Abstract
Natural killer (NK) cells play a pivotal role in the immune reaction during the bone marrow allograft rejection. Little is known, however, about the molecular mechanisms underlying the NK cell–mediated allograft recognition and rejection. In this report, we assessed the role of a recently identified NK receptor, killer cell lectinlike receptor 1 (KLRE-1), by generating knock-out mice. KLRE-1–deficient mice were born at an expected frequency and showed no aberrant phenotype on growth and lymphoid development. Nevertheless, KLRE-1–deficient cells showed a severely compromised allogeneic cytotoxic activity compared with the wild-type cells. Furthermore, allogeneic bone marrow transfer culminated in colony formation in the spleen of KLRE-1–deficient mice, whereas no colony formation was observed in wild-type recipient mice. These results demonstrate that KLRE-1 is a receptor mediating recognition and rejection of allogeneic target cells in the host immune system.
Introduction
Natural killer (NK) cells play a pivotal role in the recognition and rejection of allogeneic target cells, such as bone marrow (BM) allografts.1-3 NK cell cytotoxicity is exquisitely controlled by signals emanating from stimulatory and inhibitory NK receptors, which recognize major histocompatibility complex (MHC) class I–related molecules on the target cells.3,4 The function of NK receptor(s) in allograft rejection has been mainly probed with the aid of monoclonal antibodies (mAbs). It is shown that blocking the inhibitory receptor, CD94/NKG2A with anti-CD94 mAb enhances the cytotoxicity of C57BL/6 NK cells against BALB/c concanavalin A (Con A) blasts,5 while anti–Ly-49D mAb treatment results in suppression of the BM graft rejection.6 In both cases, however, it is still open to question whether the effect of anti-CD94 or anti-Ly49D mAbs was direct or attributed to the cross-reactivity of mAbs with other family members.5,7-10
Recently, a novel NK receptor, KLRE-1 (also known as NKG2I), belonging to the killer cell lectinlike receptors (KLRs) family has been characterized.11-13 A series of experiments using anti–KLRE-1 mAbs indicate that this receptor plays a role in the cytotoxicity mediated by NK cells in vitro.12,13 However, the definitive assessment for the functions of KLRE-1 has to await KLRE-1–deficient mice.
To this end, we have generated KLRE-1 knock-out mice and assessed the role of KLRE-1. KLRE-1–deficient cells showed little cytolytic activity against allogeneic lymphocytes. Furthermore, allogeneic bone marrow transfer resulted in significant colony formation in the spleen of KLRE-1–deficient mice. These results further confirm that KLRE-1 is a crucial NK receptor for recognition and rejection of bone marrow allograft.
Study design
Mice
C57BL/6, BALB/c mice were from Charles River Japan (Yokohama, Japan), and 129/svJ mice were from Jackson Laboratory (Bar Harbor, ME). KLRE-1 knock-out mice have been backcrossed 6 times to C57BL/6 mice. Mice were kept under the specific pathogen-free conditions, and 6- to 8-week-old mice were used for the experiments. All experiments were performed in accordance with the guidelines of Chiba University.
Construction and establishment of EGFP–KLRE-1 knock-in mice
KLRE-1 genomic clone was isolated from a mouse genomic library using the KLRE-1 cDNA as a probe (cDNA sequence, first described by Koike et al13 ). The genomic DNA fragment from NsiI site located at 1.5 kb upstream of the exon 2 (which contains ATG for first methionine) to BglII site present at 5.0 kb downstream of the exon 2 was used to construct the targeting vector (Figure 1A). Enhanced green fluorescent protein (EGFP)–poly A cassette from the pEGFP-N3 vector (Clontech Laboratories, Palo Alto, CA) was amplified by polymerase chain reaction (PCR) and inserted in frame into the EcoRI site located at 39 bp downstream of ATG, followed by the neomycin-resistant gene (neor) cassette flanked by loxP sites. The targeting vector was electroporated into R1 embryonic stem (ES) cells, and G418-resistant clones were screened by Southern blotting with the 5′ external as well as the neor internal probes. Positive clones were aggregated with BDF1 blastocysts, and chimeric mice were obtained as described.14
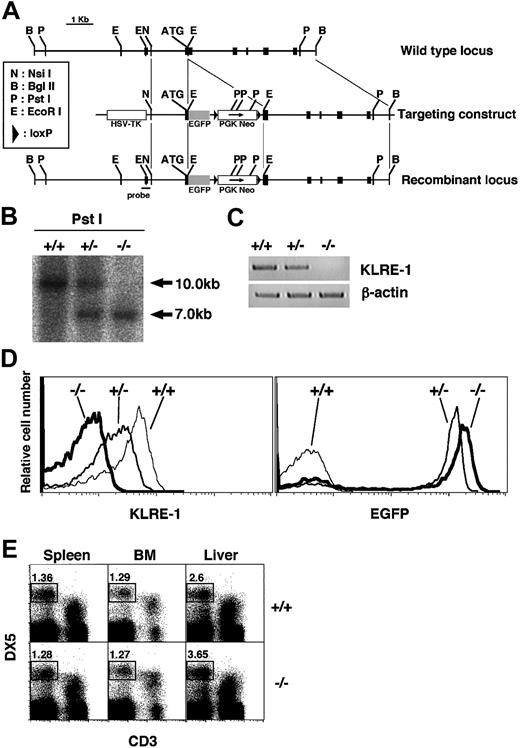
Characterization of KLRE-1 KO mice. (A) Generation of the EGFP–KLRE-1 knock-in mice. Schematic representation of the portion of the KLRE-1 gene locus (top), targeting construct (middle), and recombinant allele (bottom) including the relevant restriction sites. Probe used for Southern blot analysis is depicted. Exons are represented as black boxes. (B-E) Characterization of the EGFP–KLRE-1 knock-in mice. (B) Southern blot analysis. Southern blot analysis of Pst I-digested DNA from wild-type (WT) (+/+), heterozygous (+/–), or homozygous (–/–) KLRE-1 mice. DNA was hybridized with the probe shown in panel A to discriminate between the WT allele (10.0 kb) and the mutated allele (7.0 kb). (C) RT-PCR analysis for KLRE-1 mRNA. Total RNA prepared from DX5+ splenocytes of WT (+/+), heterozygous (+/–), or homozygous (–/–) KLRE-1 mice was subjected to RT-PCR analysis on KLRE-1 and β-actin. PCR on β-actin assured an equal amount of cDNA. (D) Expression of KLRE-1 and GFP in the DX5+TCRβ– splenocytes. The DX5+TCRβ– splenocytes from WT (+/+), heterozygous (+/–), or homozygous (–/–) KLRE-1 mice were stained with the Cy5–anti–KLRE-1 (7E8) mAb. Expression of KLRE-1 (left panel) and EGFP (right panel) is shown as a histogram. (E) Normal development of NK cells in KLRE-1 knock-out (KO) mice. Spleen, bone marrow (BM), and liver mononuclear cells from WT (+/+) and homozygous (–/–) KLRE-1 mice were stained with the PE-DX5 and the Cy5–anti-CD3ϵ. The area representing NK (DX5+ CD3ϵ–) cells is shown with percentage.
Characterization of KLRE-1 KO mice. (A) Generation of the EGFP–KLRE-1 knock-in mice. Schematic representation of the portion of the KLRE-1 gene locus (top), targeting construct (middle), and recombinant allele (bottom) including the relevant restriction sites. Probe used for Southern blot analysis is depicted. Exons are represented as black boxes. (B-E) Characterization of the EGFP–KLRE-1 knock-in mice. (B) Southern blot analysis. Southern blot analysis of Pst I-digested DNA from wild-type (WT) (+/+), heterozygous (+/–), or homozygous (–/–) KLRE-1 mice. DNA was hybridized with the probe shown in panel A to discriminate between the WT allele (10.0 kb) and the mutated allele (7.0 kb). (C) RT-PCR analysis for KLRE-1 mRNA. Total RNA prepared from DX5+ splenocytes of WT (+/+), heterozygous (+/–), or homozygous (–/–) KLRE-1 mice was subjected to RT-PCR analysis on KLRE-1 and β-actin. PCR on β-actin assured an equal amount of cDNA. (D) Expression of KLRE-1 and GFP in the DX5+TCRβ– splenocytes. The DX5+TCRβ– splenocytes from WT (+/+), heterozygous (+/–), or homozygous (–/–) KLRE-1 mice were stained with the Cy5–anti–KLRE-1 (7E8) mAb. Expression of KLRE-1 (left panel) and EGFP (right panel) is shown as a histogram. (E) Normal development of NK cells in KLRE-1 knock-out (KO) mice. Spleen, bone marrow (BM), and liver mononuclear cells from WT (+/+) and homozygous (–/–) KLRE-1 mice were stained with the PE-DX5 and the Cy5–anti-CD3ϵ. The area representing NK (DX5+ CD3ϵ–) cells is shown with percentage.
Southern blot and PCR analyses
Genotyping was performed by Southern blotting or PCR using genomic DNA from the tail.14 The probe for Southern blotting was synthesized by PCR with the primer set J03-23 (5′-AAGAGGGAATTCCAGGCACAGATG-3′) and J03-35 (5′-GGGTGCTAAACGGAAATGTAAAGC-3′). The primers used for genotyping were GFP-6 (5′-CCTCTACAAATGTGGTATGGC-3′) and GFP-8 (5′-ATGGTGAGCAAGGGCGAGGAGC-3′) for the targeted allele, and J03KO1 (5′-GATGGATGAAGCACCTGTAAC-3′) and J03KO3 (5′-TCAGAAACCCATCAGACCAACC-3′) for the wild-type allele. The primers for reverse transcriptase (RT)–PCR for KLRE-1 were J03-4 (5′-TAAGAGACAAGCAGGCACGCTGACTG-3′) and J03-7 (5′-ATGGATGAAGCACCTGTAACCCG-3′).
Cell preparation and flow cytometry
Splenic NK, spleen, bone marrow, and liver mononuclear cells were separated and stained with the appropriate antibodies as described13,15 ; phycoerythrin (PE)–anti-DX5, cyanin 5 (Cy5)–anti–T-cell receptor β (TCRβ), Cy5–anti-CD3ϵ (PharMingen, San Jose, CA), and Cy-5–labeled antibody against KLRE-1 (anti-NKG2I: 7E8) were used.13
Cytotoxic assay against the Con A blasts and BM cell engraftment
Cytotoxic assay against the Con A blasts and BM cell transfer experiments were performed essentially as described except that KLRE-1–deficient cells/mice were used in some experiments.13
Results and discussion
Establishment of EGFP–KLRE-1 knock-in mice
We have generated enhanced green fluorescent protein (EGFP)–KLRE-1 knock-in mice to rigorously assess the function of KLRE-1. Southern blot analysis with the 5′ external probe confirmed the correct recombination (Figure 1B). RT-PCR and flow cytometry analyses demonstrated that there was no detectable KLRE-1 transcript or surface expression in the homozygous mice (Figure 1C-D, –/–). A slight decrement of the expression was observed in the heterozygous mice (Figure 1D, left panel, +/–). Concomitantly, the expression of GFP was inversely correlated with the loss of KLRE-1 expression (Figure 1D, right panel). Mice homozygous for EGFP–KLRE-1 (KLRE-1–deficient mice) were born at the expected frequency and were fertile, with no apparent growth abnormality (data not shown). Regarding NK cells, no difference in the profile of CD3/DX5 expression in the spleen, bone marrow (BM), and liver mononuclear cells was noticed between wild-type littermates and knock-out mice (Figure 1E). In addition, no significant variation in the number or ratio of lymphoid subsets was detected in thymus, spleen, and BM cells, suggesting that KLRE-1 is dispensable for the development of lymphoid cells (data not shown).
KLRE-1 is critical for NK cell–mediated allorejection
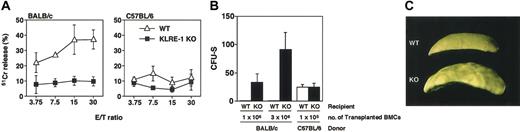
Previous reports indicate that KLRE-1 plays a pivotal role in allogeneic or redirected lysis.12,13 Therefore, the roles of KLRE-1 in cytotoxic activity against allogeneic lymphocytes were assessed using KLRE-1–deficient cells. Lymphokine activated killer (LAK) cells from C57BL/6 wild-type mice were mixed with BALB/c Con A blasts, and cytotoxic activity was examined. While wild-type LAK cells showed a significant cytolytic activity against allogeneic target cells, LAK cells from KLRE-1–deficient mice showed little activity (Figure 2A, left panel). In the syngeneic system, however, no marked difference in the cytotoxicity between wild-type and KLRE-1–deficient cells was observed (Figure 2A, right panel). These results demonstrate that KLRE-1 is crucial for NK cells to exert allogeneic cytolytic activity.
KLRE-1 plays a pivotal role in allogeneic bone marrow transfer. (A) Cytotoxic activity against allogeneic Con A blasts using KLRE-1–deficient cells. Cytolytic activity of LAK cells containing activated NK cells from wild-type C57BL/6 (WT, ▵) and KLRE-1–deficient (KLRE-1 KO, ▪) mice was assessed. Representative data from 3 independent experiments (n = 5/experiments) are shown as means ± SD. (B-C) KLRE-1 KO mice fail to reject BM allograft. (B) BALB/c BM cells (1 × 106 or 3 × 106 cells) or C57BL/6 BM cells (1 × 105 cells) were intravenously infused to lethally irradiated age-matched wild-type or KLRE-1–deficient C57BL/6 mice. At 8 days after transplantation, spleens were removed and the number of colonies on the spleen was counted. Representative data from the 3 independent experiments are shown. Data are indicated as means ± SD of 5 individuals per experiments. (C) Representative picture of allograft-induced colony formation in the spleen of KLRE-1 knock-out mice. Colony formation resulting from the BALB/c BM allograft as described in panel B can be observed on the spleen of lethally irradiated KLRE-1 null mice (KO), but not on that of C57BL/6 mice (WT) after the fixation in the Bouin solution. The photo was taken with a Nikon digital camera (Cool Pix 995) and processed with Adobe Photoshop.
KLRE-1 plays a pivotal role in allogeneic bone marrow transfer. (A) Cytotoxic activity against allogeneic Con A blasts using KLRE-1–deficient cells. Cytolytic activity of LAK cells containing activated NK cells from wild-type C57BL/6 (WT, ▵) and KLRE-1–deficient (KLRE-1 KO, ▪) mice was assessed. Representative data from 3 independent experiments (n = 5/experiments) are shown as means ± SD. (B-C) KLRE-1 KO mice fail to reject BM allograft. (B) BALB/c BM cells (1 × 106 or 3 × 106 cells) or C57BL/6 BM cells (1 × 105 cells) were intravenously infused to lethally irradiated age-matched wild-type or KLRE-1–deficient C57BL/6 mice. At 8 days after transplantation, spleens were removed and the number of colonies on the spleen was counted. Representative data from the 3 independent experiments are shown. Data are indicated as means ± SD of 5 individuals per experiments. (C) Representative picture of allograft-induced colony formation in the spleen of KLRE-1 knock-out mice. Colony formation resulting from the BALB/c BM allograft as described in panel B can be observed on the spleen of lethally irradiated KLRE-1 null mice (KO), but not on that of C57BL/6 mice (WT) after the fixation in the Bouin solution. The photo was taken with a Nikon digital camera (Cool Pix 995) and processed with Adobe Photoshop.
We have further explored the function of KLRE-1 in allogeneic BM cell transplantation using KLRE-1–deficient mice (Figure 2B-C). BALB/c BM cells infused to the lethally irradiated C57BL/6 mice were rejected, and no colony formation was seen in the spleen of C57BL/6 mice (Figure 2B-C, WT). In sharp contrast, when BALB/c BM cells were transferred into KLRE-1–deficient mice, a graft dose-dependent colony formation was observed, indicating that the rejection of grafts was severely compromised due to the absence of KLRE-1 (Figure 2B-C, KO). When syngeneic cells were used, the graft was accepted irrespective of KLRE-1 absence and resulted in the formation of a similar number of colonies (Figure 2B, rightmost column). Taken together, one can conclude that KLRE-1 is a crucial mediator for the allograft rejection in vivo.
KLRE-1 has been reported as an inhibitory NK receptor due to its association with the protein tyrosine phosphatase, Src homology domain containing tyrosine phosphatase 1 (SHP-1), and cross-linking of KLRE-1 inhibits NK cell–mediated cytotoxicity.12 In our hands, however, KLRE-1 functions as an activating receptor. In fact, cross-linking of NKG2I together with the addition of interleukin-2 (IL-2) and/or IL-12 culminates in the production of interferon γ (IFN-γ).13 The inhibition of the cytotoxicity observed in redirected lysis assay may most likely mirror the fact that the absence of KLRE-1 perturbed NK cell–mediated cytotoxicity against allogeneic cells12 (Figure 2A, left column). As KLRE-1 is devoid of any signaling motif such as immunoreceptor tyrosine-based inhibitory motif (ITIM) or immunoreceptor tyrosine-based activation motif (ITAM), further works should be necessary to decipher whether KLRE-1 functions as an inhibitory receptor or activating receptor.
In summary, we have shown that KLRE-1 is critical for NK cells to exert allogeneic recognition and rejection using KLRE-1–deficient mice.
Prepublished online as Blood First Edition Paper, April 6, 2004; DOI 10.1182/blood-2003-10-3468.
Supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology (Japan): Grants-in-Aid for Scientific Research, Scientific Research A#13307011 (M.T.), B#14370107(T.N.), and Special Coordination Funds for Promoting Science and Technology (T.N.); the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research, the Ministry of Health, Labor, and Welfare (Japan) (M.T.); and the Human Frontier Science Program Research Grant (RG00168/2000-M206) (M.T.).
E.S. and J.K. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal