Abstract
All-trans-retinoic acid (ATRA) induces growth inhibition, differentiation, and apoptosis in cancer cells, including acute promyelocytic leukemia (APL). In APL, expression of promyelocytic leukemia protein retinoic acid receptor–α (PML-RARα) fusion protein, owing to the t(15; 17) reciprocal translocation, leads to a block in the promyelocytic stage of differentiation. Here, we studied molecular mechanisms involved in ATRA-induced growth inhibition and myeloid cell differentiation in APL. By employing comprehensive high-throughput proteomic methods of 2-dimensional (2-D) gel electrophoresis and amino acid–coded mass tagging coupled with electrospray ionization (ESI) mass spectrometry, we systematically identified a total of 59 differentially expressed proteins that were consistently modulated in response to ATRA treatment. The data revealed significant down-regulation of eukaryotic initiation and elongation factors, initiation factor 2 (IF2), eukaryotic initiation factor 4AI (eIF4AI), eIF4G, eIF5, eIF6, eukaryotic elongation factor 1A-1 (eEF1A-1), EF-1-δ, eEF1γ, 14-3-3ϵ, and 14-3-3ζ/δ (P < .05). The translational inhibitor DAP5/p97/NAT1 (death-associated protein 5) and PML isoform-1 were found to be up-regulated (P < .05). Additionally, the down-regulation of heterogeneous nuclear ribonucleoproteins (hnRNPs) C1/C2, UP2, K, and F; small nuclear RNPs (snRNPs) D3 and E; nucleoprotein tumor potentiating region (TPR); and protein phosphatase 2A (PP2A) were found (P < .05); these were found to function in pre-mRNA processing, splicing, and export events. Importantly, these proteomic findings were validated by Western blot analysis. Our data in comparison with previous cDNA microarray studies and our reverse transcription–polymerase chain reaction (RT-PCR) experiments demonstrate that broad networks of posttranscriptional suppressive pathways are activated during ATRA-induced growth inhibition processes in APL.
Introduction
Acute promyelocytic leukemia (APL) is a specific type of acute myeloid leukemia (AML) and is associated with several chromosomal translations, with 98% of cases involving chromosomes 15 (PML gene) and 17 (RARα gene).1,2 The consequence of this reciprocal and balanced chromosomal translocation t(15;17)(q22, q21) is the expression of promyelocytic leukemia protein–retinoic acid receptor–α (PML-RARα) oncogenic fusion protein, which leads to disruption of PML nuclear bodies (NBs).3 Expression of PML-RARα results in differentiation arrest at the promyelocytic stage of the myeloid cell, leading to accumulation of immature hematopoietic cells in bone marrow.4
All-trans-retinoic acid (ATRA), a member of the retinoid family of molecules structurally related to vitamin A (retinol), exert profound effects on control of cell proliferation, the induction of differentiation, and apoptosis in normal cells and various cancer cells.5 The biologic effects of retinoids are modulated through nuclear receptors, including nuclear retinoic acid receptors (RARs) and retinoid X receptor (RXR), each consisting of 3 isotypes (α, β, and γ) encoded by separate genes and their isoforms (eg, α1, α2, β1-β4, γ1, or γ2), which belong to the superfamily of ligand-inducible transcription factors that includes steroid, vitamin D, thyroid hormone, peroxisome proliferator–activated receptors.6 Treatment of APL with ATRA leads to complete remission (CR) in about 90% of patients, which is attributable to proteolysis of PML-RARα, re-formation of PML NBs, and terminal differentiation of APL cells into normal granulocytes.7,8 Restored PML protein and RARα result in overcoming the differentiation block and commitment of APL cells into granulocytes, while cells deficient in PML protein are resistant to apoptosis by gamma irradiation, grow faster, and have longer survival times.4,9
PML-RARα can heterodimerize with RXR or form homodimers and subsequently bind to retinoic acid response element (RARE), located in the promoters of ATRA-responsive target genes. ATRA can bind to PML-RARα with an affinity comparable to RARα.6 In the absence of ligand, RAR-RXR in normal blasts and PML-RARα-RXR heterodimers in APL cells recruit the nuclear corepressor proteins nuclear receptor corepressor (N-CoR) or silencing mediator of retinoid and thyroid receptors (SMRT) and the transcriptional regulators Sin3A or Sin3B, which in turn form complexes with the enzymes histone deacetylase CA1 (HDAC1) or HDAC2, resulting in transcriptional repression or silencing.10-12 The transcriptional suppression occurs because deacylation of histone protein creates conformational changes, limiting access and binding of transcription factors and RNA polymerase to related genes.13 At physiologic concentrations of ATRA (10–9 to 10–8 M), the nuclear corepressor proteins and HDAC complexes are dissociated from RARα in normal blasts, which in turn results in recruitment of coactivators with histone acetyltransferase (HAT) activity, such as steroid receptor coactivator-1 (SRC-1), p300/PCAF, p300/CBP, ACTR, transcription intermediary factor 2 (TIF2), or P/CIP.12,14-16 Acetylation of lysine residues at the N-terminus of histones by HAT activity results in transactivation of responsive genes, leading to differentiation. However, the physiologic concentration of ATRA does not cause dissociation of nuclear corepressor proteins and histone deacetylase complexes from PML-RARα fusion receptors in APL blasts, leading to differentiation block. The corepressor complex is dissociated from PML-RARα only at pharmacologic concentrations (10–6 to 10–7 M) of ATRA, resulting in removal of transcriptional repression and transcription of genes related to differentiation.12,17
In addition to the release of transcriptional repression, other possible mechanisms involved in ATRA's effectiveness in myeloid cell differentiation include expression of different gene classes, including induction of p21WAF1/Cip1 cyclin-dependent kinase inhibitor18 ; C/EBP-α,β, and ϵ19,20 ; interferon regulatory factor 1 (IRF-1)21 ; and regulation of the localization of PML oncogenic domains (PODs).22 Moreover, the global analysis of multiple gene expression patterns using cDNA microarray technologies has provided additional information at the transcriptional level in response to ATRA-induced cell differentiation.23-25 These studies have established gene networks in NB4 that define phenotypic changes during decreases in cellular proliferation and promotion of granulocytic maturation by the up-regulation of p19INK4D, GADD153, BTG1, and Src-like adaptor.23
Despite the large amount of gene expression data gathered in t(15;17)–expressing APL cells, the correlation between the abundance of a specific mRNA and the abundance of the encoded protein is relatively poor and cannot elucidate the work of posttranscriptional networks involving ATRA-induced myeloid differentiation.26 The application of high-throughput proteomics can systemically identify and characterize the global protein expression profile or “proteome” and the corresponding cellular function.27 Proteomics can quantitatively analyze the differential expression patterns on a genomic scale and is particularly useful in attempts to analyze posttranscriptional networks produced in cancer cells.28
Currently, precise molecular mechanisms involved in ATRA-induced posttranscriptional regulation during myeloid cell differentiation are largely unknown. We hypothesized that identification of proteins that are modulated by ATRA through proteomic analysis leads to novel insights into ATRA-induced pathways during terminal differentiation and therapy of APL, including other retinoid sensitive cancers.
In this study, we applied the comprehensive high-throughput proteomic techniques of 2-dimensional gel electrophoresis (2-DE) and in vitro amino acid–coded mass tagging (AACT)29-32 to elucidate molecular mechanisms during ATRA-induced growth inhibition and terminal differentiation of t(15;17)–expressing APL cells. The results of this systematic study have established 3 unknown posttranscriptionally suppressed networks of proteins involving initiation factors, elongation factors, and ribonucleoproteins via ATRA treatment. The down-regulation of these ribonucleoproteins provides novel insights involving nuclear processing and export of specific mRNAs.
Materials and methods
Materials
Formic acid, acetonitrile, ammonium bicarbonate, SYPRO Ruby, and trifluoroacetic acid (TFA) were purchased from Fisher Scientific (Hampton, NH). Sequencing grade-modified trypsin was purchased from Promega (Madison, WI).
Cell culture and treatment of cells
The human acute promyelocytic leukemia (NB4) cell line was obtained from Dr Michael Andreeff (MD Anderson Cancer Center, Houston, TX). NB4 cells were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA) supplemented with 10% dialyzed fetal bovine serum (FBS). NB4 cells for 2-DE analysis were grown in RPMI 1640 media containing 50% isotopic abundance of L-leucine-5,5,5-d3 (Leu-d3). For the AACT-assisted quantitative analysis, the nontreated NB4 cells were grown in normal RPMI media, and the 1.0 μM ATRA-treated NB4 cells were grown in RPMI media containing a 99% isotopic abundance of L-leucine-5,5,5-d3 with dialyzed FBS (CDN Isotopes, Pointe-Claire, QC, Canada). For all culturing conditions, cells were seeded at 1 to 3 × 106/mL and were harvested following 48, 72, and 96 hours of ATRA treatment. (Sigma Chemical, St Louis, MO). ATRA was added to cell pools after dilution with saline from a 10 mM stock in dimethyl sulfoxide (DMSO). The maximum concentration of DMSO was maintained below 0.001% of each medium. Myeloid leukemia (HL-60) cells were obtained from American Type Culture Collection (Manassas, VA). Growth and treatment conditions were the same as described with NB4.
RNA isolation and reverse transcription–polymerase chain reaction (RT-PCR)
Total cellular RNA was isolated with Trizol reagent (Life Technologies, Gaithersburg, MD); 5.0 μg total RNA was used to synthesize cDNA. The reverse transcription reaction was performed with the use of the Superscript II RT kit (Life Technologies). Briefly, 5.0 μL of the total 20.0 μL RT product was used for polymerase chain reaction containing 1 × PCR buffer, 1.5 mM MgCl2, 250 μM deoxynucleoside triphosphate (dNTP), 0.5 U Taq polymerase (Life Technologies), and 100 ng primers: eukaryotic elongation factor 1A-1 (eEF1A-1), 5′-GGATGGAAAGTCACCCGTAAGG-3′,5′-CGCAACTGTCTGTCTCATATCACG-3′; eEF1γ,5′-AAGGAGGAAGTGAGGCGAATTC-3′, 5′-CCTCATTGGAGTACTTGCGCTTAA-3′; eEF4G, 5′-TACGGCAGAGGAACGAGAACG-3′, 5′-TTGAAGGGCTGAGAAGCGAAT-3′; eukaryotic initiation factor 4A-1 (eIF4A-1), 5′-AAGCTGACGAAATGTTAAGCCGT-3′, 5′-TGTTAATAGCCACACCTTTACGGC-3′; heterogeneous nuclear ribonucleoprotein (hnRNP) F, 5′-GAAAGCATGGGACACCGGTAC-3′, 5′-CTGTGAACTCACTGTCGCCGTAT-3′; small nuclear RNP (snRNP) D3, 5′-CGAACACCGGTGAGGTATATCG-3′, 5′-TGAAAGATGTTTCCACGTCCCA-3′; hnRNP C1/C2, 5′-TGTGGAGGCAATCTTTTCGA-3′, 5′-TGATACACGCTGACGTTTCG-3′; hnRNP K, 5′-TGGAGGAAAACCCGATAGGG-3′, 5′-AAACCAACCATGCCGTCGTA-3′; hnRNP UP2, 5′-GCGAAGATTGACGCCAGTAAGA-3′, 5′-AAAGCAGAACCCACGCCTCT-3′; snRNP E, 5′-GGTGCAGCCCATCAACCTTA-3′, 5′-TGCTAGTTGGAAACACTTTGGAGC-3; and β-actin, 5′-GTCACCAACTGGGACGACATG-3′,5′-GACAGCACTGTGTTGGCGTACA-3′. We determined the optimal number of PCR cycles for each set of primers in preliminary experiments to ensure that the amplification process was acquired during the exponential phase of amplification.33 The conditions were as follows: 1 cycle of 94° C degree for 2 minutes, 30 to 35 cycles (depending on the experiment); denaturation, 94° C for 1 minute; annealing, 55° C for 1 minute; and extension, 72° C for 1 minute. Following 30 to 35 cycles, a cycle of 72° C for 7 minutes was added for completion of the reaction. The reaction products were analyzed on a 1% agarose gel containing ethidium bromide, and cDNA synthesis was verified by detection of β-actin transcript, which was used as an internal control and kept in a linear range. Images were scanned and quantified by an Alpha Innotech densitometer using the Alpha Imager application program (Alpha Innotech, San Leandro, CA).
Western blot analysis
Whole-cell NB4 lysates were obtained from ATRA-treated exponentially growing cells. After the determination of total protein concentrations by means of the Bio-Rad DC protein assay kit (Bio-Rad, Hercules, CA), 50 μg total protein for each sample was electrophoresed and electrotransferred as previously described.34 Membranes were blocked with 5% dry milk in TBST (100 mM Tris-HCl [tris(hydroxymethyl)aminomethane]–HCl, pH 8.0; 150 mM NaCl; and 0.05% Tween-20) and probed with rabbit anti-RARα, anti-eIF4G, anti-eIF5, anti–14-3-3ϵ, anti–14-3-3ζ, anti–Rho guanosine diphosphate dissociation inhibitor (anti–Rho GDI) (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-DAP5 (death-associated protein 5) (Cell Signaling Technology, Beverly, MA) primary antibodies after the preparation of 1:1000 dilutions in TBST containing 5% dry milk, and incubated at 4° C overnight. Anti–hnRNP K antibodies, which were generously provided by Dr Pradip Raychaudhuri (University of Illinois at Chicago, Chicago, IL), were used at 1:500 dilution. After washing, membranes were incubated with horseradish peroxidase–conjugated antirabbit secondary antibody (Amersham Life Science, Arlington Heights, IL). Mouse β-actin and donkey antimouse antibodies were purchased from Sigma Chemical and used as a control for equal loading. Protein bands were visualized by enhanced chemiluminescence (KPL, Baltimore, MD). Images were scanned with a densitometer and the use of the Alpha Imager application program (Alpha Innotech).
One-dimensional SDS gel separation
Following Bradford assays, 100 μg total protein lysates for both nontreated and ATRA-treated cells were mixed at 1:1 ratios, and 100 μg mixture was separated by means of 8% to 16% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) linear gradient precast ready gels (Bio-Rad). The gel was stained with SYPRO Ruby, and each protein band was cut and digested with sequencing grade trypsin according to the manufacturer's instructions.
Two-dimensional gel electrophoresis
The NB4 cells were denatured and solubilized in a buffer containing 7 M urea, 2 M thiourea, 2% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propane sulfonic acid), 1% 3 to 10 zwittergent, and 0.8% 3 to 10 2-DE ampholytes, followed by modified Bradford assay for measuring protein concentration.35 Soluble proteins were hydrated into 17 cm pH 4 to 7 IEF strips (Bio-Rad) for 12 hours with the use of 100 μg nontreated or ATRA-treated lysate. Proteins were focused the following day for approximately 65 000 V/h total. The IEF strips were equilibrated according to the manufacturer's instructions (Bio-Rad), and each gel was further separated with the use of 8% to 18% gradient SDS-PAGE gels at 22 × 22 cm dimensions (Genomics Solutions, Ann Arbor, MI) and stained with SYPRO Ruby and imaged using an Investigator Pro Pic UV Source (Genomics Solutions) for a 70-second exposure. Four 2-DE gels were obtained for both nontreated NB4 and ATRA-treated NB4, and the comparative 2-DE gel data were obtained by means of PDQuest v7.1 (Bio-Rad). Student t test analysis was applied to the 2-DE data by means of the PDQuest software, and differences in protein expression levels between groups (nontreated versus ATRA treated) were regarded as significant for values of P less than .05. This protocol was applied to all time points. Proteins found to be up- or down-regulated were excised and digested with sequencing grade trypsin according to the manufacturer's instructions.
Microcapillary liquid chromatography (MLC) and tandem mass spectrometry (MS/MS)
In-gel tryptic digests of protein bands from 1-DE and 2-DE were run on a QSTAR Pulsar I mass spectrometer (Applied Biosystems, Framingham, MA; MDS Sciex, San Francisco, CA) coupled with LC Packings Ultimate microcapillary LC system (Dionex, Sunnyvale, CA). The PepMap C18 column (3 μm, 100 Å [10 nm], 75 μm interior diameter, 15 cm length) used for peptide separation was purchased from Dionex Packings. Ions with charge states between 2 to 4 that passed the switching criteria were selected for MS/MS collision-induced dissociation fragmentation with the rolling collision energy feature employed for fragmentation. The spray voltage was set between 1800 and 2100 V.
Results
Two-dimensional gel analysis of differentially expressed proteins via ATRA treatment in APL cells
To determine novel molecular mechanisms involved in ATRA-induced growth arrest and differentiation, we identified ATRA-modulated proteins involved in the differentiation process by examining changes in the proteome of t(15,17)–expressing APL cells.
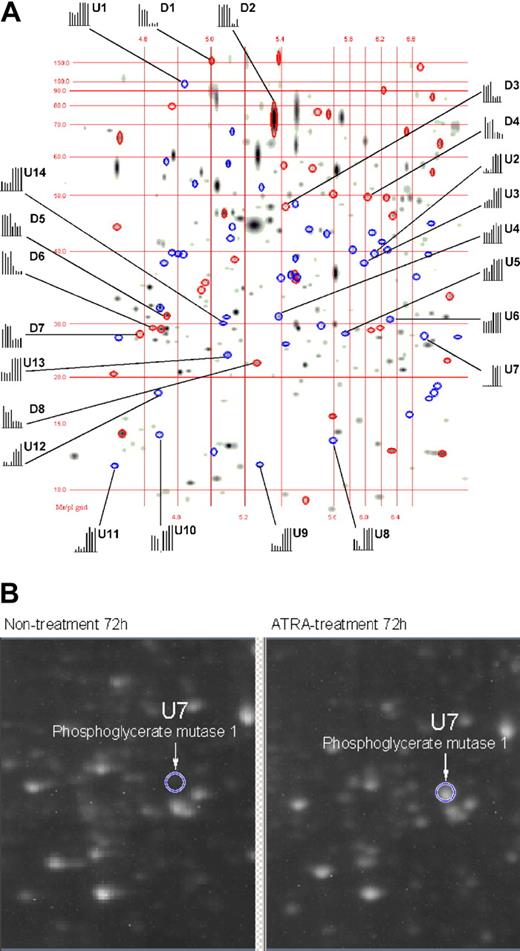
The differential expression of proteins in APL cells as a function of ATRA treatment was analyzed at 8, 48, 72, and 96 hours of ATRA treatment by means of 2-DE in a pH range of 4.0 to 7.0. With the use of PDQuest v7.1 software, 2-DE gel images for nontreated versus ATRA-treated APL cells were quantitatively analyzed as shown in Figure 1A. The 2-DE displays of the various ATRA treatment time points used in this study yielded a total of 557, 616, 630, and 658 spots that were comparatively analyzed at 8, 48, 72, and 96 hours, respectively. The PDQuest analysis of the 2-DE data found 0, 26, 38, and 37 down-regulated protein spots and 2, 31, 52, and 56 up-regulated protein spots that exhibited a 1.6-fold change or greater at 8, 48, 72, and 96 hours, respectively. The up-regulated proteins in response to ATRA treatment are displayed as blue ellipses, with down-regulated proteins displayed as red ellipses. All together, we identified 22 differentially expressed proteins from which hnRNP F, eIF4A-1, eIF4G, and eIF6 were down-regulated and which are relevant to the mechanisms of ATRA-induced repression during mRNA processing and protein synthesis events. The identity markers (D indicates down-regulation; U, upregulation) in Table 1 correspond to the labels in Figure 1A, whereas the histograms represent the comparative spot intensities present in each 2-DE gel, where the 4 left bars represent different nontreated 2-DE gels and the 4 right bars which represent different ATRA-treated 2-DE gels. These quantitative results are demonstrated in Figure 1B for spot U7, identified as phosphoglycerate mutase 1, which was dramatically up-regulated.
Matching image analysis for 2-DE. This image represents 4 nontreated 2-DE gels compared with 4 ATRA-treated 2-DE gels. The pH range was 4 to 7, with molecular weight, isoelectric point (pI) grinds, and histograms representing the quantification of identified proteins within the gels. The first 4 bars in each histogram represent the relative intensity of a protein spot from 4 individual 2-DE gels during nontreatment, and the last 4 bars represent the relative intensity of the same protein spot from 4 individual 2-DE gels during ATRA treatment. Proteins down-regulated by means of ATRA treatment are displayed as red ellipses, and up-regulated proteins are displayed as blue ellipses. Proteins labeled with “U” or “D” are listed in Table 1 along with their degree of regulation as a function of ATRA treatment. (B) A 2-DE raw image (original magnification, × 1) for nontreated and ATRA-treated 2-DE gels corresponding to the gel region where phosphoglycerate mutase 1 resolves (circle).
Matching image analysis for 2-DE. This image represents 4 nontreated 2-DE gels compared with 4 ATRA-treated 2-DE gels. The pH range was 4 to 7, with molecular weight, isoelectric point (pI) grinds, and histograms representing the quantification of identified proteins within the gels. The first 4 bars in each histogram represent the relative intensity of a protein spot from 4 individual 2-DE gels during nontreatment, and the last 4 bars represent the relative intensity of the same protein spot from 4 individual 2-DE gels during ATRA treatment. Proteins down-regulated by means of ATRA treatment are displayed as red ellipses, and up-regulated proteins are displayed as blue ellipses. Proteins labeled with “U” or “D” are listed in Table 1 along with their degree of regulation as a function of ATRA treatment. (B) A 2-DE raw image (original magnification, × 1) for nontreated and ATRA-treated 2-DE gels corresponding to the gel region where phosphoglycerate mutase 1 resolves (circle).
Proteins that are differentially expressed at 8, 48, 72, and 96 hours of ATRA treatment through 2-DE
. | Accession no. . | . | . | . | . | Peptides sequenced . | Protein expression† . | . | . | . | mRNA expression‡ . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spot . | . | Protein name . | Functional classification . | Mr, kDa* . | pl* . | . | 8 h . | 48 h . | 72 h . | 96 h . | . | |||
| D1 | Q04637 | eIF4G | Protein synthesis | 153 | 5.18 | 4 | 0.71 | 0.50 | 0.21 | 0.17 | Unchanged§ | |||
| D2 | AAH19816 | 70-kDa hsp 8 | Chaperone | 70.9 | 5.37 | 15 | 0.90 | 0.25 | 0.34∥ | 0.39 | Unknown | |||
| D3 | P04765 | eIF4A-1 | Protein synthesis | 46.1 | 5.32 | 6 | 0.95 | 0.46 | 0.34 | 0.40 | Unchanged§ | |||
| D4 | Q9Y230 | 48-kDa TATA | Transcription | 54.1 | 5.49 | 2 | 0.70 | 0.49 | 0.43 | 0.45 | Unknown | |||
| D5 | P06753-01 | Tropomyosin α3 | Cellular organization | 29.0 | 4.75 | 3 | 1.1∥ | 0.61 | 0.50 | 0.50∥ | Unknown | |||
| D6 | P42655 | 14-3-3ϵ | Signal transduction | 29.1 | 4.63 | 3 | 0.83 | 0.56 | 0.35 | 0.41 | Unknown | |||
| D7 | P56537 | eIF 6 | Protein synthesis | 26.5 | 4.56 | 2 | 0.90 | 0.60 | 0.34 | 0.38 | Unknown | |||
| D8 | P52597 | hnRNP F | mRNA splicing | 45.6 | 5.38 | 2 | 0.76 | 0.49 | 0.40 | 0.38 | Unchanged§ | |||
| U1 | Q14974 | Importin β-1 | Cellular transport | 97.2 | 4.68 | 5 | 1.0 | 1.8∥ | 2.0 | 2.4 | Repressed24 | |||
| U2 | P50213 | NAD | Metabolism | 39.5 | 6.47 | 2 | ND | 2.0 | 3.8 | 4.3 | Unknown | |||
| U3 | P47755 | CapZ α-2 | Structural | 33.0 | 5.57 | 2 | 1.0 | 2.1 | 2.7 | 3.7 | Unknown | |||
| U4 | Q9UL46 | PA28 β | Cellular defense | 27.3 | 5.44 | 4 | 1.2 | 2.1 | 1.8 | 2.3 | Induced24 | |||
| U5 | P47756 | CapZ β | Structural | 30.6 | 5.69 | 4 | 0.90∥ | 1.6 | 2.1 | 3.8 | Unknown | |||
| U6 | P78417 | GSTO 1-1 | Not defined | 27.5 | 6.23 | 7 | 0.80 | 1.7 | 1.9 | 1.7 | Unknown | |||
| U7 | P18669 | PGAM-B | Glycolysis | 28.6 | 6.75 | 7 | 1.6 | 6.5∥ | 22 | 20 | Induced25 | |||
| U8 | Q14019 | Coactosin-like | Cellular organization | 15.9 | 5.54 | 2 | 1.1∥ | 2.2 | 3.3 | 2.5 | Unknown | |||
| U9 | P29373 | CRABP-II | Retinoid metabolism | 15.6 | 5.43 | 2 | 1.5 | 2.7 | 3.7 | 4.2 | Unknown | |||
| U10 | P27482 | CLP | Signal transduction | 16.7 | 4.30 | 2 | 1.1∥ | 1.5 | 1.9∥ | 1.7 | Induced23 | |||
| U11 | P05387 | 60S P2 protein | Protein synthesis | 11.7 | 4.42 | 4 | ND | 2.5 | 3.9 | 4.7 | Unknown | |||
| U12 | P19105 | Myosin RLC | Cellular organization | 19.7 | 4.67 | 2 | 1.8 | 3.0 | 4.1 | 4.4 | Unknown | |||
| U13 | P52566 | Rho GDI 2 | Signal transduction | 22.9 | 5.10 | 4 | 1.3 | 2.0 | 2.6 | 2.6 | Unknown | |||
| U14 | P29218 | IMPase | Signal transduction | 30.2 | 5.16 | 2 | ND | 2.1∥ | 2.9 | 2.5 | Unknown | |||
. | Accession no. . | . | . | . | . | Peptides sequenced . | Protein expression† . | . | . | . | mRNA expression‡ . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spot . | . | Protein name . | Functional classification . | Mr, kDa* . | pl* . | . | 8 h . | 48 h . | 72 h . | 96 h . | . | |||
| D1 | Q04637 | eIF4G | Protein synthesis | 153 | 5.18 | 4 | 0.71 | 0.50 | 0.21 | 0.17 | Unchanged§ | |||
| D2 | AAH19816 | 70-kDa hsp 8 | Chaperone | 70.9 | 5.37 | 15 | 0.90 | 0.25 | 0.34∥ | 0.39 | Unknown | |||
| D3 | P04765 | eIF4A-1 | Protein synthesis | 46.1 | 5.32 | 6 | 0.95 | 0.46 | 0.34 | 0.40 | Unchanged§ | |||
| D4 | Q9Y230 | 48-kDa TATA | Transcription | 54.1 | 5.49 | 2 | 0.70 | 0.49 | 0.43 | 0.45 | Unknown | |||
| D5 | P06753-01 | Tropomyosin α3 | Cellular organization | 29.0 | 4.75 | 3 | 1.1∥ | 0.61 | 0.50 | 0.50∥ | Unknown | |||
| D6 | P42655 | 14-3-3ϵ | Signal transduction | 29.1 | 4.63 | 3 | 0.83 | 0.56 | 0.35 | 0.41 | Unknown | |||
| D7 | P56537 | eIF 6 | Protein synthesis | 26.5 | 4.56 | 2 | 0.90 | 0.60 | 0.34 | 0.38 | Unknown | |||
| D8 | P52597 | hnRNP F | mRNA splicing | 45.6 | 5.38 | 2 | 0.76 | 0.49 | 0.40 | 0.38 | Unchanged§ | |||
| U1 | Q14974 | Importin β-1 | Cellular transport | 97.2 | 4.68 | 5 | 1.0 | 1.8∥ | 2.0 | 2.4 | Repressed24 | |||
| U2 | P50213 | NAD | Metabolism | 39.5 | 6.47 | 2 | ND | 2.0 | 3.8 | 4.3 | Unknown | |||
| U3 | P47755 | CapZ α-2 | Structural | 33.0 | 5.57 | 2 | 1.0 | 2.1 | 2.7 | 3.7 | Unknown | |||
| U4 | Q9UL46 | PA28 β | Cellular defense | 27.3 | 5.44 | 4 | 1.2 | 2.1 | 1.8 | 2.3 | Induced24 | |||
| U5 | P47756 | CapZ β | Structural | 30.6 | 5.69 | 4 | 0.90∥ | 1.6 | 2.1 | 3.8 | Unknown | |||
| U6 | P78417 | GSTO 1-1 | Not defined | 27.5 | 6.23 | 7 | 0.80 | 1.7 | 1.9 | 1.7 | Unknown | |||
| U7 | P18669 | PGAM-B | Glycolysis | 28.6 | 6.75 | 7 | 1.6 | 6.5∥ | 22 | 20 | Induced25 | |||
| U8 | Q14019 | Coactosin-like | Cellular organization | 15.9 | 5.54 | 2 | 1.1∥ | 2.2 | 3.3 | 2.5 | Unknown | |||
| U9 | P29373 | CRABP-II | Retinoid metabolism | 15.6 | 5.43 | 2 | 1.5 | 2.7 | 3.7 | 4.2 | Unknown | |||
| U10 | P27482 | CLP | Signal transduction | 16.7 | 4.30 | 2 | 1.1∥ | 1.5 | 1.9∥ | 1.7 | Induced23 | |||
| U11 | P05387 | 60S P2 protein | Protein synthesis | 11.7 | 4.42 | 4 | ND | 2.5 | 3.9 | 4.7 | Unknown | |||
| U12 | P19105 | Myosin RLC | Cellular organization | 19.7 | 4.67 | 2 | 1.8 | 3.0 | 4.1 | 4.4 | Unknown | |||
| U13 | P52566 | Rho GDI 2 | Signal transduction | 22.9 | 5.10 | 4 | 1.3 | 2.0 | 2.6 | 2.6 | Unknown | |||
| U14 | P29218 | IMPase | Signal transduction | 30.2 | 5.16 | 2 | ND | 2.1∥ | 2.9 | 2.5 | Unknown | |||
Mr indicates molecular weight; D, down-regulation; U, up-regulation; eIF, eukaryotic translation initiation factor; hsp, heat shock protein; NAD, isocitrate dehydrogenase; ND, no data were found; CapZ, F-actin capping protein; PA28 β, proteasome activator complex subunit 2; GSTO 1-1, glutathione transferase omega 1; PGAM-B, phosphoglycerate mutase 1; CRABP-II, retinoic acid-binding protein II; CLP, calmodulin-related protein NB-1; Rho GDI 2, Rho guanosine diphosphate (GDP)-dissociation inhibitor 2; IMPase, inositol-1(or 4)-monophosphatase.
Theoretical.
ATRA-treated divided by nontreated. All values are statistically significant; P < .05 unless otherwise indicated.
The mRNA expression is as displayed in references Liu et al23 (for 0 to 72 hours), Yang et al24 (for 0 to 96 hours), or Lee et al25 (for 0 to 48 hours) or by personal communication. Unchanged mRNA expression ratios (for ATRA treated divided by nontreated) were within 0.8 to 1.2 as a function of various ATRA treatment time points.
P > .05.
See Figure 4.
Identification and quantification of differentially expressed proteins in APL via ATRA treatment using amino acid–coded mass tagging (AACT)
Although 2-DE is a powerful tool for the separation and quantification of proteins, it has limitations in resolving proteins with high isoelectric points (pIs), hydrophobic characteristics, and high molecular weights.36 In light of this, AACT was used as an additional proteomics method to overcome such limitations.
The application of AACT to the study of ATRA treatment of APL begins by culturing nontreated NB4 cells in normal culture media while ATRA-treated NB4 cells are cultured in the same media containing a deuterated form of an essential amino acid, such as Leu-d3. As a result, the proteins synthesized in Leu-d3–enriched media (99% isotopic abundance of Leu-d3) contain a deuterated leucine amino acid with a mass of 116.08 Da, as opposed to proteins expressed in normal media that have masses of 113.06 Da. After the parallel culturing of the cells in these 2 media pools, the ATRA-treated and nontreated cells are mixed at a 1:1 ratio, lysed, and further separated by SDS-PAGE. Each protein band (approximately 25 excised bands) is subject to tryptic digestion followed by MLC connected directly to the electrospray ionization source on a mass spectrometer. Since the elution times of Leu-d3–labeled peptides were essentially identical to those of the same nonlabeled peptides in a C18 reverse-phase column, both isotopic forms of the peptides elute together at corresponding retention times, but differ in mass-charge (m/z) ratios (Figure 2A). The relative intensities of the 2 isotopic peaks for nonlabeled and Leu-d3–labeled peptides correspond to the differential expression of the protein synthesized in normal NB4 cells and ATRA-treated NB4 cells, respectively. The identity of each protein is found by way of MS/MS (peptide fragmentation) experiments on each peptide, which are concurrently performed to yield peptide sequence information for protein identification search by means of Mascot (Matrix Science, Boston, MA).
AACT for a 72-hour (nontreated versus ATRA-treated) sample accounting for the quantification and identification of eEF1A-1. (A) The averaged time-of-flight (TOF) MS from C18 high-performance liquid chromatography (HPLC) retention times of 33.5 to 34.2 minutes accounting for tryptic peptide EHALLAYTLGVK. The monoisotopic m/z value of 657.85 represents the +2 charged EHALLAYTLGVK peptide produced in nontreated NB4, and 662.38 represents the EHALLAYTLGVK peptide from ATRA-treated NB4 grown in 99% Leu-d3 (116.1 Da) media. Comparison of the relative intensities of the 2 monoisotopic peaks (657.85 versus 662.38) yields a ratio (ATRA treated to nontreated) of 0.32, implying down-regulation of 3.1-fold via ATRA treatment. (B) The automated MS/MS fragmentation pattern of 657.85. The peptide sequence produced from MS/MS fragmentation yielded the identification of eEF1A-1. A few fragmentation b- and y-ions are labeled for comparison with MS/MS data from 662.38. (C) The automated MS/MS fragmentation pattern of 662.38. A 3.02 Da difference exists for each fragmented peptide containing leucine as illustrated by y7, y8, and y9, which yield m/z values of 754.4, 870.5, and 986.6, respectively. Fragments that do not contain leucine are identical to the MS/MS data for 657.85, as demonstrated by b2 and b3.
AACT for a 72-hour (nontreated versus ATRA-treated) sample accounting for the quantification and identification of eEF1A-1. (A) The averaged time-of-flight (TOF) MS from C18 high-performance liquid chromatography (HPLC) retention times of 33.5 to 34.2 minutes accounting for tryptic peptide EHALLAYTLGVK. The monoisotopic m/z value of 657.85 represents the +2 charged EHALLAYTLGVK peptide produced in nontreated NB4, and 662.38 represents the EHALLAYTLGVK peptide from ATRA-treated NB4 grown in 99% Leu-d3 (116.1 Da) media. Comparison of the relative intensities of the 2 monoisotopic peaks (657.85 versus 662.38) yields a ratio (ATRA treated to nontreated) of 0.32, implying down-regulation of 3.1-fold via ATRA treatment. (B) The automated MS/MS fragmentation pattern of 657.85. The peptide sequence produced from MS/MS fragmentation yielded the identification of eEF1A-1. A few fragmentation b- and y-ions are labeled for comparison with MS/MS data from 662.38. (C) The automated MS/MS fragmentation pattern of 662.38. A 3.02 Da difference exists for each fragmented peptide containing leucine as illustrated by y7, y8, and y9, which yield m/z values of 754.4, 870.5, and 986.6, respectively. Fragments that do not contain leucine are identical to the MS/MS data for 657.85, as demonstrated by b2 and b3.
The Leu-d3–labeled NB4 ATRA-treated cells were analyzed and compared with the nontreated NB4 cells cultured in the normal medium at 24, 72, and 96 hours of ATRA treatment. Twenty-five protein bands excised from each 1-D SDS-PAGE gel were analyzed, which resulted in approximately 3000 MS/MS sequenced peptides at these time points. A total of 501, 512, and 572 proteins were identified from the MS/MS information at the 99% confidence level at 24, 72, and 96 hours, respectively. Quantitative analysis of these data produced 26 down-regulated proteins and 14 up-regulated proteins at the various time points. This study identified a significant number of ATRA-induced down-regulated proteins that function during the initiation and elongation stages of the protein synthesis mechanism. Initiation factors IF-2, eIF5, elongation factors eEF1A-1, eEF-1-δ, and eEF1γ were found to be down-regulated. In addition, proteins involved in mRNA processing and transport such as tumor potentiating region (TPR); hnRNPs C1/C2, K, and UP2; and small nuclear ribonucleoproteins (snRNPs) D3 and E were also down-regulated, while PML-isoform 1 was found up-regulated. The magnitude at which these proteins are differentially expressed is shown in Table 2.
Proteins that are significantly modulated after ATRA treatment at 24, 72, and 96 hours through AACT analysis
. | . | . | . | . | Peptides sequenced . | Protein expression† . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession no. . | Protein name . | Functional classification . | Mr, kDa* . | pl* . | . | 24 h . | 72 h . | 96 h . | mRNA expression‡ . | ||
| P04720 | eEF1A-1 | Protein synthesis | 50.1 | 9.10 | 5 | 0.67 | 0.32 | 0.36 | Unchanged∥24 | ||
| P29692 | EF-1-δ | Protein synthesis | 31.1 | 4.90 | 2 | 0.80 | 0.53 | ND | Unchanged24 | ||
| AAH09865 | eEF1γ | Protein synthesis | 50.1 | 6.25 | 4 | 0.35 | 0.41 | 0.38 | Unchanged∥ | ||
| P46199 | IF-2 | Protein synthesis | 81.3 | 6.71 | 2 | ND | 0.47 | 0.35 | Unchanged24 | ||
| P04765 | eIF4A-1 | Protein synthesis | 46.1 | 5.32 | 3 | 0.53 | 0.34§ | 0.40§ | Unchanged∥ | ||
| P55010 | eIF5 | Protein synthesis | 49.2 | 5.41 | 2 | ND | 0.48 | ND | Unchanged24 | ||
| P42655 | 14-3-3ϵ | Signal transduction | 29.1 | 4.63 | 4 | 0.66 | 0.44 | 0.41§ | Unknown | ||
| P29312 | 14-3-3ζ/δ | Signal transduction | 27.7 | 4.73 | 2 | 0.81 | 0.58 | 0.59 | Unknown | ||
| P01112 | c-H-ras | Signal transduction | 21.2 | 5.16 | 2 | ND | ND | 0.19 | Unknown | ||
| P00540 | c-mos | Signal transduction | 37.8 | 9.15 | 3 | ND | 0.43 | 0.38 | Unknown | ||
| P30153 | PP2A | Signal transduction | 65.0 | 4.96 | 2 | 0.70 | 0.35 | 0.41 | Unknown | ||
| Q13233 | MEKK 1 | Signal transduction | 164 | 8.52 | 3 | 0.92 | ND | 0.64 | Repressed24 | ||
| P23528 | Cofilin, P18 | Structural | 18.5 | 8.22 | 9 | 0.51 | 0.64 | 0.52 | Repressed24 | ||
| P32119 | Peroxiredoxin 2 | Cellular defense | 21.9 | 5.66 | 5 | ND | 0.47 | 0.50 | Unknown | ||
| C28894 | MPO | Cellular defense | 92.4 | 9.25 | 9 | 0.35 | 0.52 | 0.48 | Repressed23 | ||
| P25763 | CDC42 homolog | Not defined | 21.3 | 6.16 | 3 | 0.35 | 0.54 | ND | Repressed24 | ||
| P10809 | 60-kDa hsp | Chaperone | 61.0 | 5.70 | 7 | 0.77 | 0.67 | 0.48 | Repressed23 | ||
| Q9NR31 | SAR1a | Transport | 22.4 | 6.22 | 4 | ND | 0.18 | 0.20 | Unknown | ||
| P12270 | Nucleoprotein TPR | mRNA transport | 266 | 5.01 | 6 | 0.88 | 0.50 | 0.48 | Unknown | ||
| P07910 | hnRNP C1/C2 | mRNA splicing | 33.7 | 4.95 | 3 | 0.64 | 0.55 | ND | Unchanged∥ | ||
| Q07244 | hnRNP K | mRNA transport | 50.9 | 5.39 | 6 | 0.52 | 0.53 | 0.50 | Unchanged∥ | ||
| P07029 | hnRNP UP2 | mRNA splicing | 24.1 | 8.02 | 1 | 1.0 | 0.68 | ND | Unchanged∥ | ||
| P43331 | snRNP D3 | mRNA splicing | 13.9 | 10.3 | 2 | ND | 0.39 | 0.41 | Unchanged∥25 | ||
| P08578 | snRNP E | mRNA splicing | 10.8 | 9.46 | 2 | 0.71 | 0.62 | 0.59 | Unchanged∥ | ||
| Q13822 | E-NPP 2 | Glycolysis | 99.0 | 7.14 | 2 | ND | ND | 0.31 | Unknown | ||
| P22712 | MBP-1 | Transcription | 37.0 | 6.79 | 2 | ND | 0.15 | ND | Unknown | ||
| Q9BWX5 | GATA-5 | Transcription | 41.3 | 9.17 | 1 | 1.5 | ND | 2.6 | Unknown | ||
| Q14974 | Importin β-1 | Transport | 97.2 | 4.68 | 2 | 1.5 | ND | 2.1 | Repressed24 | ||
| P24903 | CYPIIF1 | Cellular defense | 55.5 | 6.93 | 2 | 1.6 | ND | 1.8 | Unknown | ||
| P52565 | Rho GDI 1 | Signal transduction | 23.2 | 5.03 | 2 | 2.3 | 1.7 | ND | Unknown | ||
| P31146 | Coronin 1A | Signal transduction | 51.0 | 6.25 | 3 | 2.2 | ND | ND | Unknown | ||
| NP_000994 | 60S P1 protein | Protein synthesis | 11.5 | 4.26 | 2 | 1.0 | 1.5 | 1.8 | Unknown | ||
| P15531 | NDP kinase A | Energy | 17.1 | 5.83 | 2 | 0.9 | 1.5 | ND | Unknown | ||
| P05109 | Calgranulin A | Cellular defense | 10.8 | 6.51 | 2 | ND | 2.0 | 2.1 | Induced23 | ||
| Q99733 | NAP2 | Not defined | 42.7 | 4.60 | 3 | 1.2 | ND | 1.9 | Unknown | ||
| P47755 | CapZ α-2 | Structural | 32.9 | 5.57 | 2 | 1.8 | 2.2 | 2.6 | Unknown | ||
| P12814 | α-actinin 1 | Structural | 103 | 5.22 | 3 | 2.7 | 2.3 | 2.8 | Unknown | ||
| Q08043 | α-actinin 3 | Structural | 103 | 5.40 | 3 | 3.4 | ND | ND | Unknown | ||
| P35579 | NMMHC-A | Structural | 226 | 5.50 | 3 | 2.0 | 4.1 | 4.3 | Unknown | ||
| P29590 | isoform PML-1 | Negative regulator | 97.5 | 5.88 | 2 | 3.5 | ND | ND | Unknown | ||
. | . | . | . | . | Peptides sequenced . | Protein expression† . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession no. . | Protein name . | Functional classification . | Mr, kDa* . | pl* . | . | 24 h . | 72 h . | 96 h . | mRNA expression‡ . | ||
| P04720 | eEF1A-1 | Protein synthesis | 50.1 | 9.10 | 5 | 0.67 | 0.32 | 0.36 | Unchanged∥24 | ||
| P29692 | EF-1-δ | Protein synthesis | 31.1 | 4.90 | 2 | 0.80 | 0.53 | ND | Unchanged24 | ||
| AAH09865 | eEF1γ | Protein synthesis | 50.1 | 6.25 | 4 | 0.35 | 0.41 | 0.38 | Unchanged∥ | ||
| P46199 | IF-2 | Protein synthesis | 81.3 | 6.71 | 2 | ND | 0.47 | 0.35 | Unchanged24 | ||
| P04765 | eIF4A-1 | Protein synthesis | 46.1 | 5.32 | 3 | 0.53 | 0.34§ | 0.40§ | Unchanged∥ | ||
| P55010 | eIF5 | Protein synthesis | 49.2 | 5.41 | 2 | ND | 0.48 | ND | Unchanged24 | ||
| P42655 | 14-3-3ϵ | Signal transduction | 29.1 | 4.63 | 4 | 0.66 | 0.44 | 0.41§ | Unknown | ||
| P29312 | 14-3-3ζ/δ | Signal transduction | 27.7 | 4.73 | 2 | 0.81 | 0.58 | 0.59 | Unknown | ||
| P01112 | c-H-ras | Signal transduction | 21.2 | 5.16 | 2 | ND | ND | 0.19 | Unknown | ||
| P00540 | c-mos | Signal transduction | 37.8 | 9.15 | 3 | ND | 0.43 | 0.38 | Unknown | ||
| P30153 | PP2A | Signal transduction | 65.0 | 4.96 | 2 | 0.70 | 0.35 | 0.41 | Unknown | ||
| Q13233 | MEKK 1 | Signal transduction | 164 | 8.52 | 3 | 0.92 | ND | 0.64 | Repressed24 | ||
| P23528 | Cofilin, P18 | Structural | 18.5 | 8.22 | 9 | 0.51 | 0.64 | 0.52 | Repressed24 | ||
| P32119 | Peroxiredoxin 2 | Cellular defense | 21.9 | 5.66 | 5 | ND | 0.47 | 0.50 | Unknown | ||
| C28894 | MPO | Cellular defense | 92.4 | 9.25 | 9 | 0.35 | 0.52 | 0.48 | Repressed23 | ||
| P25763 | CDC42 homolog | Not defined | 21.3 | 6.16 | 3 | 0.35 | 0.54 | ND | Repressed24 | ||
| P10809 | 60-kDa hsp | Chaperone | 61.0 | 5.70 | 7 | 0.77 | 0.67 | 0.48 | Repressed23 | ||
| Q9NR31 | SAR1a | Transport | 22.4 | 6.22 | 4 | ND | 0.18 | 0.20 | Unknown | ||
| P12270 | Nucleoprotein TPR | mRNA transport | 266 | 5.01 | 6 | 0.88 | 0.50 | 0.48 | Unknown | ||
| P07910 | hnRNP C1/C2 | mRNA splicing | 33.7 | 4.95 | 3 | 0.64 | 0.55 | ND | Unchanged∥ | ||
| Q07244 | hnRNP K | mRNA transport | 50.9 | 5.39 | 6 | 0.52 | 0.53 | 0.50 | Unchanged∥ | ||
| P07029 | hnRNP UP2 | mRNA splicing | 24.1 | 8.02 | 1 | 1.0 | 0.68 | ND | Unchanged∥ | ||
| P43331 | snRNP D3 | mRNA splicing | 13.9 | 10.3 | 2 | ND | 0.39 | 0.41 | Unchanged∥25 | ||
| P08578 | snRNP E | mRNA splicing | 10.8 | 9.46 | 2 | 0.71 | 0.62 | 0.59 | Unchanged∥ | ||
| Q13822 | E-NPP 2 | Glycolysis | 99.0 | 7.14 | 2 | ND | ND | 0.31 | Unknown | ||
| P22712 | MBP-1 | Transcription | 37.0 | 6.79 | 2 | ND | 0.15 | ND | Unknown | ||
| Q9BWX5 | GATA-5 | Transcription | 41.3 | 9.17 | 1 | 1.5 | ND | 2.6 | Unknown | ||
| Q14974 | Importin β-1 | Transport | 97.2 | 4.68 | 2 | 1.5 | ND | 2.1 | Repressed24 | ||
| P24903 | CYPIIF1 | Cellular defense | 55.5 | 6.93 | 2 | 1.6 | ND | 1.8 | Unknown | ||
| P52565 | Rho GDI 1 | Signal transduction | 23.2 | 5.03 | 2 | 2.3 | 1.7 | ND | Unknown | ||
| P31146 | Coronin 1A | Signal transduction | 51.0 | 6.25 | 3 | 2.2 | ND | ND | Unknown | ||
| NP_000994 | 60S P1 protein | Protein synthesis | 11.5 | 4.26 | 2 | 1.0 | 1.5 | 1.8 | Unknown | ||
| P15531 | NDP kinase A | Energy | 17.1 | 5.83 | 2 | 0.9 | 1.5 | ND | Unknown | ||
| P05109 | Calgranulin A | Cellular defense | 10.8 | 6.51 | 2 | ND | 2.0 | 2.1 | Induced23 | ||
| Q99733 | NAP2 | Not defined | 42.7 | 4.60 | 3 | 1.2 | ND | 1.9 | Unknown | ||
| P47755 | CapZ α-2 | Structural | 32.9 | 5.57 | 2 | 1.8 | 2.2 | 2.6 | Unknown | ||
| P12814 | α-actinin 1 | Structural | 103 | 5.22 | 3 | 2.7 | 2.3 | 2.8 | Unknown | ||
| Q08043 | α-actinin 3 | Structural | 103 | 5.40 | 3 | 3.4 | ND | ND | Unknown | ||
| P35579 | NMMHC-A | Structural | 226 | 5.50 | 3 | 2.0 | 4.1 | 4.3 | Unknown | ||
| P29590 | isoform PML-1 | Negative regulator | 97.5 | 5.88 | 2 | 3.5 | ND | ND | Unknown | ||
eEF indicates eukaryotic translation elongation factor; c-H-ras, transforming protein p21/H-Ras-1; c-mos, proto-oncogene serine/threonine-protein kinase mos; PP2A, serine/threonine protein phosphatase 2A; MEKK 1, mitogen-activated protein kinase kinase kinase 1; MPO, myeloperoxidase; CDC42 homolog, G25K guanosine triphosphate (GTP)-binding protein; SAR1a, GTP-binding protein SAR1a; hnRNP, heterogeneous nuclear ribonucleoprotein; snRNP, small nuclear ribonucleoprotein; E-NPP 2, ectonucleotide pyrophosphatase/phosphodiesterase 2; MBP-1, C-myc promoter-binding protein; GATA-5, transcription factor GATA-5; CYPIIF1, cytochrome P450 2F1; NDP kinase A, nucleoside diphosphate kinase A; NAP2, nucleosome assembly protein 1—like 4; actinin, F-actin cross-linking protein; NMMHC-A, myosin heavy chain, nonmuscle type A; isoform PML-1, probable transcription factor PML. For other abbreviations, see Table 1.
Theoretical.
ATRA-treated divided by nontreated. All values are statistically significant, P < .05.
The mRNA expression is as displayed in references Liu et al23 (for 0 to 72 hours), Yang et al24 (for 0 to 96 hours), or Lee et al25 (for 0 to 48 hours) or by personal communication. Unchanged mRNA expression ratios (for ATRA treated divided by nontreated) were within 0.8 to 1.2 as a function of various ATRA treatment time points.
See Figure 4.
Also in Table 1.
The quantification of AACT-regulated proteins is demonstrated by eEF1A-1 (Figure 2A), where the relative intensity of a leucine-containing eEF1A-1 tryptic peptide synthesized in normal media (m/z 657.85) was compared with the same eEF1A-1 tryptic peptide synthesized in Leu-d3–rich media (m/z 662.38). Automated MS/MS fragmentation of both monoisotopic peaks (Figure 2B-C) reveals the number and authentic locations of the leucine amino acids present in the peptide. This is illustrated by comparing the MS/MS fragments from the doubly charged peptides at 657.85 and 662.38, which only separate via their m/z values when leucine is present in a fragment ion. The MS/MS 657.85 (an eEF1A tryptic peptide for residues 135 to 146, EHALLAYTLGVK) produces C-terminus fragments (y-ion series) of 751.4, 864.5, and 977.6 for y7, y8, and y9, respectively (Figure 2B). The y-ions of y7, y8, and y9 from the 662.38 ion yield m/z values of 754.4, 870.5, and 986.6 in agreement with the 3.0-Da mass increment and the location of leucine in the peptide (Figure 2C). The MS/MS fragment products of 657.85 and 662.38 are identical for ions that do not contain a leucine residue and is illustrated by b2 and b3 (N-terminus fragments) in Figure 2B-C showing identical m/z values of 267.1 and 338.1.
Validation of the 2-DE and AACT quantitative results
The validation of our high-throughput proteomic results were confirmed by analyzing protein spots excised from 2-DE using NB4 cells grown in a 99% isotopic abundance of Leu-d3 (Figure 3A), by analyzing β-actin leucine peptides produced within the 1:1 mixture with the use of AACT (Figure 3B), and by traditional Western blot analysis for some ATRA-induced AACT-regulated proteins (Figure 3C). The ability of NB4 cells grown in 99% Leu-d3 to efficiently incorporate the deuterated amino acid during protein synthesis was shown quantitatively for β-actin, heat shock protein 70, and stress-70 protein in Figure 3A. The asterisks in each plot represent a small 2% to 4% abundance of non–Leu-d3 peptide, probably as a result of initial cell seeding, whereas the highly abundant peaks of 896.0, 682.9, and 897.5 represent the same tryptic peptide, but with one Leu-d3 amino acid. The leucine containing β-actin peptide in Figure 3B confirms accurate 1:1 mixing of ATRA-treated and nontreated cells used in AACT. Also shown in Figure 3B are the ATRA-induced up-regulation of myosin heavy chain37 and the down-regulation of cofilin at 24 hours, corresponding to where these proteins resolved in the 1-D gel. The Western blot data in Figure 3C demonstrate ATRA-induced regulation of 14-3-3ϵ, 14-3-3ζ, hnRNP K, eIF4G, and Rho GDP-dissociation inhibitor 1 observed in either 2-DE or AACT proteomic approaches. Additionally, Figure 3D illustrates the down-regulation of eFI4G, eIF5, hnRNP K, and 14-3-3ζ observed in non–t(15;17)–expressing HL-60.
Validation of proteomic results. (A) A 2-DE image of the β-actin region for NB4 cultured in a 99% isotopic abundance of Leu-d3. The plots represent tryptic peptides of β-actin, heat shock protein 70, and stress-70 protein, which contain 1 leucine amino acid. The peaks of 896.0, 682.9, and 897.5 are the monoisotopic peaks for the peptides yielding the Leu-d3 amino acid, and in each plot the asterisk represents the nondeuterated form of the peptide. (B) SDS-PAGE separation of a 1:1 mixture of nontreated versus ATRA-treated NB4 at 24 hours. The ATRA-treated cells were grown in a 99% isotopic abundance of Leu-d3 media and mixed 1:1 with nontreated cells grown in normal culture media in accordance with the AACT methodology. The plots on the right represent the AACT-based quantification of tryptic peptides obtained from in-gel digests. Relative intensity comparisons of the same leucine containing peptides from nontreated and ATRA-treated NB4 yield the up-regulation of myosin heavy chain (650.7 versus 655.7), unchanged β-actin (895.9 versus 897.4), and down-regulated cofilin (669.3 versus 670.8). (C) Western blot analysis of 14-3-3ϵ, 14-3-3ζ, hnRNP K, eIF4G, Rho GDI-1, DAP5/p97/NAT1, and PML-RARα as a function of ATRA treatment, using 50 μg total protein. Antibodies to 14-3-3ϵ, 14-3-3ζ, hnRNP K, eIF4G, Rho GDI-1, DAP5/p97/NAT1, and PML-RARα were used to detect expression of specific proteins. (D) HL-60 Western blot analysis of 14-3-3ζ, hnRNP K, eIF4G, and eIF5 as a function of ATRA treatment, using 50 μg total HL-60 protein.
Validation of proteomic results. (A) A 2-DE image of the β-actin region for NB4 cultured in a 99% isotopic abundance of Leu-d3. The plots represent tryptic peptides of β-actin, heat shock protein 70, and stress-70 protein, which contain 1 leucine amino acid. The peaks of 896.0, 682.9, and 897.5 are the monoisotopic peaks for the peptides yielding the Leu-d3 amino acid, and in each plot the asterisk represents the nondeuterated form of the peptide. (B) SDS-PAGE separation of a 1:1 mixture of nontreated versus ATRA-treated NB4 at 24 hours. The ATRA-treated cells were grown in a 99% isotopic abundance of Leu-d3 media and mixed 1:1 with nontreated cells grown in normal culture media in accordance with the AACT methodology. The plots on the right represent the AACT-based quantification of tryptic peptides obtained from in-gel digests. Relative intensity comparisons of the same leucine containing peptides from nontreated and ATRA-treated NB4 yield the up-regulation of myosin heavy chain (650.7 versus 655.7), unchanged β-actin (895.9 versus 897.4), and down-regulated cofilin (669.3 versus 670.8). (C) Western blot analysis of 14-3-3ϵ, 14-3-3ζ, hnRNP K, eIF4G, Rho GDI-1, DAP5/p97/NAT1, and PML-RARα as a function of ATRA treatment, using 50 μg total protein. Antibodies to 14-3-3ϵ, 14-3-3ζ, hnRNP K, eIF4G, Rho GDI-1, DAP5/p97/NAT1, and PML-RARα were used to detect expression of specific proteins. (D) HL-60 Western blot analysis of 14-3-3ζ, hnRNP K, eIF4G, and eIF5 as a function of ATRA treatment, using 50 μg total HL-60 protein.
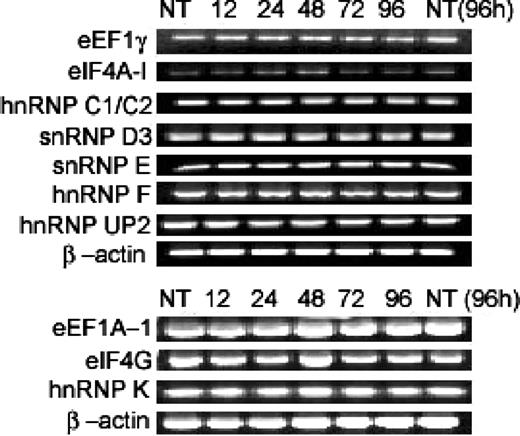
RT-PCR demonstrates posttranscriptional control of eIFs, eEFs, and sn/hnRNPs
RT-PCR results of the mRNAs that encode the suppressed eIFs, eEFs, and ribonucleoproteins included in Tables 1 and 2 illustrate that there is essentially no change in the expression of these genes as a function of ATRA treatment, despite down-regulation at the protein level (Figure 4). It was also verified by previous studies that mRNA expression of EF-1-δ, IF-2, eIF5, and snRNP D3 was unchanged from various time points ranging from 0 to 96 and 0 to 48 hours24,25 of ATRA treatment (Table 2), giving us a complete spectrum for all the eIF, eEF, and RNP mRNA expression presented in this study (9 proteins in Tables 1, 2 agreed with their differential mRNA expression, while only importin β-1 displayed the opposite results24 ).
Semiquantitative PCR data for various genes after ATRA treatment. Semiquantitative PCR data for genes EF-1-γ, eIF4A-1, hnRNP C1/C2, snRNP D3, snRNP E, hnRNP F, hnRNP UP2, eEF1A-1, eIF4G, and hnRNP K at 0, 12, 24, 48, 72, 96 hours of ATRA treatment. For equal loading, β-actin was used, and nontreated NB4 at 96 hours was also used as an additional control.
Semiquantitative PCR data for various genes after ATRA treatment. Semiquantitative PCR data for genes EF-1-γ, eIF4A-1, hnRNP C1/C2, snRNP D3, snRNP E, hnRNP F, hnRNP UP2, eEF1A-1, eIF4G, and hnRNP K at 0, 12, 24, 48, 72, 96 hours of ATRA treatment. For equal loading, β-actin was used, and nontreated NB4 at 96 hours was also used as an additional control.
PML-RARα degradation by ATRA correlates with significant changes in expression of proteins involved in posttranscriptional events
Since we identified proteins that are consistently up- or down-regulated starting with 8 hours of ATRA treatment, PML-RARα expression was monitored in response to a pharmacologic dose (1.0 μM) of ATRA at different time points (Figure 3C). This was performed to determine if ATRA-induced proteolysis of PML-RARα9 corresponds with global differential proteomic changes brought on by PML NB re-formation and RARα processes. Figure 3C illustrates that ATRA-induced proteolysis of PML-RARα begins after 24 hours of treatment and is cleaved beyond detection after 72 hours. This observation demonstrates that pharmacologic amounts of ATRA used throughout this study results in cleavage of PML-RARα, which correlates with significant protein differential expression, as very minor global protein changes were observed at the 8-hour time point (Table 1).
Discussion
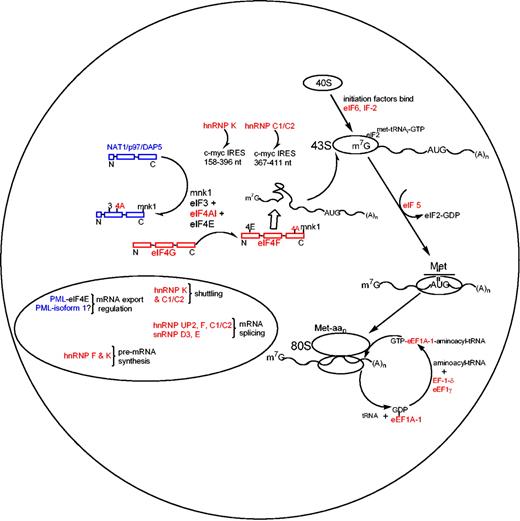
We demonstrate here that ATRA treatment of APL cells results in the regulation of posttranscriptional events involved in the initiation and elongation phases of translation and hn/snRNP proteins, which are involved in mRNA processing and export. The ATRA-induced posttranscriptional down-regulation of hnRNPs observed in this study agrees with the protein regulation observed in BCR/ABL leukemogenesis where hnRNP A, E, and P2 are controlled after transcription, but are up-regulated and thus possess oncogenic properties.38,39 Mechanistic studies have found that hnRNP E (a homolog of hnRNP K and UP2) interacts with the 5′ untranslated region (UTR) of C/EBPα (regulator of granulocytic differentiation) mRNA in chronic myelogenous leukemia and that overexpression of hnRNP E inhibits the translation of C/EBPα mRNA.40 Both hnRNP C1/C2 and hnRNP K have been shown to bind and enhance translation at the internal ribosomal entry site element (IRES) of c-myc and mutated/proto-oncogene c-myc mRNA, with hnRNP C1/C2 binding the 367-to-411 nucleotide region and hnRNP K binding the 158-to-396 nucleotide region of the c-myc IRES (Figure 5).41,42 The ATRA-induced suppression of hnRNP C1/C2 and K in APL demonstrates a mechanistic process within the complex regulation of c-myc expression as c-myc is sharply down-regulated via ATRA treatment of APL.43
The mechanisms involving ATRA-induced posttranscriptional events in APL produced from our proteomics. Red colors define ATRA-induced down-regulated proteins, and blue colors represent up-regulation (in agreement with the color code in Figure 1A).
The mechanisms involving ATRA-induced posttranscriptional events in APL produced from our proteomics. Red colors define ATRA-induced down-regulated proteins, and blue colors represent up-regulation (in agreement with the color code in Figure 1A).
The specificities of hnRNP F, UP2, snRNP E, and D3 toward mRNAs and their mechanisms in ATRA-induced differentiation in NB4 remain unknown and await further investigations. Continued mechanistic studies of hnRNP C1/C2 and K and specificity assignments of hnRNP F, UP2, snRNP E, and D3 could provide vital information regarding ATRA-induced differentiation processes.
ATRA-induced posttranscriptional suppression during initiation and elongation phases of protein synthesis
The ATRA-induced differential expression of proteins observed in this study provides novel insights into the posttranscriptional regulation of the rate-limiting initiation and elongational phases present in APL (Figure 5). The down-regulated proteins observed in this study are shown in red, with up-regulated proteins in blue. The rate-limiting phase of protein synthesis involves unwinding and transferring mRNAs by eIF4 proteins to the 43S ribosomal subunit. ATRA treatment induces a posttranscriptional suppression of eIF4A-1 and eIF4G, and up-regulation of DAP5, while eIF4E remained unchanged (Western blot; data not shown). EIF4G serves as a scaffold protein (termed eIF4F) for mRNA binding proteins eIF4A-1 and eIF4E, allowing enhanced mRNA binding properties for eIF4A-1 and eIF4E when associated with the eIF4F complex.44-51 In addition to the ATRA-induced suppression of eIF4 proteins, IF-2, eIF5, and eIF6 were also down-regulated, while NAT1/p97/DAP5, an inhibitor of translation that binds eIF3/eIF4 proteins and possesses high sequence homology to eIF4G, was shown to be up-regulated (Figures 3C and 5).52 These findings demonstrate several ATRA-induced processes that inhibit cytoplasmic mRNA transport for capped mRNAs to ribosomal subunits during this rate-limiting phase. Furthermore, it has been showed that some eIFs, such as eIF4E, selectively enhance expression of growth-promoting (eg, cyclin D1) and metastasis-related mRNAs (eg, vascular endothelial growth factor [VEGF]),53 suggesting that translation control through regulation of eIFs may play a role in proliferation and tumor growth control.
In addition to the suppression of mRNA transport mechanisms induced by ATRA, elongational processes that function in the transfer of aminoacyl-tRNA to the “A” site of the 80S ribosome have also been found to be suppressed (Figure 5). Reduced protein levels of eEF1A-1, EF-1-δ, and eEF1γ not only limit the aminoacyl-tRNA transfer rate to the 80S ribosome, but also inhibit the dissociation of inactive eEF1A-1–GDP complexes to active GTP-bound active state.54-56
Proteomic insights regarding PML-NB–mediated mRNA transport regulation and caspase activation during ATRA treatment of NB4
It has been well demonstrated that ATRA treatment of t(15;17)–expressing APL results in caspase-independent proteolysis of PML-RARα into a truncated 85-kDa product, termed ΔPML-RARα, which allows PML NB re-formation and consequential differentiation and apoptosis of APL cells.9,57,58 Following PML-RARα cleavage, PML protein has been shown to form many NBs with a number of proteins.59 One well-studied NB is PML-eIF4E, which has been observed in several leukemic lines, including U937, K562, and ATRA-treated NB4, and is able to regulate mRNA export of certain growth-stimulatory proteins, specifically, cyclin D1.60-62 In this study, it is shown that several oncogenic eIFs, eEFs, and RNPs are suppressed in a time-dependent fashion that corresponds with PML-RARα proteolysis, suggesting more diversity in terms of growth-stimulatory mRNA regulation for the PML-eIF4E NB and other PML NBs. Interestingly, overexpression of eIF4 mRNAs has been observed for eIF4A-1in melanoma cell lines,63 eIF4E mRNA in breast cancer,64 and eIF4G mRNA in squamous cell lung carcinoma,65 demonstrating the oncogenic nature of these eIF4 mRNAs and proteins in the absence of proper regulation.
The proteomics revealed in this study also provide insight into caspase-induced apoptosis pathways, where caspase up-regulation and activation is observed in response to ATRA and other retinoids for NB4 and non–t(15;17)–expressing HL-60.66-68 We have shown that ATRA induces down-regulation of 14-3-3ϵ and 14-3-3ζ/δ in NB4 (Table 2; Figure 3C) and down-regulation of 14-3-3ζ in HL-60 (Figure 3D), which was not observed in HL-60 ATRA-treated proteomic studies.68 The acidic 14-3-3ϵ, 14-3-3ζ, and 14-3-3δ proteins have a very broad range of specificities, one of which is to bind and activate protein kinase C (PKC) and certain PKC isoforms.69-73 Since the inhibition of PKC leads to activation of caspase networks and thus induction of apoptosis in APL and other leukemias,74-76 the link between diminished PKC activity via the down-regulation of 14-3-3 proteins in NB4 and HL-60 provides additional information regarding the involvement of 14-3-3 proteins toward the mechanisms of caspase activation in these leukemias.
Prepublished online as Blood First Edition Paper, May 13, 2004; DOI 10.1182/blood-2004-01-0046.
Supported by US Department of Energy (DOE) grants ERW9923 and ERW9840, and Los Alamos National Laboratory LDRD 20030508ER (X.C.). X.C. is a recipient of a Presidential Early Career Award for Scientists and Engineers (PECASE) (2000-2005). This study was supported in part by grants by Ladies Leukemia League (B.O.), IDO1-014 U54 RFA CA096300 (G.L.-B.).
M.N.H. and B.O. contributed equally to this study.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to the research groups of Lijun Yang from the Department of Pathology, Immunology, and Laboratory Medicine, University of Florida, College of Medicine, and to Myung-Ju Ahn from the Department of Internal Medicine, College of Medicine, Hanyang University, Seoul, Republic of Korea, for sharing cDNA microarray data on ATRA treatment of NB4.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal