Abstract
To investigate possible causes of the variable response to treatment in pediatric B-precursor acute lymphoblastic leukemia (ALL) and to establish potential novel therapeutic targets, we used ionizing radiation (IR) exposure as a model of DNA damage formation to identify tumors with resistance to p53-dependent apoptosis. Twenty-one of 40 ALL tumors responded normally to IR, exhibiting accumulation of p53 and p21 proteins and cleavage of caspases 3, 7, and 9 and of PARP1. Nineteen tumors exhibited apoptotic resistance and lacked PARP1 and caspase cleavage; although 15 of these tumors had normal accumulation of p53 and p21 proteins, examples exhibited abnormal expression of TRAF5, TRAF6, and cIAP1 after IR, suggesting increased NF-κB prosurvival signaling as the mechanism of apoptotic resistance. The presence of a hyperactive PARP1 mutation in one tumor was consistent with such increased NF-κB activity. PARP1 inhibition restored p53-dependent apoptosis after IR in these leukemias by reducing NF-κB DNA binding and transcriptional activity. In the remaining 4 ALL tumors, apoptotic resistance was associated with a TP53 mutation or with defective activation of p53. We conclude that increased NF-κB prosurvival signaling is a frequent mechanism by which B-precursor ALL tumors develop apoptotic resistance to IR and that PARP1 inhibition may improve the DNA damage response of these leukemias.
Introduction
B-precursor acute lymphoblastic leukemia (ALL) is the most common pediatric malignancy. In the past 2 decades, the cure rate for ALL has dramatically increased,1 and much of this success can be attributed to improved stratification of tumors paired with finely tuned treatment protocols.1-3 Further improvements in the cure of this disease, however, are hampered by a lack of understanding of molecular variability among tumors and targets for the further individualization of treatment. Treatments directed toward specific biologic pathways involved in leukemia pathology not only may improve outcome for some patients but may also reduce the toxicity and nonspecificity associated with conventional chemotherapeutic agents.
By definition, leukemic cells exhibit uncontrolled proliferation, a feature that is often facilitated by defects in apoptosis and DNA damage responses. Such defects frequently contribute to tumor progression and cytotoxic drug resistance.4-6 Indeed, the most notorious mechanism of drug resistance in human cancers is inactivation of the TP53 gene, a crucial component of the DNA damage response and apoptotic pathways. Interestingly, in pediatric ALL, the incidence of TP53 mutation is notably low, with less than 5% of tumors at diagnosis harboring detectable mutations.7,8 Although the rate of mutation at relapse is slightly higher,9 indicating a role for p53 inactivation in drug resistance, genes other than p53 that impinge on its function, such as ATM,10-14 MDM2, p21, BCL2, and BAX,15-22 might be involved in tumor progression. Furthermore, given that the balance between proapoptotic and prosurvival signals is critical in determining cell fate after DNA damage,5 distinct pathways activated in response to DNA damage may alternatively be deregulated. Indeed, it has been suggested that constitutively activated NF-κB complexes occur in almost 100% of childhood ALL tumors,23 and it is possible that such a prosurvival pathway plays a role in ALL.
To investigate the possible causes of the variable DNA damage response in pediatric B-precursor ALL and to identify potential targets for novel therapies, we assessed the proportion of tumors exhibiting resistance to p53-dependent apoptosis after exposure to ionizing irradiation (IR)–induced DNA double-strand breaks (DSBs). Surprisingly, we found that almost half the tumors exhibited resistance to IR-induced apoptosis in the presence of normal p53 activation and that this most frequently involved aberrant NF-κB prosurvival signaling, indicated by the abnormal expression of TRAF5, TRAF6, and cIAP1 after IR. PARP1 inhibition could initiate p53-dependent apoptosis after IR in these tumors through a mechanism that reduced NF-κB transcriptional activity. Impaired p53 responses caused by a TP53 mutation or by defective activation of p53 only accounted for apoptotic resistance in a small proportion of ALL tumors.
Materials and methods
B-precursor ALL
The diagnosis of presenting or relapsed B-precursor ALL was based on standard morphologic and immunophenotypic evaluation. Bone marrow samples from 40 patients (30 at diagnosis and 10 during relapse) were chosen on the basis of availability of material. Written consent was obtained from all patients before material was collected.
Cell culture and γ-irradiation
Frozen viable bone marrow mononuclear cells composed of more than 80% leukemic blasts were defrosted and cultured in RPMI (with l-glutamine, fetal calf serum [FCS], and penicillin/streptomycin). Aliquot portions of 2 × 106 cells per time point were irradiated with 5 Gy IR (cobalt Co 60) and incubated at 37° C. Only tumor samples with more than 70% viability were included in the study.
Western blot analysis
ALL cells were harvested after IR and were lysed for 30 minutes in modified RIPA buffer, as described previously.24 Standard Western blot analysis using 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was performed using sheep polyclonal antibody against p53 (donated by D.P. Lane, University of Dundee, Scotland), rabbit polyclonal antibody against p21 (Santa Cruz Biotechnology, Santa Cruz, CA), goat polyclonal antibody against PARP1 (Santa Cruz Biotechnology), and rabbit polyclonal antibodies against procaspases 3, 7, and 9 (Cell Signaling Technology, Beverly, MA) and poly(ADP-ribose) (pADPr) (Calbiochem, La Jolla, CA). Mouse monoclonal antibody against β-actin (Sigma, St Louis, MO) was used as loading control.
Inhibition of pADPr formation using 3-AB
Inhibition of PARP1 activity was performed using 3-aminobenzamide (3-AB). Cells were pretreated with 5 mM 3-AB for 2 hours and then subjected to 5 Gy IR, as described in “Cell culture and γ-irradiation.”
Mutation screening of TP53 and PARP1 by direct sequencing
Exons 4 to 8 of the TP53 gene were amplified through polymerase chain reaction (PCR) from genomic DNA and were analyzed as described previously.24 The PARP1 coding sequence was PCR amplified in 3 fragments using the following primers: fragment 1, 5′-GCA GCT AGG GGA GGA TGG CGG AGT CT-3′ and 5′-TCC CTT TGG GGT TAC CCA CTC CTT CC-3′; fragment 2, 5′-GGA AGG AGT GGG TAA CCC CAA AGG AA-3′ and 5′-AGA CAC CGC CTG CTG GAC CTC ACT-3′; and fragment 3, 5′-TGG AGT ATG AGA TCG ACC TTG AGA-3′ and 5′-CAG CCA CCG GGT GTG ACT CGG CT-3′.
Mutation analysis of ATM by denaturing high-performance liquid chromatography
All 62 coding exons of the ATM gene were PCR amplified from genomic DNA using 60 primer pairs modified from a previous protocol.25 Heteroduplex analysis was performed according to the Transgenomic Wave software (Transgenomic, Omaha, NE). Any detected changes were confirmed by direct sequencing.
Annexin V–FITC staining and FACS analysis
Apoptosis was assayed using Annexin V staining. Annexin V binds to phospholipid phosphatidylserine (PS), which, during apoptosis, is exposed to the external environment as a consequence of a loss of plasma membrane. We used the Annexin V–fluorescein isothiocyanate (FITC) apoptosis detection kit (BD PharMingen, San Diego, CA) according to the manufacturer's instructions and used a fluorescence-activated cell sorter (FACS) (Epics XL-MCL; Beckman Coulter, Fullerton, CA) to analyze samples.
Expression array analysis of apoptotic genes
Nylon arrays tetra-spotted with 96 apoptotic genes (Superarray Bioscience, Frederick, MD) were used to compare gene expression before and 8 hours after IR. RNA was labeled with biotin-16-dUTP (Roche Molecular Biochemicals, Indianapolis, IN) using the Ampolabeling–LPR kit (Superarray Bioscience), and hybridizations were carried out according to the manufacturer's instructions. Filters were converted into 16-bit images, and data were extracted. Subsequently, pUC18 background measurements were subtracted, and normalization was performed to the housekeeping gene PPIA (cyclophilin A) (ArrayVision 8.0 software; ArrayVision Imaging Research, St Catharine's, ON, Canada). Differences in median values of the tetraspot intensities between data pairs (8-0 hours after IR) were calculated, and hierarchical cluster analysis was performed using dChip V1.3 DNA Chip Analyzer software (http://www.dchip.org) under the default settings.
EMSA
Electrophoretic mobility shift assay (EMSA) was performed as previously described.26 Briefly, nuclear lysates were incubated with a radiolabeled HIV-κB oligonucleotide probe, electrophoresed, and exposed to x-ray film. Cytoplasmic lysates were subjected to Western blotting and were reacted against a mouse monoclonal IκBα antibody (Santa Cruz Biotechnology).
Results
DNA damage response analysis identifies a subgroup of B-precursor ALL with apoptotic resistance
Apoptosis can occur through 2 major pathways, the intrinsic mitochondrial-mediated apoptotic pathway and the extrinsic death receptor–mediated apoptotic pathway. After exposure to DNA-damaging agents, including IR, activated p53 initiates apoptosis predominantly through the mitochondrial pathway.27,28 This involves the release of cytochrome c, which forms an apoptosome complex with procaspase 9 and Apaf1, leading to cleavage and activation of the effector procaspases 3, 6, and 7, the converging point for both apoptotic pathways.28,29 The effector caspases initiate chromatin fragmentation and substrate cleavage. PARP1 is a protein involved in the initial response to DNA damage, which is also inactivated through its cleavage by caspase 3 in a p53-dependent manner after DNA DSBs.28,29
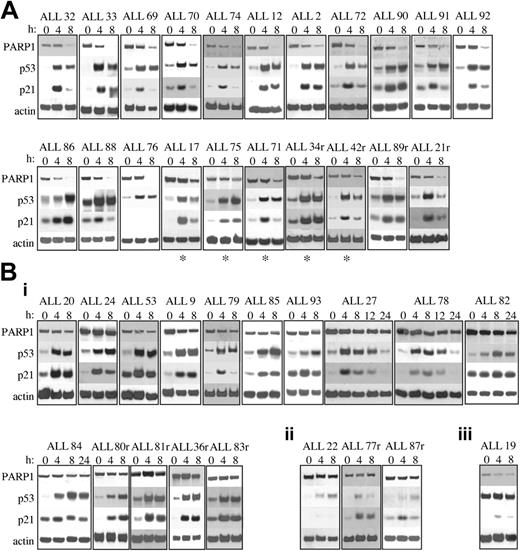
To evaluate the response to IR-induced DNA damage in 40 primary ALL tumor samples, we analyzed accumulation of the p53 and p21 proteins and cleavage of PARP1 as a marker of p53-dependent apoptotic integrity. We identified 2 major subgroups with respect to the apoptotic response to IR (Figure 1). The first subgroup, comprising 21 (52.5%) of 40 tumors, was proficient in its apoptotic response to IR. All tumors exhibited PARP1 protein cleavage by 8 hours after IR, shown by a reduction in the intensity of the band corresponding to the 116-kDa, full-length protein (Figure 1A). All tumors also exhibited normal accumulation of p53 and p21 proteins, with maximum accumulation occurring by 8 hours after IR; the exception was ALL 76, which revealed a complete absence of p21 protein induction after IR (Figure 1A). However, this did not appear to occur through a mechanism associated with IR-induced apoptosis because PARP1 was completely cleaved by 8 hours after IR. Interestingly, 5 tumors in this subgroup (denoted by asterisks) appeared to cleave PARP1 in a slightly delayed manner because some residual full-length PARP1 protein was still present 8 hours after IR (Figure 1A), suggesting a slight delay in the apoptotic response.
Western blot analysis of PARP1, p53, and p21 proteins after 5 Gy IR. (A) Twenty-one of 40 B-precursor ALL tumors exhibited normal apoptotic responses after IR showing PARP1 cleavage and normal p53 and p21 protein accumulation. ALL 76 showed no p21 protein induction but did exhibit PARP1 cleavage. (B) Nineteen of 40 with B-precursor ALL tumors exhibited defective apoptotic responses after IR. Actin shows equal loading. (i) Fifteen with ALL tumors exhibited defective PARP1 cleavage after IR despite normal p53 and p21 protein accumulation. (ii) Three ALL tumors exhibited defective PARP1 cleavage and defective activation of p53 and p21 proteins after IR, suggesting a defect upstream of p53. (iii) One ALL tumor exhibited high basal levels of p53 protein with no further accumulation of p53 or p21 proteins after IR, suggesting a TP53 mutation. r indicates relapse sample; and *, delayed PARP1 cleavage.
Western blot analysis of PARP1, p53, and p21 proteins after 5 Gy IR. (A) Twenty-one of 40 B-precursor ALL tumors exhibited normal apoptotic responses after IR showing PARP1 cleavage and normal p53 and p21 protein accumulation. ALL 76 showed no p21 protein induction but did exhibit PARP1 cleavage. (B) Nineteen of 40 with B-precursor ALL tumors exhibited defective apoptotic responses after IR. Actin shows equal loading. (i) Fifteen with ALL tumors exhibited defective PARP1 cleavage after IR despite normal p53 and p21 protein accumulation. (ii) Three ALL tumors exhibited defective PARP1 cleavage and defective activation of p53 and p21 proteins after IR, suggesting a defect upstream of p53. (iii) One ALL tumor exhibited high basal levels of p53 protein with no further accumulation of p53 or p21 proteins after IR, suggesting a TP53 mutation. r indicates relapse sample; and *, delayed PARP1 cleavage.
In contrast, the second subgroup, comprising 19 (47.5%) of 40 ALL tumors, did not exhibit PARP1 cleavage by 8 hours after IR, indicating the absence of IR-induced apoptosis (Figure 1B). Intriguingly, in most of these tumors (15 of 19), this indication of apoptotic resistance occurred in the presence of normal p53 and p21 protein accumulation (Figure 1Bi). In 4 of these (ALL 27, ALL 78, ALL 82, and ALL 84), we investigated the possibility that PARP1 cleavage was delayed, but even 24 hours after IR, the PARP1 protein remained uncleaved (Figure 1Bi). The mechanism of apoptotic resistance in these tumors, therefore, appeared to occur at a stage after p53 activation, indicated by apparently normal p53 and p21 protein accumulation yet before mitochondrial-dependent activation of procaspases, indicated by the absence of PARP1 cleavage.
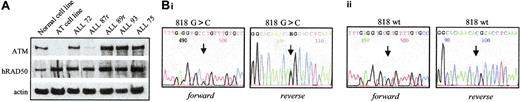
In the remaining 4 apoptosis-resistant tumors, defects in the p53 response were observed (Figure 1Bii-iii). In 3 (ALL 22, ALL 77r, and ALL 87r), virtually no p53 or p21 protein accumulation was observed after IR, suggesting a defect upstream of p53 (Figure 1Bii). Given that the ATM protein is the principle activator of p53 in the response to IR-induced DNA damage, we reasoned that the ATM gene might be inactivated in these 3 tumors. For one of these leukemias, ALL 87r, there was sufficient material for us to perform ATM protein expression analysis (Figure 2A). In contrast to 4 leukemias with normal p53 accumulation, ALL 87r revealed almost complete absence of ATM protein (Figure 2A), strongly suggesting the presence of ATM inactivation. This possibility was supported further by the fact that ALL 87r had lost 1 copy of chromosome 11 and, therefore, retained only 1 ATM allele. We carried out denaturing high-performance liquid chromatography (DHPLC) analysis (Transgenomic Wave) and direct sequencing of the 62 ATM coding exons in all 3 tumors with defective p53 activation. Although previously described polymorphisms were detected in ALL77r (exon 39 5557G>A and IVS38-8T>C) and ALL87r (exon 26 IVS26-24delT and exon 55 IVS55 + 80T>C), surprisingly no mutations altering the ATM coding sequence could be identified in any of the 3 tumors.
Confirmation of defects in the p53 pathway. (A) Western blot of ATM protein showing reduced ATM expression in ALL87r, similar to the control ataxia telangiectasia cell line. Actin shows equal loading; hRad50, a high–molecular weight protein, demonstrates an absence of sample degradation. (B) Chromatogram of the TP53 gene sequence in ALL 19. The arrow indicates the position of the mutated nucleotide. (i) A heterozygous missense 818G>C mutation was identified in the tumor DNA. (ii) The sequence was wild type at the same position in the germline DNA.
Confirmation of defects in the p53 pathway. (A) Western blot of ATM protein showing reduced ATM expression in ALL87r, similar to the control ataxia telangiectasia cell line. Actin shows equal loading; hRad50, a high–molecular weight protein, demonstrates an absence of sample degradation. (B) Chromatogram of the TP53 gene sequence in ALL 19. The arrow indicates the position of the mutated nucleotide. (i) A heterozygous missense 818G>C mutation was identified in the tumor DNA. (ii) The sequence was wild type at the same position in the germline DNA.
The final apoptosis-resistant ALL (ALL 19) had a defect consistent with the presence of a p53 mutation; we observed high basal levels of p53 protein with no subsequent accumulation of p53 or p21 proteins after IR in this tumor (Figure 1B). Consequently, we identified a heterozygous missense TP53 mutation at position 818G>C, predicted to cause amino acid change R273P (Figure 2Bi). The mutation could not be detected in the nontumor DNA (Figure 2Bii). Interestingly, ALL19 exhibited low basal levels of full-length PARP1 protein (Figure 1B), which was cleaved after IR. Given that p53 mutations confer apoptotic resistance and, therefore, would not be expected to initiate PARP1 cleavage, it was likely that in ALL 19 an association occurred between TP53 inactivation and loss of baseline PARP1 protein expression.
We next wanted to confirm that our apoptotic assay involving PARP1 cleavage did indeed reflect p53-dependent activation of apoptosis (Figure 3). First, we used Western blot analysis to directly assess cleavage of procaspases 3, 7, and 9 in 8 representative leukemias (Figure 3A). Four ALL tumors with normal responses to IR (ALL 17, ALL 75, ALL 89r, and ALL 91) showed complete cleavage of the procaspases by 24 hours after IR (Figure 3A). Two of these, ALL 17 and ALL 75, which were previously shown to have slightly delayed kinetics of PARP1 cleavage (Figure 1A), also exhibited slightly delayed caspase activation (Figure 3Ai). In contrast, 4 apoptosis-resistant tumors (ALL 20, ALL 82, ALL 84, and ALL87r) revealed an almost complete absence of detectable cleavage of the procaspases by 24 hours after IR (Figure 3Aii). These findings were confirmed by FACS analysis of Annexin V–stained cells: 3 apoptosis-proficient ALL tumors (ALL 92, ALL 90, and ALL 88) clearly underwent IR-induced apoptosis, whereas 3 apoptosis-resistant ALL tumors (ALL 20, ALL 82, and ALL 85) did not exhibit notable apoptosis by 8 hours after IR (Figure 3B).
PARP1 cleavage correlates with apoptosis. (A) Western blot analysis showing cleavage of procaspases 3, 7, and 9 after 5 Gy IR to indicate apoptosis. Cleavage of full-length procaspases 7 and 9 and appearance of the largest cleavage product of caspase 3 are shown. Actin shows equal loading. r indicates relapse; *, delayed PARP1 cleavage. (i) Apoptosis-proficient ALL tumors showed complete cleavage of caspases 3, 7, and 9 after IR. (ii) Apoptosis-resistant ALL tumors showed absence of cleavage of caspases 3, 7, and 9 up to 24 hours after IR. (B) FACS analysis of Annexin V apoptosis assay indicating loss of plasma membrane and showing the percentage of cells undergoing IR-induced apoptosis at 0 and 8 hours after 5 Gy IR. Background apoptosis has been excluded. Data represent the average and SE from 3 apoptosis-proficient ALL tumors (ALL 92, ALL 90, and ALL 88; solid line) and 3 apoptosis-resistant ALL tumors (ALL 20, ALL 82, and ALL 85; broken line). (C) Annexin V apoptosis assay and FACS analysis showing the percentage of cells undergoing spontaneous apoptosis at 0, 8, and 24 hours. Data represent the average and SE from 3 apoptosis-proficient ALL tumors (ALL 92, ALL 90, and ALL 88; solid line) and 3 apoptosis-resistant ALL tumors (ALL 20, ALL 82, and ALL 85; broken line).
PARP1 cleavage correlates with apoptosis. (A) Western blot analysis showing cleavage of procaspases 3, 7, and 9 after 5 Gy IR to indicate apoptosis. Cleavage of full-length procaspases 7 and 9 and appearance of the largest cleavage product of caspase 3 are shown. Actin shows equal loading. r indicates relapse; *, delayed PARP1 cleavage. (i) Apoptosis-proficient ALL tumors showed complete cleavage of caspases 3, 7, and 9 after IR. (ii) Apoptosis-resistant ALL tumors showed absence of cleavage of caspases 3, 7, and 9 up to 24 hours after IR. (B) FACS analysis of Annexin V apoptosis assay indicating loss of plasma membrane and showing the percentage of cells undergoing IR-induced apoptosis at 0 and 8 hours after 5 Gy IR. Background apoptosis has been excluded. Data represent the average and SE from 3 apoptosis-proficient ALL tumors (ALL 92, ALL 90, and ALL 88; solid line) and 3 apoptosis-resistant ALL tumors (ALL 20, ALL 82, and ALL 85; broken line). (C) Annexin V apoptosis assay and FACS analysis showing the percentage of cells undergoing spontaneous apoptosis at 0, 8, and 24 hours. Data represent the average and SE from 3 apoptosis-proficient ALL tumors (ALL 92, ALL 90, and ALL 88; solid line) and 3 apoptosis-resistant ALL tumors (ALL 20, ALL 82, and ALL 85; broken line).
To exclude the possibility that the difference in IR-induced apoptosis was a consequence of a difference in spontaneous apoptosis between the 2 subgroups of tumors, we used Western blot analysis and densitometry and found that there was no significant difference in the amount of reduction of procaspase 7 protein (relative to actin) at 8 hours and 24 hours between 2 apoptosis-proficient (ALL 75 and ALL 17) and 2 apoptosis-resistant ALL tumors (ALL 82 and ALL 20) (data not shown). Annexin V staining also showed that there was no difference in spontaneous apoptosis at 0, 8, and 24 hours between 3 apoptosis-proficient (ALL 92, ALL 90, and ALL 88) and 3 apoptosis-resistant (ALL 20, ALL 82, and ALL 85) ALL tumors (Figure 3C).
Overall, therefore, we have identified a subgroup of 19 ALL tumors that exhibited resistance to IR-induced apoptosis. Strikingly, this resistance most frequently occurred in the presence of normal p53 activation, suggesting the presence of a mechanism that could suppress p53-dependent caspase activation.
Apoptotic response to IR-induced DNA damage and clinical response
Next, we addressed the clinical significance of the apoptotic response to IR (Table 1). The most striking correlation related to the 4 apoptosis-resistant ALL tumors with defective p53 activation. These were all clearly clinically aggressive: 3 of the patients experienced relapses, and the fourth could not achieve remission on standard therapy (Table 1).
Clinical information of patients with apoptosis-proficient and apoptosis-resistant ALL tumors
ALL . | Age at diagnosis, y . | WBC count at diagnosis, × 109/L . | Length of first CR, mo . | Clinical outcome . | Prognostic cytogenetic abnormality . |
|---|---|---|---|---|---|
| Apoptosis-proficient | |||||
| 32 | 12 | 210 | 56 | CR | — |
| 33 | 7 | 195 | 10 | R | — |
| 69 | 4 | 19 | 11 | CR | Hyperdiploid (55) |
| 70 | 6 | 56 | 11 | Regimen change | — |
| 74 | 2 | 7 | 62 | CR | Hyperdiploid (55) |
| 12 | 4 | 96 | 19 | R | — |
| 2 | 7 | 49 | 41 | CR | t(12;21) |
| 72 | 3 | 16 | 8 | CR | Hyperdiploid (58-59) |
| 90 | 2 | 11 | 0 | CR | Hyperdiploid (56) |
| 91 | 7 | 7 | 4 | CR | Hyperdiploid (57) |
| 92 | 6 | 47 | 0 | CR | t(12;21) |
| 86 | 11 | 4 | 1 | D | Hyperdiploid (55) |
| 88 | 5 | 6 | 1 | CR | — |
| 76 | 5 | 8 | 36 | CR | TEL-AML ×2 |
| 17 | 3 | 12 | 34 | CR | t(12;21) |
| 75 | 9 | 330 | 7 | CR | — |
| 71 | 2 | 37 | 24 | CR | Hyperdiploid (54-56) |
| 34r | 3 | 41 | 65 | R | Hyperdiploid (58) |
| 42r | 3 | 68 | 33 | R | — |
| 89r | 15 | 3 | 35 | R | Hyperdiploid (53) |
| 21r | 15 | 55 | 9 | R | — |
| Apoptosis-resistant (normal p53 response) | |||||
| 20 | 18 | 118 | 42 | CR (BMT as standard) | — |
| 24 | 4 | 15 | 28 | Regimen change | TEL-AML 1(complex translocation) |
| 53 | 12 | 106 | 11 | R | — |
| 9 | 10 | 6 | 56 | CR | t(12;21) |
| 27 | 15 | 13 | 59 | CR | t(1;19) |
| 78 | 15 | 18 | 9 | CR | t(12;21) |
| 79 | 4 | 8 | 25 | R | — |
| 82 | 3 | 6 | 5 | CR | Hyperdiploid (50) |
| 84 | 7 | 21 | 7 | CR | — |
| 85 | 14 | 202 | 4 | CR | — |
| 93 | 5 | 3 | 1 | CR | — |
| 80r | 3 | 35 | 41 | R | t(12;21) |
| 81r | 4 | 18 | 72 | R | Hyperdiploid (56) |
| 36r | 8 | 20 | 43 | R | t(1;19) |
| 83r | 4 | 237 | 19 | R | Hyperdiploid (53) |
| Apoptosis-resistant (defective p53 response) | |||||
| 19* | 13 | 69 | 16 | R and D | — |
| 22† | 2 | 140 | 31 | Regimen change | t(1;19) |
| 77r† | 9 | 2 | 38 | R | t(12;21) |
| 87r† | 11 | 16 | 11 | R | Near haploid (27) |
ALL . | Age at diagnosis, y . | WBC count at diagnosis, × 109/L . | Length of first CR, mo . | Clinical outcome . | Prognostic cytogenetic abnormality . |
|---|---|---|---|---|---|
| Apoptosis-proficient | |||||
| 32 | 12 | 210 | 56 | CR | — |
| 33 | 7 | 195 | 10 | R | — |
| 69 | 4 | 19 | 11 | CR | Hyperdiploid (55) |
| 70 | 6 | 56 | 11 | Regimen change | — |
| 74 | 2 | 7 | 62 | CR | Hyperdiploid (55) |
| 12 | 4 | 96 | 19 | R | — |
| 2 | 7 | 49 | 41 | CR | t(12;21) |
| 72 | 3 | 16 | 8 | CR | Hyperdiploid (58-59) |
| 90 | 2 | 11 | 0 | CR | Hyperdiploid (56) |
| 91 | 7 | 7 | 4 | CR | Hyperdiploid (57) |
| 92 | 6 | 47 | 0 | CR | t(12;21) |
| 86 | 11 | 4 | 1 | D | Hyperdiploid (55) |
| 88 | 5 | 6 | 1 | CR | — |
| 76 | 5 | 8 | 36 | CR | TEL-AML ×2 |
| 17 | 3 | 12 | 34 | CR | t(12;21) |
| 75 | 9 | 330 | 7 | CR | — |
| 71 | 2 | 37 | 24 | CR | Hyperdiploid (54-56) |
| 34r | 3 | 41 | 65 | R | Hyperdiploid (58) |
| 42r | 3 | 68 | 33 | R | — |
| 89r | 15 | 3 | 35 | R | Hyperdiploid (53) |
| 21r | 15 | 55 | 9 | R | — |
| Apoptosis-resistant (normal p53 response) | |||||
| 20 | 18 | 118 | 42 | CR (BMT as standard) | — |
| 24 | 4 | 15 | 28 | Regimen change | TEL-AML 1(complex translocation) |
| 53 | 12 | 106 | 11 | R | — |
| 9 | 10 | 6 | 56 | CR | t(12;21) |
| 27 | 15 | 13 | 59 | CR | t(1;19) |
| 78 | 15 | 18 | 9 | CR | t(12;21) |
| 79 | 4 | 8 | 25 | R | — |
| 82 | 3 | 6 | 5 | CR | Hyperdiploid (50) |
| 84 | 7 | 21 | 7 | CR | — |
| 85 | 14 | 202 | 4 | CR | — |
| 93 | 5 | 3 | 1 | CR | — |
| 80r | 3 | 35 | 41 | R | t(12;21) |
| 81r | 4 | 18 | 72 | R | Hyperdiploid (56) |
| 36r | 8 | 20 | 43 | R | t(1;19) |
| 83r | 4 | 237 | 19 | R | Hyperdiploid (53) |
| Apoptosis-resistant (defective p53 response) | |||||
| 19* | 13 | 69 | 16 | R and D | — |
| 22† | 2 | 140 | 31 | Regimen change | t(1;19) |
| 77r† | 9 | 2 | 38 | R | t(12;21) |
| 87r† | 11 | 16 | 11 | R | Near haploid (27) |
Shown for each patient are age and WBC at diagnosis, length of first continuous remission (CR), clinical outcome (R, relapse; D, death), and prognostic cytogenetic abnormalities (—, absence of prognostic cytogenetic abnormality; number of chromosomes is given in parentheses alongside ploidy status).
TP53 mutant.
Defective activation of p53 (possible ATM inactivation).
Because many of the patients in this study were only in short-term follow-up, we focused on the association between the in vitro apoptotic response to IR and prognostic features in apoptosis-proficient and apoptosis-resistant tumors with normal p53 activation. Although the presence of an adverse white blood cell (WBC) count (greater than 50 × 109/L) at diagnosis occurred at a comparable frequency between these 2 subgroups of tumors, an adverse age (older than 10 years) at diagnosis did appear to correlate more strongly with apoptosis resistance, present in 6 (40%) of 15 apoptosis-resistant tumors with normal p53 activation and only 4 (20%) of 21 apoptosis-proficient tumors (Table 1). Prognostic cytogenetic abnormalities also segregated differentially between the 2 subgroups of tumors. Most strikingly, good prognostic hyperdiploid karyotypes were more frequently associated with apoptosis-proficient tumors, present in 9 (43%) of 21 patients, compared with only 3 (20%) of 15 apoptosis-resistant tumors with normal p53 activation. Interestingly, the good prognostic t(12;21) did not segregate differentially between the subgroups of tumors, whereas the poor prognostic marker, t(1;19), was detectable only in apoptosis-resistant tumors. No t(9;22) or MLL gene rearrangements were present in our cohort of patients (Table 1).
Therefore, tumors that exhibited apoptotic resistance and normal p53 activation appeared to have poorer prognostic features than apoptosis-proficient tumors, whereas ALL tumors with defects in p53 activation were uniformly clinically aggressive.
Array expression analysis indicates increased NF-κB activity in apoptosis-resistant tumors with normal p53 activation after IR
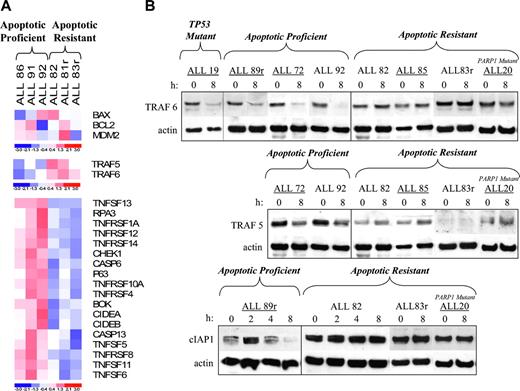
To establish the cause of apoptotic resistance in the 15 ALL tumors with normal p53 activation, we used mini–gene expression analysis. We analyzed the effect of IR on the expression of 96 apoptotic genes in 3 apoptosis-proficient leukemias (ALL 86, ALL 91, and ALL 92) and 3 apoptosis-resistant leukemias (ALL 82, ALL 81r, and ALL 83r) with normal p53 activation. Eight hours after IR, as expected, both subgroups revealed similar transcriptional responses of the p53-responsive genes MDM2, BCL2, and BAX (Figure 4A). In contrast, compared with the apoptosis-proficient tumors, the apoptosis-resistant tumors exhibited higher expression of the TRAF5 and TRAF6 genes after IR and reduced expression of several caspases, tumor necrosis family-super family (TNF-SF) genes, and proapoptotic genes. These included CASPASES 6 and 13, the TNF-SF genes TNFRSF12 (Apo3/DR3), TNFRSF1A (TNF-R1), TNFRSF14, TNFRSF10A (TRAIL-R1/DR4), TNFRSF4 (OX40), TNFRSF8 (CD30), TNFSF11 (TRANCE), TNFSF13 (APRIL), TNFSF6 (FAS-L), and TNFSF5 (CD40L/CD154), the proapoptotic genes CIDEA, CIDEB, and BOK, and the DNA repair/replication genes CHEK1, RPA3, and P63 (Figure 4A).
Mini–gene expression analysis indicating increased prosurvival pathway signaling in apoptosis-resistant ALL tumors. (A) Cluster analysis of the difference in expression of apoptotic genes before and 8 hours after 5 Gy IR comparing apoptosis-proficient and apoptosis-resistant tumors. Three patterns of gene expression were observed. (B) Western blot analyses showing defective expression of TRAF5, TRAF6, and cIAP1 proteins in apoptosis-resistant ALL tumors compared with apoptosis-proficient ALL tumors and the p53 mutant leukemia, ALL 19, 8 hours after IR. Actin serves as a loading control. (Underlined ALL tumors were not previously analyzed by array expression analysis.)
Mini–gene expression analysis indicating increased prosurvival pathway signaling in apoptosis-resistant ALL tumors. (A) Cluster analysis of the difference in expression of apoptotic genes before and 8 hours after 5 Gy IR comparing apoptosis-proficient and apoptosis-resistant tumors. Three patterns of gene expression were observed. (B) Western blot analyses showing defective expression of TRAF5, TRAF6, and cIAP1 proteins in apoptosis-resistant ALL tumors compared with apoptosis-proficient ALL tumors and the p53 mutant leukemia, ALL 19, 8 hours after IR. Actin serves as a loading control. (Underlined ALL tumors were not previously analyzed by array expression analysis.)
Consistent with the gene expression data, TRAF5 and TRAF6 were also aberrantly expressed at the protein level in the apoptosis-resistant tumors. All apoptosis-proficient tumors, and the p53 mutant leukemia (ALL 19) in the case of TRAF6, exhibited complete down-regulation of TRAF5 and TRAF6 proteins 8 hours after IR, whereas the apoptosis-resistant leukemias either maintained or exhibited a slight increase in the level of these proteins (Figure 4B). Notably, the initial levels of TRAF5 appeared to be lower in the apoptosis-resistant ALL tumors; furthermore, ALL 83r expressed no detectable TRAF5 protein either before or after IR (Figure 4B).
TRAF5 and TRAF6 can mediate prosurvival signals. TRAF6 in particular is known to be a potent inducer of NF-κB activity.30-33 Therefore, it was possible that the aberrant high expression of TRAF5 and TRAF6 proteins after IR caused an increase in NF-κB transcriptional activity in the apoptosis-resistant leukemias. To address this possibility, we investigated the expression of an NF-κB-responsive, antiapoptosis protein, cIAP1,34,35 after IR. In an apoptosis-proficient tumor, cIAP1 protein expression peaked at 2 hours and then decreased by 8 hours after IR (Figure 4B). In contrast, in 3 apoptosis-resistant tumors, the basal levels of cIAP1 appeared to be constitutively high, and no down-regulation was observed by 8 hours after IR (Figure 4B), suggesting the presence of increased NF-κB transcriptional activity.
Taken together, it appeared that in the apoptosis-resistant ALL tumors with normal p53 activation, NF-κB activity was aberrantly increased, suggesting that overriding survival signals might provide the mechanism of resistance to IR-induced apoptosis by suppressing p53-transduced apoptotic signals.
Normal p53-mediated, IR-induced apoptosis can be restored in apoptosis-resistant tumors through PARP1 inhibition
The targeted inactivation of PARP1 during apoptosis suggests it has regulatory functions in the response to DNA damage. Indeed, PARP1 activity is induced by DNA strand breaks, leading to the synthesis of pADPrs.36,37 Consequently, several proteins, including PARP1 itself, are posttranslationally modified by pADPrs,38,39 leading to changes in protein–DNA interactions involved in chromatin modeling and DNA repair.38,39 Before the completion of DNA repair or apoptosis, hydrolysis of pADPr is required to reinstate cellular energy levels, and, in the event of apoptosis, PARP1 is inactivated by its cleavage.39,40 The antiapoptotic function of PARP1 is implied further by the recent findings that PARP1 is a transcriptional coactivator of NF-κB, a function that is mediated by its direct binding and (ADP-ribose) polymerization activity.41,42
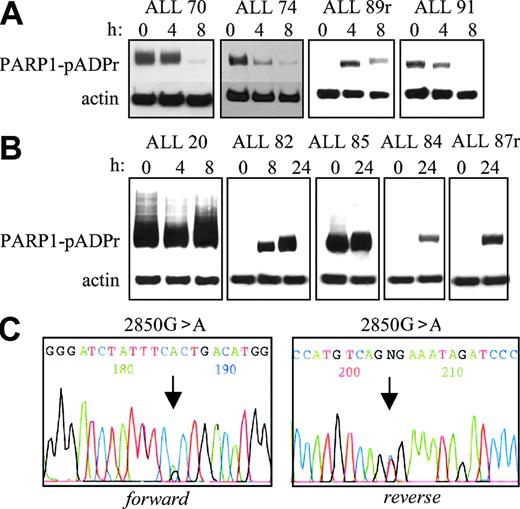
Therefore, we next investigated whether abnormal PARP1 activity might contribute to the increased NF-κB transcriptional activity and, hence, apoptosis resistance to IR in the 15 ALL tumors with normal p53 activation. To address this possibility, we examined the levels of pADPr attached to PARP1 itself in 9 tumors (Figure 5). Consistent with the concomitant degradation of pADPr and the inactivation of PARP1, in 4 apoptosis-proficient leukemias (ALL 70, ALL 74, ALL 89r, and ALL 91), pADPr could no longer be detected by 8 hours after IR (Figure 5A). In contrast, and consistent with the absence of PARP1 inactivation, in 5 apoptosis-resistant leukemias, 4 with normal p53 activation (ALL 20, ALL 82, ALL 84, and ALL 85) and 1 with defective p53 activation (ALL87r), pADPr could still be detected 24 hours after IR (Figure 5B). Because PARP1 is a transcriptional coactivator of NF-κB,41,42 it seemed likely that such prolonged PARP1 activity might contribute to the increase in NF-κB activity in these tumors. Intriguingly, in ALL 20 (Figure 5B), the basal levels of pADPr detected were strikingly high and were maintained throughout the response to IR, suggesting the presence of a hyperactive PARP1 protein.
Western blot analysis showing levels of pADPr attached to the PARP1 protein (PARP-pADPr) as an indication of PARP1 activity after 5 Gy IR. (A) Apoptosis-proficient ALL tumors showed pADPr degradation by 8 hours after IR. (B) Apoptosis-resistant ALL tumors showed an absence of pADPr degradation up to 24 hours after IR. (C) Chromatogram of the PARP1 gene sequence in ALL 20. Forward and reverse primer sequencing identified a heterozygous missense 2850G>A mutation in the tumor DNA. The arrow indicates the position of the mutated nucleotide.
Western blot analysis showing levels of pADPr attached to the PARP1 protein (PARP-pADPr) as an indication of PARP1 activity after 5 Gy IR. (A) Apoptosis-proficient ALL tumors showed pADPr degradation by 8 hours after IR. (B) Apoptosis-resistant ALL tumors showed an absence of pADPr degradation up to 24 hours after IR. (C) Chromatogram of the PARP1 gene sequence in ALL 20. Forward and reverse primer sequencing identified a heterozygous missense 2850G>A mutation in the tumor DNA. The arrow indicates the position of the mutated nucleotide.
Because the activity or ability of the PARP1 protein to become cleaved might be affected by PARP1 gene mutation, we directly sequenced the coding region of PARP1 in 11 leukemias. All revealed wild-type PARP1 alleles, with one exception: in ALL 20, we detected a PARP1 heterozygous missense mutation at nucleotide 2850G>A (Figure 5C). This sequence change, predicted to cause an amino acid alteration A898T, was also present in a remission bone marrow sample, suggesting its germline origin. The mutation was not present in 100 normal mitoses (data not shown); furthermore, it was located in the core catalytic domain of the protein, a conserved region that is crucial for pADPr formation,43,44 reinforcing the likelihood that this mutation carried a pathologic consequence. It is possible that this hyperactive PARP1 protein not only contributed to the increase in NF-κB activity but also played a role in the initiation of leukemogenesis in ALL 20.
We next reasoned that inhibiting PARP1 might reduce NF-κB activity and induce apoptosis after IR in tumors with apoptotic resistance and normal p53 activation. It has been reported that PARP1 inhibitors sensitize cells to damage-induced apoptosis in several cellular contexts.39 We investigated the effect of 5 mM 3-AB, which competes with ADP-ribose units at the catalytic site of PARP1 and prevents polymer formation (Figure 6A), in 4 apoptosis-resistant tumors (Figure 6A). Strikingly, in 3 tumors with normal p53 activation (ALL 20, ALL 82, and ALL 84), 3-AB completely abrogated the formation of pADPr on PARP1 itself 24 hours after IR and enabled complete activation of procaspases 3, 7, and 9 (Figure 6Ai). In contrast, in ALL 87r, which had defective activation of p53 and virtual absence of ATM protein, treatment with 3-AB did not appear to enable complete activation of the procaspases even 24 hours after IR (Figure 6Aii). However, the presence of some cleavage of procaspases 3 and 7, but not of procaspase 9, may indicate some degree of death receptor–mediated apoptosis in this tumor.
PARP1 inhibition using 5 mM 3-AB restored IR-induced apoptosis in apoptosis-resistant ALL tumors with increased NF-κB signaling by reducing the transcriptional activity of NF-κB. (A) Western blot analysis showing inhibition of PARP1 activity with 3-AB after 5 Gy IR. pADPr attached to the PARP1 protein (PARP1-pADPr) indicates PARP1 activity. Cleavage of procaspases 7 and 9 and the appearance of the largest cleavage product of caspase 3 was measured before and after IR and PARP1 inhibition. Actin shows equal loading. (i) Treatment with 3-AB and IR initiated procaspase cleavage in 3 apoptosis-resistant ALL tumors with abnormal NF-κB signaling. (ii) Treatment with 3-AB and IR failed to initiate procaspase cleavage in an apoptosis-resistant ALL tumor with defective p53 activation. (B) Adding 3-AB reduced the expression of the NF-κB–responsive protein cIAP1 and of TRAF6 but not of TRAF5, and it induced caspase 3 cleavage after 5 Gy IR in an apoptosis-resistant leukemia (ALL 82). Actin serves as a loading control. (C) EMSA showing the effect of 3-AB on NF-κB activation and binding to a 29-bp HIV-κB oligonucleotide (see “Materials and methods”) after 5 Gy IR. (i) Adding 3-AB abrogated NF-κB DNA binding 8 hours after IR in the nuclear protein. (ii) Adding 3-AB had no effect on the degradation of the NF-κB inhibitory protein IκBα in the cytoplasmic protein.
PARP1 inhibition using 5 mM 3-AB restored IR-induced apoptosis in apoptosis-resistant ALL tumors with increased NF-κB signaling by reducing the transcriptional activity of NF-κB. (A) Western blot analysis showing inhibition of PARP1 activity with 3-AB after 5 Gy IR. pADPr attached to the PARP1 protein (PARP1-pADPr) indicates PARP1 activity. Cleavage of procaspases 7 and 9 and the appearance of the largest cleavage product of caspase 3 was measured before and after IR and PARP1 inhibition. Actin shows equal loading. (i) Treatment with 3-AB and IR initiated procaspase cleavage in 3 apoptosis-resistant ALL tumors with abnormal NF-κB signaling. (ii) Treatment with 3-AB and IR failed to initiate procaspase cleavage in an apoptosis-resistant ALL tumor with defective p53 activation. (B) Adding 3-AB reduced the expression of the NF-κB–responsive protein cIAP1 and of TRAF6 but not of TRAF5, and it induced caspase 3 cleavage after 5 Gy IR in an apoptosis-resistant leukemia (ALL 82). Actin serves as a loading control. (C) EMSA showing the effect of 3-AB on NF-κB activation and binding to a 29-bp HIV-κB oligonucleotide (see “Materials and methods”) after 5 Gy IR. (i) Adding 3-AB abrogated NF-κB DNA binding 8 hours after IR in the nuclear protein. (ii) Adding 3-AB had no effect on the degradation of the NF-κB inhibitory protein IκBα in the cytoplasmic protein.
Next, we investigated whether PARP1 inhibition initiated IR-induced apoptosis by reducing the expression of NF-κB–responsive genes after IR (Figure 6B). In an apoptosis-resistant tumor (ALL 82), the basal level of cIAP1 protein was notably high and was maintained until 8 hours after IR (Figure 6B). After treatment with 3-AB, however, the overall expression level of the cIAP1 protein was significantly reduced, and expression decreased by 8 hours after IR, coinciding with the appearance of the cleaved caspase 3 fragment (Figure 6B). In addition, in response to IR, both TRAF5 and TRAF6 proteins, though initially down-regulated, were subsequently aberrantly up-regulated by 24 hours. After the addition of 3-AB, however, TRAF6, but not TRAF5, remained down-regulated by 24 hours after IR (Figure 6B). This suggested that TRAF6 may be positively regulated by NF-κB.
We used EMSA analysis to investigate the effect of 3-AB on the IR-dependent binding of NF-κB to target DNA (Figure 6Ci). We found that IR induced binding of NF-κB from the nuclear fraction to an HIV-κB oligonucleotide at 4 and 8 hours after IR (Figure 6Ci), whereas addition of 3-AB abrogated the DNA-binding response 8 hours after IR. This effect was not dependent on the sequestration of NF-κB in the cytoplasm by its inhibitory protein, IκBα, as IR-induced degradation of IκBα in the cytoplasmic fraction, shown by a reduction in the intensity of the corresponding band at 4 and 8 hours after IR, was not affected by the addition of 3-AB (Figure 6Cii).
Therefore, we found that PARP1 inhibition could initiate IR-induced apoptosis in the apoptosis-resistant ALL tumors with normal p53 activation by reducing the DNA binding and transcriptional activity of NF-κB and the expression of downstream prosurvival proteins such as cIAP1.
Together, our observations support the role of increased NF-κB prosurvival signaling in apoptotic resistance to IR in a subgroup of ALL tumors, and this mechanism appears to be more frequent than defects in p53 activation. Prolonged PARP1 activity appears to contribute to this increased NF-κB activity because PARP1 inhibition can initiate p53-dependent apoptosis in these tumors by reducing the ability of NF-κB to bind to and induce the transcription of its target genes.
Discussion
In the current study we found that a significant proportion of B-precursor ALL tumors exhibited apoptotic resistance to IR-induced DNA damage. In most of these tumors, apoptotic resistance appeared to occur because of increased NF-κB activity, indicated by the aberrant expression of TRAF5, TRAF6, and cIAP1 proteins. Intriguingly, PARP1 inhibition could restore IR-induced apoptosis in these tumors by reducing NF-κB binding and transcriptional activity. Surprisingly, only 4 apoptosis-resistantALL tumors exhibited defects in p53 activation, the key event in DNA damage–induced apoptosis.
Our study revealed that in most ALL tumors, apoptotic resistance to IR involved increased NF-κB–dependent prosurvival signals that appeared to be capable of suppressing p53-transduced apoptotic signals and preventing mitochondrial-dependent caspase activation after DNA damage. Interestingly, array analysis showed that in the normal response to IR, ALL cells activated molecules involved in death receptor–mediated and p53-mediated apoptosis, an observation previously reported after treatment with cytotoxic drugs.5,45,46 However, TNF signaling appeared to be aberrant in the apoptosis-resistant tumors, suggesting that though increased NF-κB prosurvival signals may be the driving force for apoptotic resistance in these tumors, the process may be facilitated by the concomitant down-regulation of additional proapoptotic genes.
It was not possible from the current study to conclude whether the defective expression of TRAF5 and TRAF6 was the initial cause of the aberrant increase in NF-κB activity in the apoptosis-resistant leukemias. However, given our observation that the inhibition of PARP1, a transcriptional coactivator of NF-κB, led to a reduction in TRAF6 expression after IR, it is possible that NF-κB can regulate TRAF6 expression through a positive feedback loop, a mechanism also proposed for other NF-κB–responsive proteins.35 Although future studies are required to elucidate the precise mechanisms governing deregulation of these survival pathways in pediatric ALL, the role of other TRAF members in drug resistance has been suggested in several types of cancers, including B-cell chronic lymphoblastic leukemia (B-CLL) and Hodgkin disease.47-49
We found that prolonged PARP1 activity contributed to the increase in NF-κB transcriptional activity. Consequently, inhibiting PARP1 activity abolished NF-κB DNA binding, leading to the down-regulation of its target proteins, cIAP1 and TRAF6, events that would facilitate p53-dependent apoptosis.5,34,35 Therefore, PARP1 inhibition may well be a valuable tool with which to improve the killing of ALL cells exhibiting apoptotic resistance mediated through increased survival signals.
The apparently hyperactive mutation we identified in the core of the catalytic domain of PARP1,43,44 a highly conserved region crucial for proper pADPr formation,43,50,51 most likely contributed to the increase in NF-κB activity in ALL 20. However, it was unclear whether the mutant protein was also involved in tumor initiation, though a distinct mechanism of tumor etiology may apply to this patient, who presented at 18 years of age. Although PARP1 mutations have not previously been reported in any human malignancy to our knowledge, interestingly, increased PARP1 activity through gene amplification or increased transcription has previously been reported in several other cancers, including high-grade lymphoma.52-55
In contrast to apoptotic resistance associated with NF-κB, TP53 gene mutation only accounted for apoptotic resistance in a single patient; similarly, defective activation of p53 after IR was not a frequent event and was detected in only 3 tumors. Despite the latter type of DNA damage response being reminiscent of the defect in ATM mutant B-CLL tumors56 and the clear reduction in ATM protein expression in ALL 87r, we could not detect any changes in the ATM gene that were predicted to alter the amino acid sequence in any of the 3 tumors. Because our screening method was sensitive enough to detect polymorphisms and because epigenetic silencing of ATM gene expression was unlikely given our ability to obtain ATM reverse transcription–PCR (RT-PCR) products, it was possible that ATM inactivation in these tumors occurred as a consequence of aberrant gene splicing resulting from the presence of undetected changes in the intronic sequence. To date, reports on the frequency of ATM mutation in pediatric ALL are not consistent; they vary from complete absence to an incidence of 20%.10-14 Although our findings suggest that defects in the p53 response, through defective activation or mutation, are relatively rare in pediatric ALL, they cause a severe clinical phenotype.
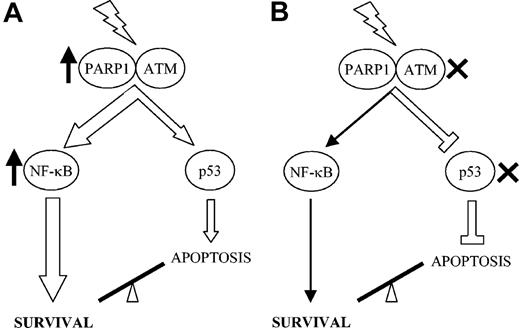
Taken together, our findings imply 2 distinct models by which B-precursor ALL cells can become resistant to damage-induced apoptosis (Figure 7). The most frequent mechanism involves an aberrant increase in NF-κB prosurvival signals, which is sufficient to override the p53-dependent apoptotic signal and to prevent mitochondrial-dependent caspase activation (Figure 7A). Prolonged or increased PARP1 activity and a concomitant defect in the up-regulation of various proapoptotic genes may contribute to this mechanism. A second mechanism of apoptotic resistance, occurring infrequently in pediatric ALL, involves inactivation of the p53 response, either by mutation of p53 itself or by inactivation of a gene upstream of p53, such as ATM, leading specifically to strong impairment of apoptosis after DNA damage (Figure 7B). The tumor cells, therefore, survive because of the presence of default prosurvival (eg, NF-κB–dependent) DNA damage signals.
Two alternative models describing 2 mechanisms of IR-induced apoptotic resistance in pediatric B-precursor ALL. (A) In most apoptosis-resistantALL tumors, the p53 pathway is functional, but increased prosurvival signals caused by hyperactive PARP1 or increased NF-κB activity outweigh the apoptotic signal transduced by p53. (B) In some tumors the apoptotic pathway is impaired by the mutation of TP53 or by inactivation of a gene upstream of p53. Therefore, tumor cells survive as a consequence of default prosurvival (eg, NF-κB–dependent) signals.
Two alternative models describing 2 mechanisms of IR-induced apoptotic resistance in pediatric B-precursor ALL. (A) In most apoptosis-resistantALL tumors, the p53 pathway is functional, but increased prosurvival signals caused by hyperactive PARP1 or increased NF-κB activity outweigh the apoptotic signal transduced by p53. (B) In some tumors the apoptotic pathway is impaired by the mutation of TP53 or by inactivation of a gene upstream of p53. Therefore, tumor cells survive as a consequence of default prosurvival (eg, NF-κB–dependent) signals.
In contrast to the uniformly clinically aggressive ALL tumors with defective p53 activation, for most tumors we were unable to establish a clear relationship between in vitro IR–induced apoptosis and clinical behavior. Clinical resistance can obviously arise through additional mechanisms not addressed in this study. However, our aim here was not to directly test drug sensitivity in ALL. Rather we used IR to induce a specific type of DNA damage that activates a well-defined DNA damage response pathway through p53 and the mitochondria with the intention of identifying molecular events that might cause biologic variability in ALL. Indeed, though increased NF-κB–mediated survival signals may not be involved in leukemia initiation, this defect appears to contribute to the pathology of a subgroup of tumors and, therefore, is likely to reflect the molecular heterogeneity of pediatric ALL with respect to treatment outcome. Most important, we have identified potential novel therapeutic targets that may well be exploited to further individualize the treatment of patients with ALL.
Prepublished online as Blood First Edition Paper, May 13, 2004; DOI 10.1182/blood-2003-11-4039.
Supported by the United Birmingham Hospitals Endowment Fund, the Leukaemia Research Fund, and Birmingham Children's Hospital Research Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Aris Eliopoulos and Dr Clare Davies, CRUK Institute for Cancer Studies, Birmingham University, for providing reagents for the EMSA analysis and for useful discussions.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal