Abstract
Retinoblastoma (Rb) and family members have been implicated as key regulators of cell proliferation and differentiation. In particular, accumulated data have suggested that the Rb gene product pRb is an important controller of erythroid differentiation. However, current published data are conflicting as to whether the role of pRb in erythroid cells is cell intrinsic or non–cell intrinsic. Here, we have made use of an in vitro erythroid differentiation culture system to determine the cell-intrinsic requirement for pRb in erythroid differentiation. We demonstrate that the loss of pRb function in primary differentiating erythroid cells results in impaired cell cycle exit and terminal differentiation. Furthermore, we have used coculture experiments to establish that this requirement is cell intrinsic. Together, these data unequivocally demonstrate that pRb is required in a cell-intrinsic manner for erythroid differentiation and provide clarification as to its role in erythropoiesis.
Introduction
The retinoblastoma (Rb) gene product pRb and family members p107 and p130 have been implicated as key regulators of cell proliferation and differentiation.1,2 In particular, extensive accumulated data have suggested that pRb is an important controller of erythroid differentiation. Rb–/– mice die of ineffective erythropoiesis on embryonic day 13.5 (E13.5), with decreased numbers of definitive erythrocytes observed in the peripheral blood.3-5 Rb–/– fetal livers (FLs) contained decreased numbers of erythroid colony-forming units (CFU-Es) and erythroid blast-forming units (BFU-Es) that underwent poor hemoglobinization in vitro, suggesting impaired erythroid differentiation.4
Recent findings have cast doubt as to whether these defects are intrinsic to the erythroid lineage (for reviews, see Whyatt and Grosveld6 and Liu et al7 ). Analysis of chimeric mice generated by injections of Rb–/– embryonic stem (ES) cells into wild-type blastocysts showed significant contributions of Rb–/– cells to the erythroid compartment with apparently normal differentiation of Rb–/– erythroid cells in vivo.8,9 Significantly, recent studies have identified a novel requirement for pRb in regulating placental architecture and function.10 Using tetraploid aggregation analysis and embryo-specific loss of pRb, it was shown that providing a wild-type placenta to an Rb–/– embryo was sufficient to alleviate the anemia and embryonic lethality of Rb–/– embryos.10,11 These data, therefore, suggest that the erythroid defects described in Rb–/– embryos are non–cell intrinsic.
Nonetheless, certain lines of evidence suggest that there is indeed a cell-intrinsic requirement for pRb in erythropoiesis. First, although the anemia appeared to be alleviated in the above experiments, increased numbers of nucleated erythroid cells were found in the peripheral blood of placenta-rescued Rb–/– animals, indicating some impairment in erythropoiesis.10,11 In another study, hematopoietic reconstitution of wild-type, lethally irradiated mice with Rb–/– FL cells resulted in the persistence of nucleated erythroid cells in peripheral blood and in extramedullary erythropoiesis.12 These combined data are consistent with a cell-intrinsic role for pRb in regulating the differentiation of the erythroid lineage. In view of these conflicting reports, we have used an erythroid primary culture assay together with coculture experiments to examine directly whether pRb has a cell-intrinsic role in erythropoiesis.
Study design
Erythroid cell cultures
Rb+/+ and Rb–/– embryos were obtained from Rb+/– matings 12.5 days after coitus. Erythroid cultures were grown essentially as previously described.13 After 3 to 4 days in expansion media, cultures were depleted of Ter119+ cells by magnetic-activated cell separation, and cells were resuspended at 1 × 106 cells/mL in differentiation media.13 For coculture experiments, Ter119-depleted cultures were carboxyfluorescein diacetate succinimidyl ester (CFSE) labeled as described14 before differentiation. Fluorescence-activated cell sorter (FACS) analysis was performed as previously described.15 Microscopy was performed using a Zeiss Axioskop 2 microscope (Carl Zeiss Microimaging, Thornwood, NY) with a Zeiss Plan-Neoflur × 40 objective (numerical aperture 0.75). Digital images were obtained using a Diagnostic Instruments Spot RT Slider camera (Sterling Heights, MI) and analyzed with Spot Software Windows version 4.0.2 imaging software.
Cell cycle analysis
Live cells were fixed in ice-cold 95% ethanol and stained for propidium iodide (PI) as described16 or were directly stained with 10 μM DRAQ5 (Alexis Biochemicals, Lausen, Switzerland) in phosphate-buffered saline (PBS) and incubated at room temperature for 10 minutes. DNA analysis was performed as previously described.16 For in vitro analysis of 5-bromodeoxyuridine (BrdU), 5 μL of 10 mg/mL BrdU was added to 500 μL culture for 1 hour. BrdU was detected by following the manufacturer's specifications (BrdU Flow Kit; BD PharMingen, San Diego, CA) and analyzed using FACSCalibur (BD Biosciences, San Jose, CA).
Results and discussion
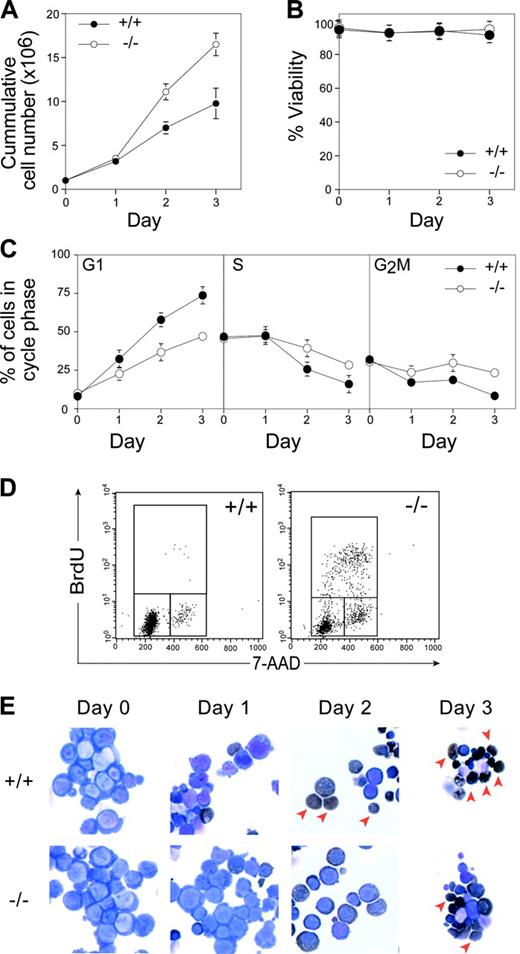
To examine the cellular requirement for pRb in the erythroid lineage, we took advantage of a recently developed primary culture system that allows expansion and differentiation of erythroid progenitors in vitro.13 We cultured E12.5 wild-type and Rb–/– FLs to allow the expansion of erythroid progenitors in the presence of erythropoietin, dexamethasone, and stem cell factor. No differences were observed in proliferation, viability, cell size, or cell cycle distribution between Rb–/– and wild-type cells in exponentially expanding cultures (data not shown). After 3 to 4 days of expansion, purified wild-type and Rb–/– erythroid progenitors were made to differentiate in the presence of high levels of erythropoietin and insulin. In these conditions, wild-type erythroid progenitors terminally differentiate synchronously over 3 days, resulting in a burst of proliferation followed by cell cycle exit, hemoglobinization, and enucleation to produce mature erythrocytes (Figure 1). In contrast to wild-type cells, Rb–/– cells showed increased numbers of erythroid cells produced over the time course of differentiation (Figure 1A). The increased production appeared to be caused solely by increased proliferation because Rb–/– and wild-type cultures showed equivalent viability over this period (Figure 1B). Indeed, Rb–/– cells showed impaired cell cycle exit compared with wild-type cultures, with significantly increased proportions of Rb–/– cells in S phase and continued BrdU incorporation on day 2 (Figure 1C-D).
In vitro differentiation of Rb–/– erythroid progenitors. (A) Cumulative proliferation was assessed by measuring the number of live cells in each culture using trypan blue exclusion assay. (B) Level of viability determined by trypan blue exclusion. (C) Cell cycle profiles were measured by PI analysis, and the percentage of cells in each cell cycle stage was determined by Modfit LT Software (Verity Software House, Topsham, ME). (D) FACS determination of BrdU incorporation after 1-hour in vitro BrdU pulse at 2 days after differentiation induction. (E) Morphologic analysis of cytospins after erythroid differentiation by May-Grünwald-Giemsa and benzidine (dark brown) staining. Arrowheads denote enucleated cells. Error bars indicate SD. These data are representative of 4 independent experiments.
In vitro differentiation of Rb–/– erythroid progenitors. (A) Cumulative proliferation was assessed by measuring the number of live cells in each culture using trypan blue exclusion assay. (B) Level of viability determined by trypan blue exclusion. (C) Cell cycle profiles were measured by PI analysis, and the percentage of cells in each cell cycle stage was determined by Modfit LT Software (Verity Software House, Topsham, ME). (D) FACS determination of BrdU incorporation after 1-hour in vitro BrdU pulse at 2 days after differentiation induction. (E) Morphologic analysis of cytospins after erythroid differentiation by May-Grünwald-Giemsa and benzidine (dark brown) staining. Arrowheads denote enucleated cells. Error bars indicate SD. These data are representative of 4 independent experiments.
Concomitant with the cell cycle exit defects, Rb–/– erythroid cells showed defective differentiation. Although a significant proportion of wild-type erythroid cells enucleated by day 2 of culture, Rb–/– cells showed fewer benzidine-positive cells (a marker of hemoglobinization) and impaired enucleation, similar to that observed in vivo10,12 (Figure 1E arrows; Figure 2A). The enucleation defects appeared to reflect a delay rather than a block in differentiation because a proportion of Rb–/– erythroid cells can eventually enucleate (Figure 1E). Consistent with this notion, there was a delay in the appearance of erythroid differentiation markers such as Ter119, ALAS2, Band 3, and globin genes in differentiating Rb–/– cultures (data not shown). Together, these data unequivocally demonstrate that pRb is required for erythroid differentiation and, by virtue of the in vitro system used, that this requirement is intrinsic to the erythroid lineage.
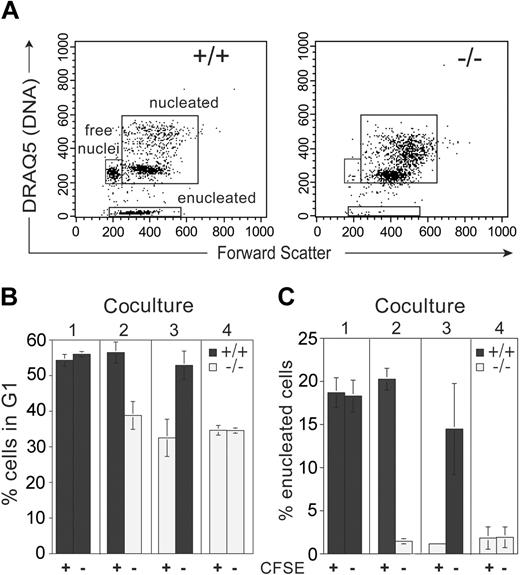
Analysis of wild-type and Rb–/– erythroid cocultures.Rb–/– or wild-type erythroid progenitors were CFSE labeled and mixed with combinations of Rb–/– or wild-type unlabeled cells (combinations 1 to 4). (A) FACS scheme used to discriminate nucleated erythroid cells, enucleated erythrocytes, and extruded “free” nuclei based on the DNA dye DRAQ5 and forward scatter on day 2 of differentiation. Note the near absence of enucleated and free nuclei in differentiated Rb–/– erythroid cells. CFSE-labeled (+) and unlabeled (–) cells were separated based on fluorescence intensity. (B) Cell cycle profiles for each population measured by DRAQ5 analysis of nucleated erythroid cells. (C) Enucleated cell percentage based on gate indicated in panel A. Error bars indicate SD. These data are representative of 2 independent experiments.
Analysis of wild-type and Rb–/– erythroid cocultures.Rb–/– or wild-type erythroid progenitors were CFSE labeled and mixed with combinations of Rb–/– or wild-type unlabeled cells (combinations 1 to 4). (A) FACS scheme used to discriminate nucleated erythroid cells, enucleated erythrocytes, and extruded “free” nuclei based on the DNA dye DRAQ5 and forward scatter on day 2 of differentiation. Note the near absence of enucleated and free nuclei in differentiated Rb–/– erythroid cells. CFSE-labeled (+) and unlabeled (–) cells were separated based on fluorescence intensity. (B) Cell cycle profiles for each population measured by DRAQ5 analysis of nucleated erythroid cells. (C) Enucleated cell percentage based on gate indicated in panel A. Error bars indicate SD. These data are representative of 2 independent experiments.
To explain the apparent contradictions in the literature regarding the role of pRb in erythropoiesis, a model has been proposed by which pRb would have a cell-intrinsic, but a non–cell-autonomous, role in erythroid differentiation (ie, the defects are intrinsic to the erythroid compartment but result from a defective homotypic cell-signaling function).6 To address this model and to formally discount the possibility that the presence or absence of a population present at a small frequency in the cultures contributes to the Rb–/– phenotype (eg, defective heterotypic cell-signaling function), we performed coculture experiments. Rb–/– or wild-type erythroid progenitors were pulse-labeled with the vital dye CFSE and were mixed with unlabeled wild-type or Rb–/– cells. Cell cycle exit and enucleation were monitored using the DNA dye DRAQ5 on day 2 of erythroid differentiation (Figure 2A), with each genotype discriminated by CFSE fluorescence. CFSE labeling did not significantly alter the behavior of wild-type or Rb–/– cells compared with unlabeled controls (Figure 2B, combinations 1 and 4). Importantly, the presence of wild-type cells did not alter the cell cycle or enucleation phenotype of Rb–/– cells (compare Rb–/– cells in combination 4 with those in combinations 2 and 3). These experiments demonstrate that the requirement for pRb is cell intrinsic to the erythroid lineage.
In this report, we provide definitive evidence that the erythroid differentiation defects that result from loss of pRb function are cell intrinsic. Interestingly, though we observed both impaired cell cycle exit and delayed terminal differentiation of erythroid progenitors, we did not observe any decreased survival, as reported in vivo,4 suggesting that altered survival may be non–cell intrinsic and most likely caused by the hypoxic environment of Rb–/– FL in vivo. The in vitro system described here provides an ideal setting in which to identify and functionally characterize the downstream components regulated by pRb that control terminal erythroid differentiation.
Prepublished online as Blood First Edition Paper, May 20, 2004; DOI 10.1182/blood-2004-02-0618.
Supported by a project grant from the National Health and Medical Research Council of Australia and by a Melbourne University Research Scholarship (K.M.D.). P.O.H. is a Special Fellow of the Leukemia and Lymphoma Society of America.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Hartmut Beug for helpful advice in setting up the erythroid culture system; Olivia Cakebread for technical assistance; Jodie Hayes and Mary Thorpe for animal husbandry; Andrew Fryga and Ralph Rossi for assistance with FACS; and Jian-Guo Zhang and Helene Martin for kind gifts of reagents. We also thank David Thomas and Luke Dow for comments and critical reading of the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal