis identified a 6-nucleotide element corresponding to sequences +164 to +169 as the core-binding region for the formation of this complex. Transfection analysis demonstrated that the mutation, which abolished DNA-protein interaction, resulted in significant inhibition of the VWF promoter activity. DNA pull-down analysis, mass spectrometry, and Western blot analysis demonstrated that a 32-kDa polypeptide with homology to histone H1 constituted the endothelial-specific DNA binding protein, or a DNA binding subunit of this protein complex. On the basis of these results, we hypothesize that an H1-like protein functions as an endothelial cell-specific transcriptional activator of the VWF promoter. (Blood. 2004;104: 1725-1732)
Introduction
The process of cell type-specific gene regulation is complex and involves interaction of transacting factors and cis-acting elements that are present throughout the gene sequences.1,2 Despite increasing knowledge of the mechanism of cell type-specific gene regulation in several cell lineages, including erythroid and skeletal myocytes,3,4 the molecular mechanisms that regulate endothelial-specific gene expression are not determined.
Von Willebrand factor is a glycosylated protein that is involved in primary hemostasis.5 It functions as a mediator of platelet adhesion to damaged subendothelium and acts as a carrier and stabilizer of coagulation factor VIII in the plasma.6 Von Willebrand factor (VWF) expression is restricted to endothelial cells and megakaryocytes, and this highly restrictive expression pattern has been used as a marker to distinguish endothelial cells from other cell types.7 Determining the mechanism of transcriptional regulation of the VWF is important from 2 aspects: (1) It will provide insights into the mechanism of endothelial-specific gene regulation, and (2) it will provide necessary information toward determining the molecular nature of the diseases that result from either high or low levels of VWF.
We had previously demonstrated that a 734-bp region of the VWF gene spanning sequences -487 to +247 activated gene expression in endothelial cells in culture.8 A highly endothelial cell-restricted activation pattern of the VWF promoter sequences -487 to +247 was also confirmed by 2 independent groups.8-10 Deletion and base substitution mutation analyses of the VWF promoter by others and us have resulted in identification of several transacting factors that positively and negatively regulate the activity of this promoter. GATA, Ets, and NFY (interacting with the CCAAT sequence) transcription factors function as activators,8,11,12 whereas Oct1, NF1, and NFY (binding to a novel sequence) function as repressors of the VWF promoter activity.12-14
We have demonstrated that a cell type-specific mechanism regulates the interaction of NFY with histone deacetylase 1 (HDAC1) and GATA6.15 In nonendothelial cells recruitment of GATA6-NFY-HDAC1 complex to the VWF promoter results in hypoacetylation of associated histone H4 and inhibition of promoter activity. In endothelial cells, GATA6 is not found in a complex with NFY. Furthermore, in endothelial cells HDACs are not recruited to the VWF promoter because of significantly reduced association of NFY and HDACs. Thus, VWF promoter-associated histone H4 is hyperacetylated, leading to promoter activation.15
We have now further extended our investigation of the mechanism of endothelial cell-specific regulation of the VWF promoter. Here, we report the identification of a histone H1-like protein that specifically interacts with the VWF promoter sequences in endothelial cells and participates in transcriptional activation of the promoter.
Materials and methods
Cell culture, transfection, and plasmid generation
HEK293 cells and human umbilical vein endothelial cells (HUVECs) were grown and maintained as previously described.8,12 Generation of plasmids HGH-K, HGH-KY, and HGH-Krm3 were previously described.8,12,14 To generate the plasmid HGH-KH, fragments -487 to +183 and +153 to +247 of the VWF promoter were first amplified by polymerase chain reaction (PCR). Specific 30-base oligonucleotide primers that corresponded to the 5′ and 3′ regions of each fragment were used to obtain 2 double-stranded fragments that contained the +155 to +184 sequences in common. The base substitutions TTT (or AAA) instead of GCC (or CGG) in the sequences +165 to +167 were incorporated into both +153 to +183 complementary primer sequences. The 2 mutant fragments (-487 to +184 and +155 to +247) were then combined and used as a PCR template to generate the -487 to +247 fragment, containing the base substitution mutations in sequences +168 to +170 element. For this second PCR, the primers that corresponded to -487 to -457 and +213 to +247 with an additional 9 bases that contained SalI sites were used. The PCR and cloning of the mutant -487 to +247 sequences into PφHGH to generate plasmid HGH-KH were as previously described for the generation of the wild-type HGH-K plasmid.8 Stable and transient transfection of bovine aortic endothelial (BAE) cells and growth hormone analysis were carried out as previously described.8,12 Briefly, BAE cells in 6-well culture dishes were transfected with VWF-HGH plasmids (2.91 μg/well) for transient transfection or VWF-HGH plasmids (2.91 μg) and a plasmid containing neomycin resistance gene under the regulation of Rous sarcoma virus (RSV) promoter (0.75 μg/well). Transfections were performed with use of calcium phosphate precipitation technique as previously described.8,12 Growth hormone analyses were performed 48 hours after transfection for transiently transfected cells. Stably transfected cells (48 hours after transfection) were grown in the presence of neomycin drug analog G418 (400 μg) for 3 weeks, and then either colonies were selected or cells were allowed to grow to confluency to obtain heterogeneous population of stably transfected cells.
Gel mobility and supershift assays
Nuclear extracts from HUVECs and HEK293 cells were prepared by the method of Schreiber et al.16 Double-stranded wild-type and mutant oligonucleotides corresponding to various regions of the VWF promoter (shown in Figures 2A and 3A) were radioactively labeled with use of 32P-ATP (adenosine 5′-triphosphate) and polynucleotide kinase as previously described.8 Nuclear extracts (5 μg) were incubated with oligonucleotide probes (10 000 cpm) on ice for 15 minutes in a 20-μL reaction mixture containing 50 mM KCl, 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (pH = 7.9), 5 mM MgCl2, 1 mM EDTA (ethylenediaminetetraacetic acid), 5% glycerol, and 1 μg poly(dI-dC). Complexes were resolved on a 5% nondenaturing polyacrylamide gel in 1 × Tris (tris(hydroxymethyl)aminomethane)-borate EDTA buffer (pH = 7.4) as previously described.8 For supershift assays, extracts were preincubated with antibodies (1 μg) under the conditions described earlier for 1 hour at 4°C prior to the addition of the probe and then further incubated at room temperature for 15 minutes. Complexes were resolved on a 5% nondenaturing polyacrylamide gel as described earlier.
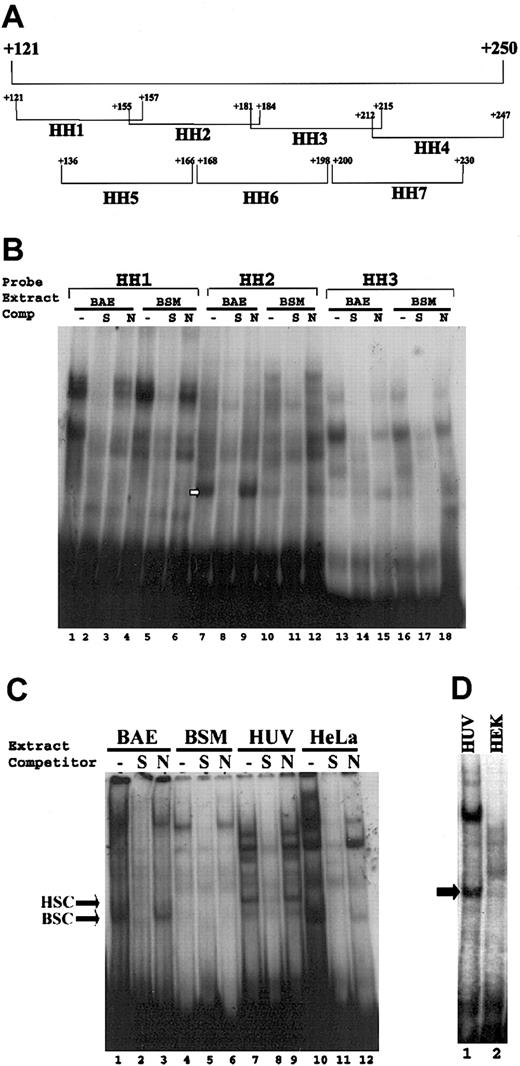
Analyses of endothelial-specific protein-DNA complex formation with sequences +155 to +184. (A) Represents the oligonucleotide probes that collectively span sequences +121 to +247 region of the VWF gene. Oligonucleotide probes (bracketed) are designated HH1 to HH7, with beginning and ending nucleotides shown. (B) Gel mobility assays were performed with use of HH1, HH2, and HH3 double-stranded oligonucleotide probes (10 000 cpm) and nuclear extracts (5 μg) prepared from bovine aortic endothelial (BAE) and bovine aortic smooth muscle (BSM) cells. (C-D) Gel mobility assays were performed with use of HH2 probe (10 000 cpm) corresponding to sequences +155 to +184 and nuclear extracts (5 μg) prepared from (C) HUVECs and HeLa cells and (D) HUVEC (HUV) and HEK293 (HEK) cells. The absence of competitor is represented by “-”. S (double-stranded unlabeled oligonucleotide +155 to +184) and N (double-stranded unlabeled oligonucleotide -490 to -460) represent specific and nonspecific oligonucleotide competitors. The arrows represent the endothelial specific complex in each panel.
Analyses of endothelial-specific protein-DNA complex formation with sequences +155 to +184. (A) Represents the oligonucleotide probes that collectively span sequences +121 to +247 region of the VWF gene. Oligonucleotide probes (bracketed) are designated HH1 to HH7, with beginning and ending nucleotides shown. (B) Gel mobility assays were performed with use of HH1, HH2, and HH3 double-stranded oligonucleotide probes (10 000 cpm) and nuclear extracts (5 μg) prepared from bovine aortic endothelial (BAE) and bovine aortic smooth muscle (BSM) cells. (C-D) Gel mobility assays were performed with use of HH2 probe (10 000 cpm) corresponding to sequences +155 to +184 and nuclear extracts (5 μg) prepared from (C) HUVECs and HeLa cells and (D) HUVEC (HUV) and HEK293 (HEK) cells. The absence of competitor is represented by “-”. S (double-stranded unlabeled oligonucleotide +155 to +184) and N (double-stranded unlabeled oligonucleotide -490 to -460) represent specific and nonspecific oligonucleotide competitors. The arrows represent the endothelial specific complex in each panel.
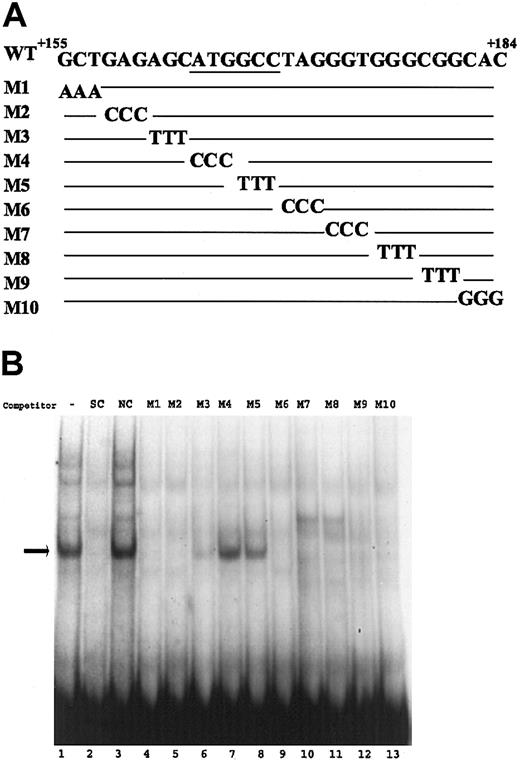
Mutational analysis of sequences +155 to +184. (A) The sequences of the wild-type and mutant oligonucleotides (M1-M10) corresponding to region +155 to +184 of the VWF promoter are shown. The base substitution mutations in each probe are shown, with the wild-type sequence represented as a line. (B) Gel mobility experiments were carried out with HUVEC nuclear extracts as described in Figure 2. The probes used in all samples were the wild-type oligonucleotide, and mutant oligonucleotides were used (100 × excess) as competitors. The - represents the lack of competitor. Arrow indicates the specific complex.
Mutational analysis of sequences +155 to +184. (A) The sequences of the wild-type and mutant oligonucleotides (M1-M10) corresponding to region +155 to +184 of the VWF promoter are shown. The base substitution mutations in each probe are shown, with the wild-type sequence represented as a line. (B) Gel mobility experiments were carried out with HUVEC nuclear extracts as described in Figure 2. The probes used in all samples were the wild-type oligonucleotide, and mutant oligonucleotides were used (100 × excess) as competitors. The - represents the lack of competitor. Arrow indicates the specific complex.
DNA pull-down assays and Western blot analysis
For DNA pull-down analysis, concatemeric oligonucleotides (trimeric) that corresponded to wild-type sequences +155 to +184 and sequences containing 6-base pair substitution mutations in element ATGGCC (combined mutations m4 and m5 shown in Figure 3A to generate sequence CCCTTT) were synthesized. These oligonucleotides (referred to as HH2wt [wild type] and HH2mut [mutant]) were biotinylated at one end and conjugated to magnetic beads (Dynabeads M-280 streptavidin) according to manufacturer's protocol (Dynal ASA, Oslo, Norway). HH2wt and HH2mut conjugated beads (each containing 2 μg double-stranded biotinylated oligonucleotide) were blocked with 0.5% nonfat milk to reduce nonspecific binding. Nuclear extracts (500 μg) from HUVECs and HEK293 cells were first incubated with the HH2mut conjugated beads (80 μL) in 200 μL buffer that was used in gel mobility assay described in “Gel mobility and supershift assays.” Nuclear proteins that were bound to HH2mut conjugated beads were separated from the nonbound proteins with use of the Dynal MPC-S magnetic particle concentrator. Proteins bound to the HH2mut conjugated beads were eluted with use of elution buffer that was optimized for protein elution (1M KCl, 20 mM HEPES pH 7.9, 10% glycerol, 0.5 mM EDTA). The supernatants (the entire samples) containing the unbound proteins were then incubated with the oligonucleotides containing the wild-type HH2 sequences (HH2wt), and the DNA-bound proteins were similarly eluted with use of the optimized elution buffer described earlier. The eluted proteins (half of entire sample) as well as the flow-through proteins (9.6 μg) from HH2wt conjugated beads were run on 12% sodium dodecylsulfate (SDS)-polyacrylamide gel, and the gel was either stained with Coomassie Blue to detect proteins or transferred to polyvinylidene diflouride (PVDF) membrane for Western blot analysis. Western blot analyses were performed as previously described.17
Antibodies and purified histone H1
The antibodies used for Western blot and supershift analyses were anti-histone H1 (AE4) mouse monoclonal antibody and immunoglobulin G (IgG). Antibodies and purified histone H1 were obtained from Upstate Biotechnology, Lake Placid, NY.
Mass spectrometry and protein identification
The Coomassie Blue-stained protein (apparent molecular weight [MW], 32 kDa) was excised from SDS-PAGE (polyacrylamide gel electrophoresis) gel under sterile tissue culture conditions to minimize contamination. The gel slice was destained with 50% methanol/50 mM NH4HCO3 in water and then cut into 1-mm3 pieces, reduced, alkylated with iodoacetamide, and digested in gel with trypsin.18 The peptides were extracted, dried under vacuum, and dissolved in 20 μL 2% acetonitril/0.1% formic acid in water. Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) analyses were carried out with use of Waters (Milford, MA) CapLC and Q-TOF mass spectrometer along with a PicoTip needle (New Objective, Woburn, MA) and a precolumn flow splitter, as described previously.19 The analytical column was 75-μm internal diameter (ID) × 15 cm, PepMap C18 (LC Packings, Sunnyvale, CA). Mobile phase A was 2% acetonitrile/0.1% formic acid, whereas mobile phase B was 90% acetonitril/0.1% formic acid. The gradient was 5% to 60% B in 90 minutes, and the flow rate was approximately 200 nL/min. The MS/MS spectra were processed, and the deconvoluted MS/MS spectra were directly used to search the National Center for Biotechnology Information (NCBI) nonredundant protein database with use of the mascot search program (Matrix Science, London, United Kingdom).
Results
Identification of an endothelial cell-specific protein-DNA complex formation
The results of deletion analyses by others and us have lead to the identification of a region of the VWF promoter, spanning sequences +155 to +247, which is necessary for endothelial cell-specific activation of the VWF promoter (shown by a bracket in Figure 1).8,13 This region contains a GATA6 binding sequence that functions as an activator and a novel NFY binding site, which functions as a repressor.12,15 We have demonstrated that interaction of GATA6 with its cognate binding site is necessary for the VWF promoter activation in endothelial cells.8 However, this does not exclude the possibility that other transcription factors can also interact with the DNA sequences that confer endothelial cell-specific activity to the VWF promoter and, thus, participate in promoter activation.
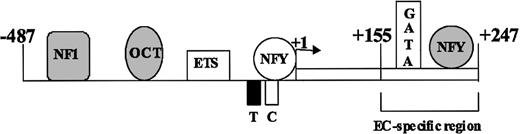
Schematic model of VWF promoter activation. The schematic representation of the endothelial cell-specific VWF promoter sequences -487 to +247 and transacting factors that interact with these sequences are shown. The line represents the 5′-flanking region, and the open box represents the first exon, with transcription start site shown by the arrow. The T and C represent TATA and CCAAT sequences, respectively. Factors NF1, Oct1, and NFY, which function as repressors, are shown as shaded symbols. Factors Ets, NFY, and GATA, which function as activators, are shown as open symbols. The region conferring endothelial-specific activity to the promoter (EC-specific region) is shown by the bracket.
Schematic model of VWF promoter activation. The schematic representation of the endothelial cell-specific VWF promoter sequences -487 to +247 and transacting factors that interact with these sequences are shown. The line represents the 5′-flanking region, and the open box represents the first exon, with transcription start site shown by the arrow. The T and C represent TATA and CCAAT sequences, respectively. Factors NF1, Oct1, and NFY, which function as repressors, are shown as shaded symbols. Factors Ets, NFY, and GATA, which function as activators, are shown as open symbols. The region conferring endothelial-specific activity to the promoter (EC-specific region) is shown by the bracket.
To determine whether there are endothelial cell-specific proteins that directly interact with the sequences +155 to +247 of the VWF promoter, we carried out gel mobility assays. For these analyses oligonucleotide probes that collectively spanned this entire region were used (Figure 2A). These oligonucleotide probes were incubated with nuclear extracts from bovine aortic endothelial cells (BAECs), bovine smooth muscle cells (BSMCs), HUVECs, HeLa cells, and HEK293 cells. Gel mobility experiments were performed as previously described.8 Bovine aortic endothelial cells were routinely used as a model of a VWF-expressing cell type in the previous transfection analyses that determined the endothelial-specific function of the VWF sequences +155 to +247. Thus, we first compared the pattern of DNA-protein complex formation in BAE and BSM cells. The results of these analyses demonstrated that the oligonucleotide probe corresponding to sequences +155 to +184 (referred to as HH2 probe) formed a complex (shown by white arrow in Figure 2B, lane 7) that was observed preferentially with nuclear proteins from BAE but not with BSM cells. Formation of this complex was abolished in the presence of 100 × excess of the unlabeled HH2 oligonucleotide (used as specific competitor), whereas a 100 × excess of a nonspecific competitor had no effect, thus demonstrating that this was a specific complex (Figure 2B, lanes 7-12). Other oligonucleotides when used as probes either did not form a specific complex or the complexes formed had a similar pattern in both endothelial and nonendothelial cells (data shown for HH1 [Figure 2B, lanes 1-6] and HH3 [Figure 2B, lanes 13-18] as representatives). The presence of a minor amount of a similar complex with HH3 probe, as well as a minor amount of complex with HH2 probe and BSM nuclear extracts, was not reproducible in multiple experiments. These results demonstrated that only the HH2 probe specifically formed a consistent and significant protein-DNA complex with nuclear extracts of BAECs.
To determine whether the specific complex that is formed with the HH2 probe and BAEC nuclear extracts was also formed with other cell types, we carried out similar analysis with use of HH2 probe and nuclear extracts prepared from HUVECs, HeLa cells, and HEK293 cells. The results demonstrated that the HH2 probe also formed a specific complex with nuclear extracts prepared from HUVECs but not HeLa or HEK293 cells (Figure 2C-D). The endothelial-specific complexes formed with BAECs and HUVECs did not migrate exactly to the same position in the gel mobility assay (Figure 2C, compare HSC and BSC shown by arrows). However, these results demonstrate that sequences +155 to +184 of the VWF promoter form an endothelial cell-specific protein-DNA complex with nuclear extracts of both BAECs and HUVECs.
DNA sequences ATGGCC spanning nucleotides +164 to +169 are necessary for endothelial cell-specific protein-DNA complex formation
To further characterize the DNA sequences that are necessary for the formation of the endothelial-specific protein-DNA complex, base substitution mutation analyses were carried out. For these analyses, 10 oligonucleotides corresponding to sequences +155 to +184 were generated; each contained a different 3-base substitution mutation, and the oligonucleotides were used as competitors in gel mobility experiments (Figure 3A). The results demonstrated that the oligonucleotides M4 and M5 failed to compete with the wild-type probe, whereas other mutant oligonucleotides successfully competed (Figure 3B). In addition, when oligonucleotides M4 and M5 were used as probes, they failed to form the specific complex that was observed with the wild-type probe (data not shown). These results demonstrated that the base substitution mutations in oligonucleotides M4 and M5 abolished protein-DNA complex formation. The mutations in M4 and M5 collectively spanned the sequences +164 to +169 and corresponded to nucleotides ATGGCC. Thus, we concluded that this ATGGCC element contained the core sequence necessary for the formation of the endothelial-specific protein-DNA complex.
Mutation in the ATGGCC element results in inhibition of the VWF promoter activity in endothelial cells
To determine whether formation of the endothelial-specific DNA-protein complex is necessary for the VWF promoter activation, we performed mutation and transfection analyses in BAE cells. The base substitution mutations that were used to generate the M5 oligonucleotide (shown in Figure 3A) were incorporated into the VWF promoter fragment corresponding to sequences -487 to +247. The resulting fragment was fused to human growth hormone gene to generate plasmid HGH-KH. The activity of this plasmid was compared with that of the wild-type HGH-K in both transient and stable transfections. The results of transient transfection analyses demonstrated that the mutation in ATGGCC element resulted in an approximately 50% reduction in the promoter activity, whereas mutations of the repressor elements NF1 and NFY had no significant effect on the VWF promoter activity in endothelial cells as previously demonstrated14,12 (Figure 4B). Results of stable transfection analyses with individually selected clones demonstrated that the level of activity of the mutant promoter in HGH-KH plasmid was significantly lower than that of the wild-type HGH-K plasmid. Similar results were obtained when heterogeneous populations of stably transfected cells were analyzed (data not shown). These results demonstrated that the mutation, which inhibited the complex formation, also significantly reduced the promoter activity in both transiently and stably transfected BAE cells (Figure 4). The reduction in the promoter activity (as a result of the mutation in ATGGCC element) was greater in stably transfected compared with transiently transfected cells, thus suggesting that chromatin structure may be associated with the function of this protein complex. These results demonstrated that the endothelial-specific protein complex that interacted with ATGGCC element in the +155 to +184 region of the VWF promoter functionally participated in activation of the VWF promoter.
Expression of VWF promoter fragment with base substitution mutations in endothelial-specific protein-DNA binding site. (A) DNA sequence of the VWF region that forms the endothelial-specific DNA-protein complex with the protein(s) binding site underlined. Arrows show the base substitution mutations that are incorporated into the VWF promoter to generate the HGH-KH plasmid. (B) Growth hormone expression is shown from transiently transfected BAE cells with VWF-HGH plasmid containing no mutation ([HGH-K] wild type), mutation in GCC sequence as shown in panel A, and 2 plasmids with mutations in NFY (HGH-KY) and NF1 (HGH-Krm3) repressor binding sites. For these analyses, 12 independent transfection experiments were carried out for each plasmid. (C) Growth hormone expressions are shown from BAE cells stably transfected with (HGH-K) wild-type and HGH-KH. For these experiments, 36 individual clones of stably transfected cells (G418-resistant clones) were selected for each transfected plasmid, and the results represent the mean value of growth hormone from these clones. Solid triangle labeled M5 represents the 3-base substitution mutation shown in panel A. Open triangles in panels B and C represent mutations in NFY or NF1 binding site. Percentage of growth hormone activities was determined as previously described for transient8 and stable transfection.12 The error bars represent standard error.
Expression of VWF promoter fragment with base substitution mutations in endothelial-specific protein-DNA binding site. (A) DNA sequence of the VWF region that forms the endothelial-specific DNA-protein complex with the protein(s) binding site underlined. Arrows show the base substitution mutations that are incorporated into the VWF promoter to generate the HGH-KH plasmid. (B) Growth hormone expression is shown from transiently transfected BAE cells with VWF-HGH plasmid containing no mutation ([HGH-K] wild type), mutation in GCC sequence as shown in panel A, and 2 plasmids with mutations in NFY (HGH-KY) and NF1 (HGH-Krm3) repressor binding sites. For these analyses, 12 independent transfection experiments were carried out for each plasmid. (C) Growth hormone expressions are shown from BAE cells stably transfected with (HGH-K) wild-type and HGH-KH. For these experiments, 36 individual clones of stably transfected cells (G418-resistant clones) were selected for each transfected plasmid, and the results represent the mean value of growth hormone from these clones. Solid triangle labeled M5 represents the 3-base substitution mutation shown in panel A. Open triangles in panels B and C represent mutations in NFY or NF1 binding site. Percentage of growth hormone activities was determined as previously described for transient8 and stable transfection.12 The error bars represent standard error.
Characterization of the protein complex that interacts with ATGGCC sequence
To characterize the endothelial cell-specific protein complex that interacted with the ATGGCC sequence, we performed DNA pull-down analysis as described in “Materials and methods.” For this analysis, concatemeric oligonucleotides (trimeric), corresponding to the wild-type sequences +155 to +184, and sequences containing 6-base pair substitution mutations in the ATGGCC element (combined M4 and M5 shown in Figure 3A) were synthesized. We used the mutant oligonucleotide to reduce nonspecifically bound proteins and also to confirm that the endothelial-specific protein complex did not interact with the mutant oligonucleotide.
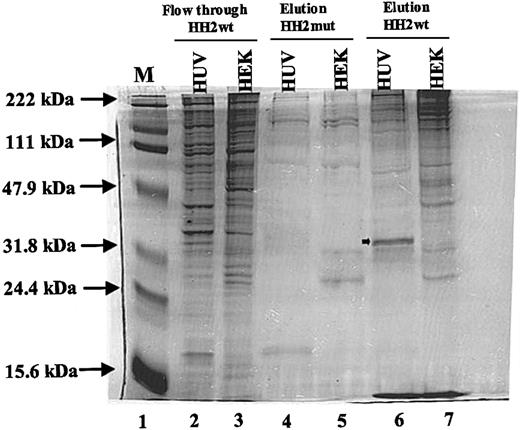
The results of these analyses demonstrated that there was a polypeptide with an approximate molecular weight of 32 kDa (shown by arrow in Figure 5, lane 6) exclusively detected among proteins that were eluted from HH2wt bound HUVEC nuclear extracts (Figure 5, lane 6). No similar polypeptide was detected in nuclear proteins that were bound to the mutant probe (Figure 5, lane 4). A reduced amount of a similarly sized polypeptide was detectable in the flow-through fraction from the HH2wt probe (Figure 5, lane 2). This may be due to incomplete removal of the specific 32-kDa polypeptide by the HH2wt probe. The 32-kDa polypeptide was not detected in the HEK293 cell nuclear proteins that were eluted from the HH2wt or HH2mut probes (Figure 5, lanes 5 and 7). However, there were some other polypeptides that were detected in HEK293 nuclear proteins bound to the HH2wt probe that were not detected in HUVEC nuclear proteins bound to the same probe (Figure 5, lanes 5 and 7).
Analysis of the nuclear proteins bound to sequences +155 to +184. Nuclear extracts (500 μg) from HUVEC (HUV) and HEK293 (HEK) cells were used in DNA pull-down analyses as described in “Materials and methods.” Lane 1 represents the protein molecular weight marker. Lanes 2 and 3 represent flow-through proteins that were not bound to HH2 wild-type (HH2wt) probe. Lanes 4 and 5 represent eluted proteins bound to HH2 mutant (HH2mut) probe. Lanes 6 and 7 represent eluted proteins bound to HH2wt probe. Arrow in lane 6 represents the endothelial-specific protein. Positions of protein molecular weight markers are shown by arrows.
Analysis of the nuclear proteins bound to sequences +155 to +184. Nuclear extracts (500 μg) from HUVEC (HUV) and HEK293 (HEK) cells were used in DNA pull-down analyses as described in “Materials and methods.” Lane 1 represents the protein molecular weight marker. Lanes 2 and 3 represent flow-through proteins that were not bound to HH2 wild-type (HH2wt) probe. Lanes 4 and 5 represent eluted proteins bound to HH2 mutant (HH2mut) probe. Lanes 6 and 7 represent eluted proteins bound to HH2wt probe. Arrow in lane 6 represents the endothelial-specific protein. Positions of protein molecular weight markers are shown by arrows.
These results were consistent with the results of gel mobility experiments that demonstrated the formation of an endothelial-specific DNA-protein complex with sequences +155 to +184. The data demonstrated that at least one endothelial-specific nuclear protein with a molecular weight of 32 kDa interacted with sequences ATGGCC in the VWF promoter. These results also suggested that VWF sequences +155 to +184 might interact with other nuclear proteins specifically in nonendothelial cells.
Endothelial-specific 32-kDa protein is a histone H1-like protein
To identify the 32-kDa protein that interacted with the VWF promoter in an endothelial-specific manner, we performed LCMS/MS analysis of a tryptic digest of the 32-kDa band obtained from the Coomassie-stained SDS-PAGE gel of the DNA pull-down analysis. Several peptides were sequenced (Table 1) and were shown to correspond to sequences spanning amino acids 37 to 82 of histone H1 member 5 (Figure 6).
Peptide sequences obtained by LC-MS/MS and matched to histone H1 member 5
. | . | Monoisotopic molecular mass . | . | |
|---|---|---|---|---|
| Peptide sequence . | Precursor ions, m/z . | Observed . | Calculated . | |
| NGLSLAALKK | 507.832+ | 1013.65 | 1013.62 | |
| ALAAGGYDVEK | 547.292+ | 1092.57 | 1092.55 | |
| ATGPPVSELITK | 606.862+ | 1211.70 | 1211.68 | |
| KATGPPVSELITK | 670.902+ | 1339.78 | 1339.77 | |
| ALAAGGYDVEKNNSR | 782.883+ | 1563.75 | 1563.76 | |
. | . | Monoisotopic molecular mass . | . | |
|---|---|---|---|---|
| Peptide sequence . | Precursor ions, m/z . | Observed . | Calculated . | |
| NGLSLAALKK | 507.832+ | 1013.65 | 1013.62 | |
| ALAAGGYDVEK | 547.292+ | 1092.57 | 1092.55 | |
| ATGPPVSELITK | 606.862+ | 1211.70 | 1211.68 | |
| KATGPPVSELITK | 670.902+ | 1339.78 | 1339.77 | |
| ALAAGGYDVEKNNSR | 782.883+ | 1563.75 | 1563.76 | |
Sequence of histone H1 member 5. Amino-acid sequence of histone H1 member 5 is shown. The bold sequences represent the peptides identified by mass spectrometry of the 32-kDa endothelial-specific polypeptide. Asterisks represent the boundaries of globular domain.
Sequence of histone H1 member 5. Amino-acid sequence of histone H1 member 5 is shown. The bold sequences represent the peptides identified by mass spectrometry of the 32-kDa endothelial-specific polypeptide. Asterisks represent the boundaries of globular domain.
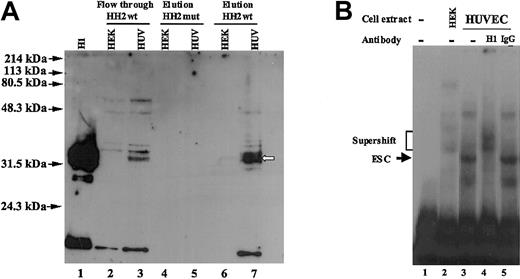
On the basis of the above-mentioned MS result, we used an anti-histone H1 antibody in a Western blot analysis to determine whether this antibody recognized the 32-kDa protein. The antibody used was a mouse monoclonal antibody (clone AE-4) raised against nuclei of myeloid leukemia cells (Upstate Biotechnology). This antibody was reported by the manufacturer to recognize histones H1, (Mr ∼32-33 kDa) and detect a doublet because of hyperphosphorylation of a subpopulation of histones H1. Cross-reactivity with other histone isoforms, leading to detection of bands at 17 kDa, was also reported. This antibody is cross-reactive to human and bovine histones H1. For this analysis a DNA pull-down assay was performed exactly as described for Figure 5, except that after SDS-PAGE analysis of the pulled down protein, they were transferred to nylon membrane and incubated with the anti-histone H1 antibody. As a control for the activity of the antibody, we also included a lane containing purified histone H1 (Figure 7A, lane 1). The results demonstrated that the anti-histone H1 antibody specifically recognized the endothelial-specific 32-kDa polypeptide that was eluted from the HH2wt probe-bound HUVEC nuclear proteins (Figure 7A, lane7). There was no signal detected in the lane that contained the HEK293 polypeptides eluted from the HH2wt probe (Figure 7A, lane 6). In addition there were no signals in the lanes that contained the polypeptides pulled down with the HH2mut probe incubated with either HUVEC or HEK293 nuclear extracts. However, the flow-through fractions (HH2wt-probe unbound proteins) from both HUVEC and HEK293 samples showed polypeptides (distinct from 32-kDa polypeptide) detected by the antibody that were common to both cell types (Figure 7A, lanes 2-3). In addition to these, the flow-through fraction from HUVECs showed a signal similar to that observed in the HH2wt-eluted fraction (Figure 7A, compare lanes 3 and 7). As discussed in the previous section, this may represent the incomplete pull down of the 32-kDa protein by the HH2wt probe. Supershift analysis demonstrated that the DNA endothelial-specific protein complex that was observed in gel mobility experiments was also recognized and specifically supershifted by this anti-histone H1 antibody (Figure 7B).
Western blot analyses of the DNA pull-down proteins from HUVEC and HEK293 cells and supershift of the gel mobility complex. (A) DNA pull-down experiment was performed exactly as described for Figure 5. The eluted and flow-through polypeptides were analyzed on SDS-PAGE gel followed by transfer to a nylon membrane and incubation with anti-histone H1 antibody as described in “Materials and methods.” Purified histone H1 was used as positive control (shown in lane 1). Lanes 2 and 3 represent flow-through proteins that were not bound to HH2 probe. Lanes 4 and 5 represents eluted proteins bound to HH2mut probe. Lanes 6 and 7 represent eluted proteins bound to HH2wt probe. White arrow represents the endothelial-specific protein that is recognized by anti-histone H1 antibody. Position of protein molecular weight markers are shown by black arrows. (B) Nuclear extracts prepared from HUVECs (lanes 3-5) were incubated in the absence (lane 3) and presence of 1 μg anti-histone H1 (lane 4) or IgG (lane 5) antibodies prior to addition of oligonucleotide probe (sequences +155 to +184). The anti-histone H1 antibody used was a mouse monoclonal antibody (clone AE-4), and the IgG used was normal mouse IgG. Nuclear extracts from HEK293 cells (lane 2) were used as control to distinguish the position of endothelial-specific complex from other complexes. Lane 1 represents probe only. The positions of the endothelial specific-complex (ESC), and supershifted complexes are shown by arrows and bracket, respectively.
Western blot analyses of the DNA pull-down proteins from HUVEC and HEK293 cells and supershift of the gel mobility complex. (A) DNA pull-down experiment was performed exactly as described for Figure 5. The eluted and flow-through polypeptides were analyzed on SDS-PAGE gel followed by transfer to a nylon membrane and incubation with anti-histone H1 antibody as described in “Materials and methods.” Purified histone H1 was used as positive control (shown in lane 1). Lanes 2 and 3 represent flow-through proteins that were not bound to HH2 probe. Lanes 4 and 5 represents eluted proteins bound to HH2mut probe. Lanes 6 and 7 represent eluted proteins bound to HH2wt probe. White arrow represents the endothelial-specific protein that is recognized by anti-histone H1 antibody. Position of protein molecular weight markers are shown by black arrows. (B) Nuclear extracts prepared from HUVECs (lanes 3-5) were incubated in the absence (lane 3) and presence of 1 μg anti-histone H1 (lane 4) or IgG (lane 5) antibodies prior to addition of oligonucleotide probe (sequences +155 to +184). The anti-histone H1 antibody used was a mouse monoclonal antibody (clone AE-4), and the IgG used was normal mouse IgG. Nuclear extracts from HEK293 cells (lane 2) were used as control to distinguish the position of endothelial-specific complex from other complexes. Lane 1 represents probe only. The positions of the endothelial specific-complex (ESC), and supershifted complexes are shown by arrows and bracket, respectively.
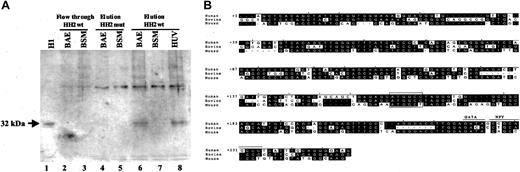
Because the formation of an endothelial-specific DNA complex was originally determined with use of bovine endothelial nuclear extracts, we performed similar DNA pull-down/Western blot analyses with BAEC and BSMC nuclear extracts with use of the wild-type and the mutant HH2 probes. As a control we also used eluted proteins from the HUVEC extracts that were bound to the wild-type HH2 probe. The results demonstrated that a similar polypeptide of 32-kDa molecular weight that was recognized by anti-histone H1 antibody was pulled down from BAEC (and HUVECs) nuclear extracts with the wild-type HH2wt but not the mutant probe (Figure 8A, lanes 4, 6, and 8). This polypeptide was not detected in BSMC nuclear extracts that were pulled down with either mutant or the wild-type probe (Figure 8A, lanes 5 and 7). These results demonstrated that a similar histone H1-like 32-kDa protein in both human and bovine endothelial cells specifically interacted with the ATGGCC sequences in the VWF promoter.
Western blot analysis of the DNA pull-down proteins from BAECs, BSMCs, and HUVECs. (A) DNA pull-down experiment and Western blot analyses were performed exactly as described for Figure 7A. Purified histone H1 was used as positive control (lane 1). Lanes 2 and 3 represent flow-through proteins that were not bound to HH2wt probe. Lanes 4 and 5 represents eluted proteins bound to HH2mut probe. Lanes 6 and 7 represent eluted proteins bound to HH2wt probe. Lane 8 represents the eluted protein from HUVECs bound to HH2wt probe. Arrow represents the endothelial-specific protein migrating approximately at 32-kDa position (shown by molecular weight marker) that is recognized by anti-histone H1 antibody. (B) Sequences of the human, bovine, and mouse VWF genes corresponding to nucleotides +1 to +250 (exon 1) are shown. Nucleotides in black boxes represent the regions of homology. The GATA and repressor NFY binding sites are shown. The ATGGCC sequence that comprises the binding site of the 32-kDa H1-like protein is shown in the open boxes.
Western blot analysis of the DNA pull-down proteins from BAECs, BSMCs, and HUVECs. (A) DNA pull-down experiment and Western blot analyses were performed exactly as described for Figure 7A. Purified histone H1 was used as positive control (lane 1). Lanes 2 and 3 represent flow-through proteins that were not bound to HH2wt probe. Lanes 4 and 5 represents eluted proteins bound to HH2mut probe. Lanes 6 and 7 represent eluted proteins bound to HH2wt probe. Lane 8 represents the eluted protein from HUVECs bound to HH2wt probe. Arrow represents the endothelial-specific protein migrating approximately at 32-kDa position (shown by molecular weight marker) that is recognized by anti-histone H1 antibody. (B) Sequences of the human, bovine, and mouse VWF genes corresponding to nucleotides +1 to +250 (exon 1) are shown. Nucleotides in black boxes represent the regions of homology. The GATA and repressor NFY binding sites are shown. The ATGGCC sequence that comprises the binding site of the 32-kDa H1-like protein is shown in the open boxes.
Characterization of the ATGGCC element as a binding site for histone H1-like protein in both human and bovine endothelial cells raised the possibility that this element may be commonly conserved in the VWF sequences of both these species. A comparative analysis of the VWF sequences corresponding to the 5′ untranslated regions of human,20 bovine,21 and mouse22 demonstrated that the ATGGCC sequence is highly conserved (Figure 8B)
Discussion
Analyses of the mechanism of transcriptional regulation of a number of endothelial cell-specific promoters have demonstrated that commonly expressed transcription factors, as well as transcription factors with endothelial preferential pattern of expression, participate in the regulation of one or more endothelial cell-specific promoters.8,11-13,15,23-29
Characterization of a small region of the VWF promoter (corresponding to sequences +155 to +247) that was shown to be necessary for endothelial-specific activation of the promoter provides a reasonably small DNA target for investigation into the mechanism of its endothelial-specific function. We had previously demonstrated that this region contains binding sites for commonly expressed GATA6 and NFY transcription factors.8,12,15 We have now demonstrated that this region of the VWF promoter contains a binding site for an endothelial cell-specific protein with homology to histone H1.
With use of extensive gel mobility analysis of the entire +155 to +247 region, we have shown clearly that the sequences +155 to +184 form an endothelial cell-specific protein-DNA complex. Our analysis identified an exact binding sequence that constitutes the ATGGCC element at position +164 to +169 as the binding site for this endothelial-specific protein. Analysis of the VWF promoter containing mutations in the ATGGCC element demonstrated that the functional consequence of inhibition of the endothelial-specific protein-DNA complex formation is a significant reduction in the promoter activity, thus, demonstrating that protein(s), which binds to the ATGGCC element, participates in the transcriptional activation of the VWF promoter. The observation that this sequence is highly conserved in the VWF gene among human, bovine, and mouse is consistent with the role of this sequence as a cis-acting element that participates in regulation of the VWF gene expression.
DNA pull-down analysis demonstrated that a polypeptide with a molecular weight of 32 kDa that interacts with the wild-type VWF sequences +155 to +184 could be isolated specifically from the nuclear extracts of endothelial cells. A mutation that abolished the DNA-protein interaction in the gel mobility assay also inhibited interaction of the 32-kDa protein with the DNA probe in the DNA pull-down assay. Mass spectrometry analysis of the 32-kDa protein demonstrated that this protein contained regions of homology with H1 histone family member 5.30,31 Western blot analysis with use of an anti-histone H1 antibody further confirmed the homology of the 32-kDa polypeptide to the histone H1. Supershift analyses demonstrated that the histone H1 antibody, which recognized the 32-kDa protein, also recognized the endothelial-specific protein-DNA complex that was observed in gel mobility experiment. These results strongly suggested that the 32-kDa histone H1-like protein that was pulled down by DNA chromatography was either the DNA binding protein or a component of the endothelial-specific protein complex that was shown by gel mobility assay to interact with sequences +155 to +184 of the VWF promoter.
The gel mobility assays with use of HH2 probe with BAE and HUVEC nuclear extracts indicated that the endothelial-specific complexes of these 2 cell types do not comigrate exactly to the same position, whereas DNA-Western blot analyses demonstrated that a similarly migrating 32-kDa protein was recognized by anti-histone H1 antibody in both human and bovine endothelial cells. The observed differences in gel mobility assay may be due to differences in the components of the complexes in these 2 endothelial cells. Complexes observed in gel mobility may include other proteins in addition to histone H1 that differentially contribute to the complex formation in the endothelial cell types of these 2 species. Alternatively, a minor band (localized under the major complex) that was observed in gel mobility assay of human endothelial nuclear extracts and appeared to comigrate to the same position as the complex observed in bovine endothelial cells (Figure 2C-D) may correspond to the histone H1-DNA complex. Thus, the major band may represent a complex of histone H1 and an additional low molecular weight component as suggested earlier. The observation that the major complex is supershifted by anti-histone H1 antibody (Figure 7B) supports the hypothesis that the major complex contained histone H1.
On the basis of these results we hypothesize that the anti-histone H1 antibody detected histone H1 variants that are commonly expressed in all cell types, but in addition this antibody also recognized a 32-kDa protein that is specifically expressed in endothelial cells. However, we cannot exclude the possibility that another (as yet unidentified) endothelial-specific protein may bind to the ATGGCC element and then specifically recruit a H1 histone to the VWF promoter DNA sequences. This possibility is consistent with a role for the histone H1-like 32-kDa protein as an endothelial cell-specific participant in the VWF promoter activation.
Histones H1 have been generally proposed to play a structural role in appropriate assembly of the chromatin structure.32 This function of the histone H1 had lead to the hypothesis that they generally play a repressive role in transcription regulation.33 However, recently histone H1 was shown to participate in transcriptional activation of certain genes in yeast, tetrahymena, and higher eukaryotes.34-39 Analyses of the proteins in 2D-PAGE gels from H1-deleted mutant chicken DT40 B-cell lines compared with the wild type demonstrated that each histone H1 variant uniquely participates in both up- and down-regulation of a subset of proteins.36 A detailed analysis of the specific role of histone H1 on the regulation of mouse mammary tumor virus (MMTV) promoter demonstrated that histone H1 participates in basal and induced activation of MMTV and mediates increased synergy between progesterone receptor and NF1 binding to the promoter.37,38
In mammals, the presence of multiple variants of histone H1 that are differentially expressed during development, in spermatocytes and oocytes, as well as differential level of expression of subtypes in somatic cells, has lead to the hypothesis that they may participate in selective gene regulation.40,41 To date at least 8 variants of histone H1 that include testis- and oocyte-specific histone H1 variants (H1t and H1oo), somatic variants (H1a, H1b, H1c, H1d, and H1e), and replacement H1 histones (H10 and its subtype H5) are recognized.41-43 The complexity of assigning exact function to the histone H1 may be partially due to the presence of multiple subtypes, which from the knock-out studies appear to compensate for each other.44-46 However, specific histone H1 variants were demonstrated to influence expression of a subset of genes as well as transgenes.47,48 In addition, correlation of specific histone H1 subtypes to cell differentiation suggests that cell type-specific gene regulation may be influenced by H1.49,50
Histones H1 consist of a tripartite structure that includes a central globular domain that is highly conserved among somatic subtypes and 2 random coil tails at C- and N-terminal region.51 The peptide sequences of the H1-like 32-kDa polypeptide that was shown to interact with the VWF promoter corresponded to that of histone H1a (also referred to as histone H1 member 5) and specifically mostly to the globular domain.52 The globular domains of histones H1 mediate the interactions of these histones with DNA and contained winged helix motifs that were found in some transcription factors.53-55
Despite the emerging concept that histones H1 participate in both negative and positive regulation of specific genes, there are very few reports of specific DNA binding sequences that recognize histones H1. Histones H1 have been reported to interact with a NF1 recognition sequence in the mouse α2(I) collagen promoter, and a specific sequence in the rat albumin gene promoter as well as Ω element in histone H3.2 gene promoter.56-58 Among these, both mouse α2(I) collagen and rat albumin H1 binding sequences included TGGC elements (in forward and reverse orientation, respectively) that are similar to the core region of the sequence ATGGCC in the VWF promoter. On the basis of these observations we hypothesize that DNA sequences corresponding to TGGC may be a potential specific binding site for some histone H1 variants.
Histones H1 undergo posttranslational modifications, including poly (ADP-ribosyl)ation, acetylation, and phosphorylation.51 This may account for observation of histones H1 at 30- to 32-kDa molecular weights as reported by others and observed in our analyses.56 The observation of 2 bands with anti-histone H1 antibody in our DNA pull-down SDS-PAGE and Western blot analyses is also consistent with previous reports that somatic histones H1 are generally detected as 2 bands in SDS-PAGE, a faster migrating band consisting of the H1-1 and a slower migrating band that consists of remaining subtypes.51 Among the various somatic histones H1, the histone H1a has the most restricted pattern of expression. Its expression was restricted to thymus, testis, spleen, as well as lymphocytic and neuronal cells.51 The reported lack of expression of histone H1a in HEK293 cells is consistent with our results, which did not demonstrate the 32-kDa H1-like protein-DNA binding activity in HEK293 cells.52,59 To our knowledge there has been no analysis of the expression pattern of H1a gene specifically in endothelial cells; however, H1a was reported to be present in various organs, and this was attributed to contamination of these organs by lymphocytic cells.51 On the basis of our results we hypothesize that, if the 32-kDa histone H1-like protein is the H1a, the detection of this protein in various organs may also be due to the presence of vascular networks containing endothelial cells in various organs.
On the basis of these observations it is tempting to hypothesize that highly differentiated endothelial cells may express a specific subtype of replacement histone H1, potentially H1a, that could participate in determining the differentiation state of endothelial cells through regulation of endothelial-specific gene expression. On the basis of our observations we propose that the 32-kDa endothelial-specific protein may represent an endothelial-specific variant of histone H1 or an endothelial-specific transcription factor with high homology to histone H1 that participates in transcription regulation of the VWF promoter. Further analysis of this protein through cloning will determine the exact nature of this protein and its role in VWF gene expression in particular and endothelial-specific gene regulation in general.
Prepublished online as Blood First Edition Paper, May 18, 2004; DOI 10.1182/blood-2004-01-0082.
Supported by research grant from the National Institute of Health (HL-67729 to N.J.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 4. Expression of VWF promoter fragment with base substitution mutations in endothelial-specific protein-DNA binding site. (A) DNA sequence of the VWF region that forms the endothelial-specific DNA-protein complex with the protein(s) binding site underlined. Arrows show the base substitution mutations that are incorporated into the VWF promoter to generate the HGH-KH plasmid. (B) Growth hormone expression is shown from transiently transfected BAE cells with VWF-HGH plasmid containing no mutation ([HGH-K] wild type), mutation in GCC sequence as shown in panel A, and 2 plasmids with mutations in NFY (HGH-KY) and NF1 (HGH-Krm3) repressor binding sites. For these analyses, 12 independent transfection experiments were carried out for each plasmid. (C) Growth hormone expressions are shown from BAE cells stably transfected with (HGH-K) wild-type and HGH-KH. For these experiments, 36 individual clones of stably transfected cells (G418-resistant clones) were selected for each transfected plasmid, and the results represent the mean value of growth hormone from these clones. Solid triangle labeled M5 represents the 3-base substitution mutation shown in panel A. Open triangles in panels B and C represent mutations in NFY or NF1 binding site. Percentage of growth hormone activities was determined as previously described for transient8 and stable transfection.12 The error bars represent standard error.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/6/10.1182_blood-2004-01-0082/6/m_zh80180466520004.jpeg?Expires=1765006002&Signature=VlcrCY32BNxLZHIpPy9-7uib4yZKNhG6LB64dz9nt3MbyaKndTU9tH-lmv1PkufUJhBpgxQowIVe0MD4jK7zbqXlr4k5ReF8CX0BcVaceVEV93dUyNNlho2epz1WPUtkTHTk8rKtIvo44xHDh0UOb5czNXGCdW6LKs8XFvDMYtb2HS6myOujK06mxR~Bpom9XlwMh7IQD~po3HxjLWCYMd3-VGtbqvofyixh~dBpUoWcVWC76Rd61C5fUZyMFmc2HiUK2mg4EAxnulwVXCRhIsXqRpdil7GNplvIEvao467GLoV8HAG7fN4OEAnmHuzxyDV6FtsJxeYum08i4LYSRQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal