Abstract
The glycoprotein Ib-IX-V (GP Ib-IX-V) complex mediates platelet binding to von Willebrand factor (VWF) through its largest polypeptide, GP Ibα. Of the many GP Ibα monoclonal antibodies described, AP1 is of particular interest because it blocks static VWF binding induced by 2 modulators, ristocetin and botrocetin, and platelet adhesion to VWF surfaces under flow. We mapped the AP1 binding site to a region encompassing Arg218 to Tyr228, comprising the α1 helix and β13 strand defined by the GP Ibα crystal structure. AP1 binding absolutely required Arg218, Asp222, and Glu225. We evaluated the ability of cells expressing mutants of this region to bind VWF under static conditions in the presence of modulators, and to attach to and roll on a VWF matrix under flow. These data indicate that 2 regions within the sequence Arg218 to Tyr228 have important roles in VWF binding: the α1 helix has a regulatory role and the β turn and β13 strand bind VWF directly. Despite this, the only effect of a synthetic peptide corresponding to Leu214 to Val229 was to slightly increase the rolling velocity of GP Ibα-expressing Chinese hamster ovary (CHO) cells on VWF. This region thus appears to be more important for maintaining the regional conformation of GP Ibα, thereby facilitating the interaction with VWF. (Blood. 2004;104:3971-3978)
Introduction
The formation of platelet thrombi at high shear stress is initiated by the binding of the platelet receptor, the glycoprotein Ib-IX-V (GP Ib-IX-V) complex, to its ligand in the subendothelium, von Willebrand factor (VWF). This receptor-ligand interaction is indispensable for tethering platelets to the injured vessel wall as a prerequisite for integrin-mediated firm arrest.1
The GP Ib-IX-V complex contains 4 polypeptides, each of which spans the platelet plasma membrane once: the 2 disulfide-linked components of GP Ib (GP Ibα and GP Ibβ), GP IX, and GP V.2 The largest of these, GP Ibα, contains the VWF binding domain. Glycoprotein Ibα comprises, in order, an N-terminal flanking sequence (His1-Ile35) containing a disulfide loop between Cys4 and Cys17, 8 tandem leucine-rich repeats (LRR) (Leu36-Ala200),3,4 a C-terminal flanking sequence (Phe201-Gly268) with 2 disulfide loops (Cys209-Cys248 and Cys211-Cys264), a region of dense negative charge (Gly268-Glu282) containing 3 sulfated tyrosine residues at positions 276, 278, and 279, and an elongated, heavily O-glycosylated sialomucin core, termed the macroglycopeptide region. Between the macroglycopeptide and the plasma membrane is a stretch of approximately 30 amino acids that is virtually devoid of sites of O-glycosylation, which is followed by a single transmembrane domain and a cytoplasmic domain of approximately 100 amino acids that plays an important role in linking the whole complex to a submembrane network of actin filaments5 and to signal transducer and adapter molecules.6
Most of the current understanding of how VWF binds to the GP Ib-IX-V complex has come from studies in which the binding was induced by the nonphysiologic modulators of VWF activity, ristocetin7 and botrocetin.8 More recent investigations have been carried out using systems that allow the interaction to be studied under flow, systems such as parallel-plate flow chambers and viscometers.9,10 These studies have shown that the interaction induced by ristocetin more closely resembles shear-induced binding of VWF to GP Ibα than does the botrocetin-induced interaction.11 Through a number of techniques, which include the use of synthetic peptides,12 phage-display,13 proteolytic or mutated fragments of GP Ibα,14,15 canine-human GP Ibα chimeras,16 and GP Ibα monoclonal antibody epitope mapping,13 3 regions within the GP Ibα N-terminus have been implicated in either binding or regulating the binding of VWF, the leucine-rich repeats, the C-terminal flank, and the negatively charged sulfated tyrosine sequence.
The C-terminal flanking domain (Phe201-Gly268) contains 2 loops formed by disulfide bonds between Cys209 to Cys248 and Cys211 to Cys264 (Figure 1A). The first disulfide loop is affected by the gain-of-function mutations, Gly233Val and Met239Val, that cause platelet-type von Willebrand disease.17 There are 2 additional artificial mutations, Asp235Val and Lys237Val, that also induce gain of VWF binding function.18 We previously demonstrated that cells expressing any of 4 gain-of-function mutants showed increased ristocetin- and botrocetin-dependent binding of VWF and, relative to wild-type cells, rolled more slowly on immobilized VWF at similar shear stresses, a finding indicative of decreased off rates of the association. A role for the second loop was suggested by Vicente et al,19 who showed that a peptide spanning residues Ser251 to Tyr279 efficiently blocked ristocetin-induced VWF binding but only partially inhibited botrocetin-induced binding. Likewise, Katagiri et al12 found that ristocetin-induced VWF binding was inhibited most by a peptide encompassing residues Asp235 to Lys262. Taken together, these studies suggest that the VWF binding site within the C-terminal flank comprises more than merely simple linear sequences and that the proper conformation of this region is necessary for VWF binding.
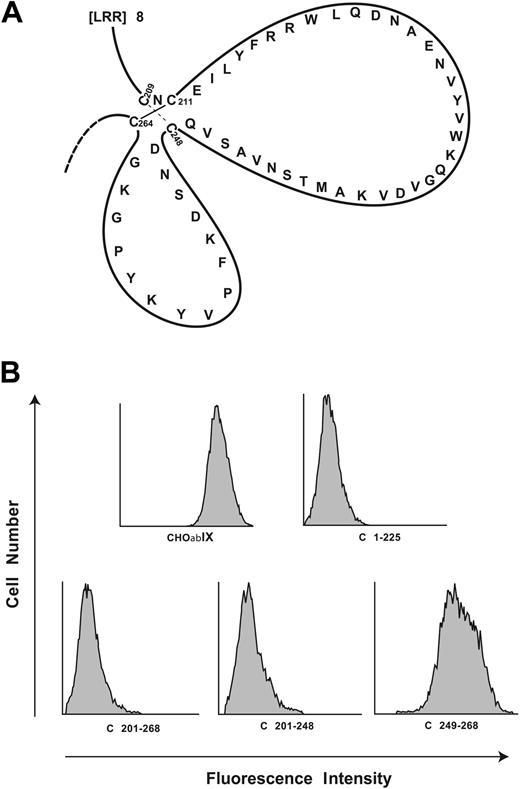
Localization of the AP1 epitope to the C-terminal flank domain. (A) Disulfide loops carboxyl terminal to the GP Ibα leucine-rich repeats. The loops are formed by disulfide bonds between Cys209 and Cys248 and between Cys211 and Cys264. (B) Binding of monoclonal antibody AP1 to canine-human GP Ibα chimeras. C plus the number denotes the sequence of human GP Ibα replaced by the corresponding canine sequence. These studies narrow the epitope to residues 201 to 225 of human GP Ibα.
Localization of the AP1 epitope to the C-terminal flank domain. (A) Disulfide loops carboxyl terminal to the GP Ibα leucine-rich repeats. The loops are formed by disulfide bonds between Cys209 and Cys248 and between Cys211 and Cys264. (B) Binding of monoclonal antibody AP1 to canine-human GP Ibα chimeras. C plus the number denotes the sequence of human GP Ibα replaced by the corresponding canine sequence. These studies narrow the epitope to residues 201 to 225 of human GP Ibα.
Studies with monoclonal antibodies also contributed to defining the VWF binding sites within the C-terminal flank of GP Ibα. Several antibodies, AP1,16 VM16d,11,16 TM60,16 and 6B4,13 bind within this region. AP1 and 6B4 block VWF binding induced by both ristocetin and botrocetin, and also potently inhibit the attachment of GP Ib-IX-expressing cells to a VWF surface under flow, whereas VM16d preferentially inhibits botrocetin-dependent binding of VWF, with minimal effects on the ristocetin-induced interaction.11 TM60 partially inhibits VWF binding induced by both ristocetin and botrocetin. Of interest, AP1, VM16d, and TM60 all also inhibit thrombin-induced binding to GP Ibα,20-23 suggesting that this region contains an important thrombin binding site. This had been suggested previously by Katagiri et al,12 who showed that the peptide Phe216 to Thr240 inhibited high-affinity thrombin binding and by Wicki and Clemetson,24 who showed that an important thrombin binding site on GP Ibα lies N-terminal to Lys237.21 This presumption has now been confirmed in 2 crystal structures of the GP Ibα N-terminus in complex with thrombin, which show this region of GP Ibα bound by thrombin exosite I.25,26 Interestingly, AP1 and VM16d also block the interaction of GP Ibα with the leukocyte integrin Mac-1.27
Of all the GP Ibα antibodies, only AP1 can block the interaction of GP Ibα with VWF, thrombin, and Mac-1, rendering its epitope of intense interest for understanding the ligand-binding functions of GP Ibα. The AP1 epitope has been narrowed to the sequence Phe201 to Glu225,28 which comprises the α1 helix followed by the β13 strand.3,4 In this study, our intent was to map the epitope in detail and to test the effects on VWF binding function of amino acid substitutions within the epitope.
Materials and methods
Antibodies and peptides
The following GP Ibα monoclonal antibodies were used: AK2 (Research Diagnostics, Flanders, NJ), AN51 (Dako, Carpinteria, CA), AP1 (a gift from Dr Dermot Kenny, The Royal College of Surgeons, Dublin, Ireland), VM16d (Monosan, Uden, the Netherlands), SZ2 (Research Diagnostics), and WM23. All but WM23 bind within the N-terminal 282 residues of GP Ibα. AK216 and AN5116,29 block VWF binding induced by both ristocetin and botrocetin and bind within the first leucine-rich repeat (amino acid residues 36-59). AP1 also blocks both ristocetin- and botrocetin-induced VWF binding (epitope 201-268)16 ; VM16d (epitope 201-268)11,16 and SZ2 (epitope 269-282)16 both block only botrocetin-induced VWF binding. WM23 binds within the GP Ibα macroglycopeptide and does not interfere with the binding of any GP Ibα ligands.30 A polyclonal antibody against VWF conjugated to horseradish peroxidase (HRP) was purchased from Dako. An N-terminal biotinylated peptide corresponding to residues Leu214 to Val229 of mature GP Ibα was synthesized by Mimotopes (Clayton, Victoria, Australia).
Site-directed mutagenesis
Mutagenesis was performed directly on the GP Ibα cDNA cloned into the mammalian expression vector pDX31 using a commercial polymerase chain reaction-based mutagenesis kit (QuikChange; Stratagene, La Jolla, CA). Mutated GP Ibα cDNAs were sequenced to verify the mutations.
Cell lines and transfections
Chinese hamster ovary (CHO) cells expressing GP Ibα, GPIbβ, and GP IX (CHOαβIX)31 were grown in α-minimal essential medium (α-MEM; Life Technologies, Bethesda, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 400 μg/mL G418, 80 μg/mL methotrexate, 400 μg/mL hygromycin, 100 U/mL penicillin, and 100 U/mL streptomycin. To develop stable cell lines expressing GP Ibα mutants, the mutated GP Ibα cDNAs were transfected (LipofectAMINE2000; Life Technologies) into CHO βIX cells, which stably express GP Ibβ and GP IX,32 along with a hygromycin-resistance gene. Transfected cells were grown in progressively increasing hygromycin concentrations to a final concentration of 400 μg/mL. The cells were then repeatedly sorted for GP Ib-IX expression with WM23-coupled magnetic beads according to the procedure described by the manufacturer (Dynabeads; Dynal, Lake Success, NY).
Flow cytometry
Surface expression and conformation of mutant GP Ibα polypeptides were examined by flow cytometry after labeling individual aliquots of cells with one of several GP Ibα monoclonal antibodies. Surface expression was determined using WM23, as it is not affected by mutations in the GP Ibα N-terminus.14,30 In brief, adherent cells were detached with versene (Life Technologies), then washed with and resuspended in Tris (tris(hydroxymethyl)aminomethane)-buffered saline (TBS) containing 0.5% bovine serum albumin (BSA). The cells (5 × 105) were then incubated with 2 μg/mL antibody for 60 minutes at room temperature, washed twice, then incubated with a fluorescein isothiocyanate-conjugated rabbit anti-mouse secondary antibody (Zymed Laboratories, South San Francisco, CA) for 30 minutes. The cells were then washed again twice and analyzed on a flow cytometer (Epics XL; Beckman Coulter, Hialeah, FL) stimulating with laser light at 488 nm and collecting light emitted at wavelengths above 520 nm.
Peptide binding assay
Aliquots (50 μL) of VWF, A1 domain, or AP1 monoclonal antibody, each at 3 μg/mL in TBS, pH 7.4, were coated on the wells of 96-well plates (Immulon 2HB; Dynex, Chantilly, VA) overnight at 4°C. The wells were then washed 5 times with TBS and coated with 3% polyvinylpyrrolidone for 90 minutes at 37°C, rinsed in TBS, and incubated with the synthetic GP Ibα peptide plus 10% FBS and 0.05% Tween 20 for 1 hour at 37°C. The wells were then washed 5 times with TBS, 10% FBS, 0.05% Tween 20, and peptide binding was revealed by reaction with avidin-conjugated horseradish peroxidase (BioRad, Hercules, CA), using 3,3′,5,5′-tetramethylbenzidine (Sigma, St Louis, MO) for detection. Absorbance at 450 nm was measured on a Spectra Max Plus 96-well plate enzyme-linked immunosorbent assay reader (ELISA; Molecular Devices, Sunnyvale, CA).
ELISA for ristocetin- and botrocetin-mediated VWF binding
CHOαβIX or control cells (2 × 105) were plated on wells of a 24-well plate coated with 0.01% poly-L-lysine. After 36 hours, the cells were washed twice with phosphate-buffered saline (PBS)/0.5% BSA and incubated for 20 minutes at room temperature with either GP Ibα antibodies (1 μg/mL) or increasing concentrations of VWF (0-4 μg/mL) in the presence of 1.5 mg/mL ristocetin or 2.0 μg/mL botrocetin. Nonspecific binding was determined in the absence of modulator or in the presence of saturating concentrations of GP Ibα antibodies (AK2, SZ2, or AP1). Unbound VWF was removed by washing 4 times with 0.1 M sodium acetate/0.5 × PBS. The cells were then incubated in α-MEM/5% FBS with either HRP-conjugated rabbit anti-human VWF (diluted 1:4000) or HRP-conjugated goat anti-mouse antibody diluted (1:4000) for 10 minutes at 4°C. The cells were then washed twice with TBS/0.5% BSA. To normalize for the slight differences in GP Ibα expression between the cell lines, the data are expressed as a ratio of VWF binding to GP Ibα antibody binding.
Parallel-plate flow chamber
The system includes a parallel-plate flow chamber and an inverted-stage microscope (Eclipse TE300; Nikon, Garden City, NY) equipped with a high-speed digital camera (Model Quantix; Photometrics, Tucson, AZ). The parallel-plate flow chamber was composed of a polycarbonate slab, a silicon gasket, and a glass coverslip held together by vacuum such that the coverslip forms the bottom of the chamber. The coverslips were coated with 20 μg/mL human VWF. Cell suspensions in TBS were drawn through the chamber with a Harvard syringe pump (Harvard Apparatus, Holliston, MA), with the wall shear stress being proportional to the flow rate.33
In studies of GP Ib-mediated cell adhesion and rolling on VWF, suspensions of either mutant CHO αβIX, wild-type CHO αβIX, or CHO βIX cells (1 × 106 cells/mL) were injected into the chamber, and the cells allowed to settle for 1 minute onto the immobilized VWF. In the peptide inhibition assay, the coverslips were postcoated with 50 μg/mL peptides for 1 hour at 37°C. The chamber was then perfused with TBS containing 0.5% BSA, generating wall shear stresses of 2.5, 10, or 20 dynes/cm2. The rolling of cells was recorded for 2 minutes on videotape. Acquired images were analyzed offline using MetaMorph software (Universal Images, West Chester, PA) to quantify the number of adherent cells and their rolling velocities.34 Cells considered to be rolling were those that translocated over the matrix while maintaining constant contact. The rolling velocity was defined as the distance a cell traveled during a defined period (μm/s).
Results
Characterization of the AP1 epitope
We used a 3-step approach to map in detail the AP1 epitope: first, we used chimeric canine-human GP Ibα polypeptides to roughly locate the binding region; second, we mutated to alanine individual amino acids in the region of human GP Ibα identified in step 1; and, third, we mutated those charged amino acids identified in step 2 to uncharged residues or to residues with the same charge to test whether the contributions of these residues to binding were purely electrostatic.
We first analyzed 2 sets of canine-human chimeras. The first set includes C1-225, C201-268, and C201-248,28 the name denoting the portion of the human sequence replaced by the corresponding canine sequence. Representative histograms for AP1 binding to the canine-human chimeras are shown in Figure 1B. AP1 binding was lost in the first 3 chimeras, but regained in C249-268. Each of the chimeras bound the control antibody, WM23 (not shown).
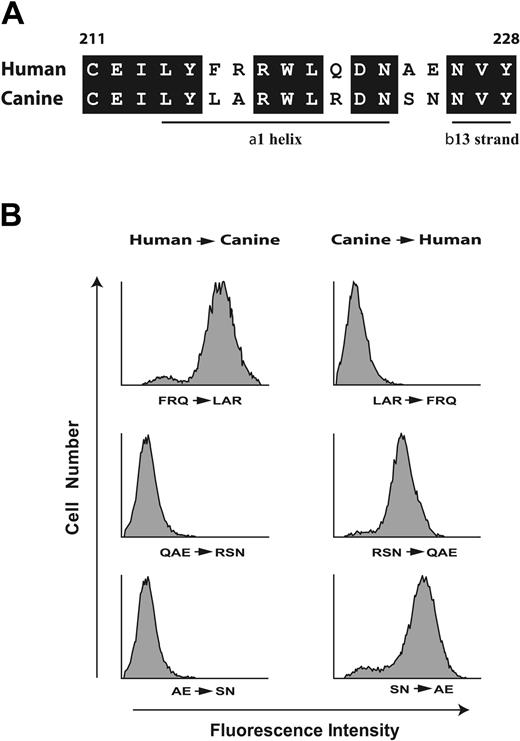
These findings narrowed the AP1 epitope to the sequence Phe201 to Glu225. In constructing the second set of chimeras, we took advantage of the fact that human and canine GP Ibα differ at only 5 sites within the sequence Cys211 to Tyr228 (Figure 2A), which are likely responsible for the difference in AP1 binding. The first type of chimeras (Figure 2B) had the human amino acids converted to the corresponding canine residues (Figure 2A), the second had the opposite changes, using C1-225 as the template (Figure 2B). AP1 binding was abolished when human Gln221/Ala224/Glu225 (QAE) or even only Ala224/Glu225 (AE) were replaced in combination by their canine counterparts. C1-225 acquired AP1 binding when the canine residues were converted to their human counterparts (Figure 2B). Furthermore, interchange of human Phe216/Arg217/Gln221 (FRQ) with canine Leu216/Ala217/Arg221 (LAR) has no effect on AP1 binding (Figure 2A). Taking into account the potential role of the conserved amino acids within this region, these results suggest that the AP1 epitope lies within the sequence Arg218 to Tyr228.
The AP1 epitope lies within the region spanning R218 to Y228. (A) Alignment of the human and canine sequences between Cys211 and Tyr228. Conserved amino acids are boxed and the locations of the α1 helix and β13 strands are noted. This designation is from the structure published by Uff et al,4 where they designated the β13 strand as extending from residue 226 to residue 228. In the 2 structures of the GP Ibα N-terminus in complex with the VWF A1 domain,3,35 the 13 β strand extends from residue 227 to residue 232, although it is not formally designated as β13. (B) Binding of the GP Ibα monoclonal antibody AP1 to CHO cells expressing chimeric GP Ibα was assessed by flow cytometry. Left panels show nonconserved residues of wild-type human GP Ibα within the sequence depicted in panel A mutated to corresponding canine residues; right panels, nonconserved canine residues mutated to the corresponding human residues using C1-225 as the template.
The AP1 epitope lies within the region spanning R218 to Y228. (A) Alignment of the human and canine sequences between Cys211 and Tyr228. Conserved amino acids are boxed and the locations of the α1 helix and β13 strands are noted. This designation is from the structure published by Uff et al,4 where they designated the β13 strand as extending from residue 226 to residue 228. In the 2 structures of the GP Ibα N-terminus in complex with the VWF A1 domain,3,35 the 13 β strand extends from residue 227 to residue 232, although it is not formally designated as β13. (B) Binding of the GP Ibα monoclonal antibody AP1 to CHO cells expressing chimeric GP Ibα was assessed by flow cytometry. Left panels show nonconserved residues of wild-type human GP Ibα within the sequence depicted in panel A mutated to corresponding canine residues; right panels, nonconserved canine residues mutated to the corresponding human residues using C1-225 as the template.
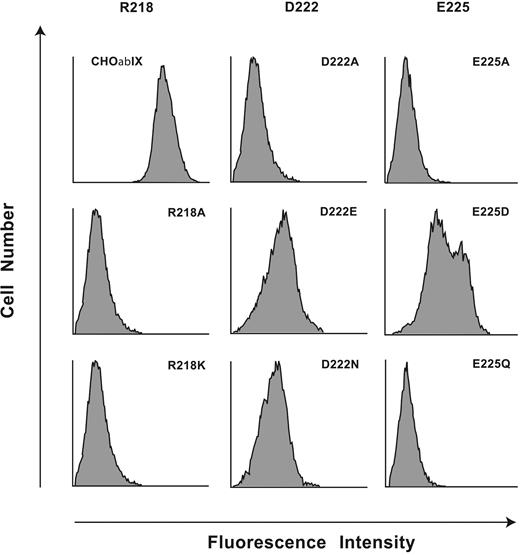
To reveal amino acids required for AP1 binding, we generated 11 mutants of human GP Ibα within the region Arg218 to Tyr228: 9 with individual amino acids converted to alanine (Arg218Ala, Leu220Ala, Gln221Ala, Asp222Ala, Asn223Ala, Glu225Ala, Asn226Ala, Val227Ala, Tyr228Ala), 1 alanine converted to serine (Ala224Ser), and Trp219 mutated to Phe because the Trp219Ala mutant was not expressed on the cell surface. All other mutants were expressed normally as part of recombinant GP Ib-IX complexes and displayed normal binding of the conformation-sensitive antibodies (data not shown). Only the replacement of Arg218, Asp222, or Glu225 with alanine abolished AP1 binding, which was retained by the other 8 mutants (Figure 3). This result raised the possibility that binding of AP1 to GP Ibα is largely electrostatic, as is the VWF-GP Ibα interaction.3,18
The charged amino acids Arg218, Asp222, and Glu225 are indispensable for the AP1 epitope. Flow cytometry histograms of AP1 binding to mutations of the 3 charged residues, Arg218, Asp222, and Glu225, replaced with either Ala, with a residue of the same charge, or in the case of the 2 acidic residues, with the corresponding amides. Results are presented as histograms of the log fluorescence intensities of 104 cells from the representative of 3 to 5 individual experiments.
The charged amino acids Arg218, Asp222, and Glu225 are indispensable for the AP1 epitope. Flow cytometry histograms of AP1 binding to mutations of the 3 charged residues, Arg218, Asp222, and Glu225, replaced with either Ala, with a residue of the same charge, or in the case of the 2 acidic residues, with the corresponding amides. Results are presented as histograms of the log fluorescence intensities of 104 cells from the representative of 3 to 5 individual experiments.
We then mutated the 3 charged amino acids to contain side chains with either no charge or with the same charge, the mutations being Arg218Lys, Asp222Asn, Asp222Glu, Glu225Gln, and Glu225Asp. Neither the Arg218Lys mutant nor the Arg218Ala mutant bound AP1 (Figure 3), reflecting an absolute requirement for Arg at this site. Mutation of Asp222 to either Asn or Glu markedly reduced AP1 binding, although a small amount of residual binding was retained in both mutants. Also, Glu225Gln showed no binding, while Glu225Asp partially restored AP1 binding, indicating that the carboxylic acid functional group at residue 225 was an important component of the AP1 epitope. Taken together, the data implicate both 3-dimensional conformation and electrostatic interactions as important for the AP1 epitope.
Botrocetin- and ristocetin-mediated VWF binding to GP Ibα
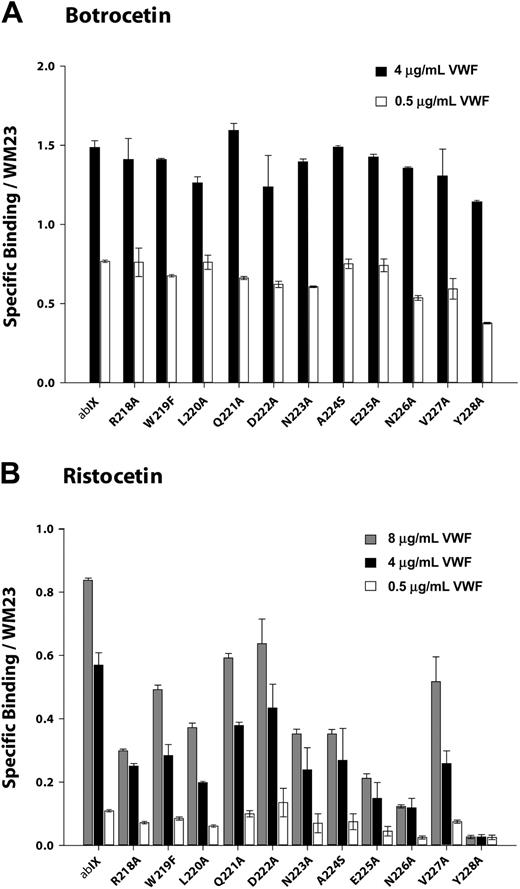
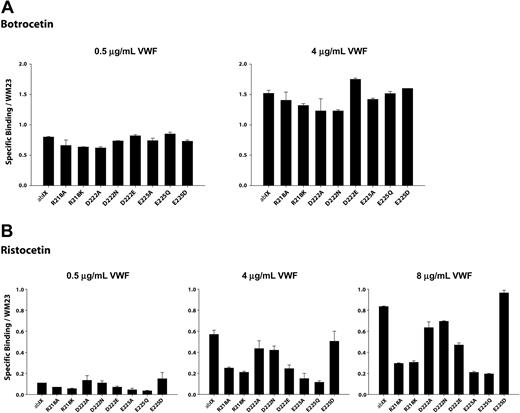
We measured VWF binding to cells expressing the GP Ibα mutants at several VWF concentrations in the presence of either 1.5 mg/mL ristocetin or 2 μg/mL botrocetin. The binding was normalized for differences in GP Ibα expression. As shown in Figures 4A and 5A, none of the point mutants was markedly deficient in botrocetin-induced binding, indicating that they did not substantially alter GP Ibα conformation.
Modulator-induced VWF binding to alanine mutants. Cells expressing either wild-type GP Ibα or alanine mutants were incubated with increased concentrations of VWF in the presence of (A) 2.0 μg/mL botrocetin or (B) 1.5 mg/mL ristocetin. Cells were then incubated with HRP-conjugated anti-human VWF antibody. Results were normalized based on GP Ibα surface expression, as determined by the binding of the antibody WM23. Error bars indicate SEM.
Modulator-induced VWF binding to alanine mutants. Cells expressing either wild-type GP Ibα or alanine mutants were incubated with increased concentrations of VWF in the presence of (A) 2.0 μg/mL botrocetin or (B) 1.5 mg/mL ristocetin. Cells were then incubated with HRP-conjugated anti-human VWF antibody. Results were normalized based on GP Ibα surface expression, as determined by the binding of the antibody WM23. Error bars indicate SEM.
Effect of charged mutations on modulator-induced VWF binding. Cells expressing either wild-type or mutant GP Ibα were incubated with increased concentration of VWF in the presence of (A) 2.0 μg/mL botrocetin or (B) 1.5 mg/mL ristocetin. The binding of VWF to mutant GP Ibα was assessed as described in the legend for Figure 4. Error bars indicate SEM.
Effect of charged mutations on modulator-induced VWF binding. Cells expressing either wild-type or mutant GP Ibα were incubated with increased concentration of VWF in the presence of (A) 2.0 μg/mL botrocetin or (B) 1.5 mg/mL ristocetin. The binding of VWF to mutant GP Ibα was assessed as described in the legend for Figure 4. Error bars indicate SEM.
Ristocetin-induced VWF binding studies were performed at a fixed concentration of ristocetin (1.5 mg/mL) and at VWF concentrations ranging from 0.5 μg/mL to 8 μg/mL. As seen in Figure 4B, VWF binding tended to increase as mutations progressed from Arg218Ala to Asp222Ala and to decrease between Asp222Ala to Val228Ala. This pattern was more obvious at higher VWF concentrations.
Mutations of the charged residues Arg218, Asp222, and Glu225 revealed an interesting pattern in ristocetin-induced VWF binding (Figure 5B). Mutations of R218 diminished binding to a similar extent (70%) whether the Arg residue was converted to Ala or the more conservative Lys. Mutations of Asp222 had the same effect regardless of whether the change was to Ala or Asn, reducing VWF binding by about 25%. Of interest, substitution of Asp222 with Glu, which maintains both charge and pKa, reduced binding to an even greater extent than the less conservative mutations. Mutation of Glu225 to either Ala or Gln reduced binding markedly, to approximately 20% of wild type. Unlike the situation with Asp222, however, replacement of the Asp with the carboxylic acid-containing side chain of Glu maintained full VWF binding. The 2 acidic residues thus appear to have different roles in VWF binding.
A peptide based on Leu214 to Val229 binds both AP1 and VWF and does not block GP Ibα-VWF interaction under flow
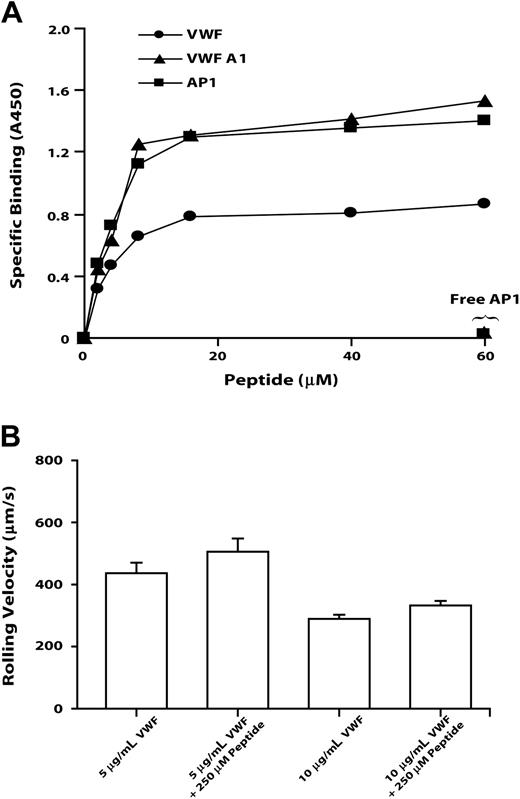
To verify that the sequence identified by AP1 epitope mapping directly binds both the antibody and VWF, we examined the binding of a synthetic peptide based on GP Ibα Leu214 to Val229 to native VWF, to the VWF A1 domain, and to AP1. The peptide bound the 3 ligands saturably, with half-maximal binding for all 3 occurring at approximately 2 μM (Figure 6). Peptide binding to each of the ligands was abolished by excess AP1. This peptide did not block the GP Ibα-VWF interaction, decreasing neither the number of CHO cells adherent on VWF (not shown) nor their rolling velocity (Figure 6B).
The GP Ibα AP1 peptide binds VWF, VWP A1, and AP1, but it does not inhibit GP Ib-mediated cell rolling on VWF. GP Ibα peptide encompassing Leu214 to Val229 binds immobilized VWF, VWF A1 domain, and AP1 (A) and affects the rolling of GP Ibα-expressing cells on immobilized VWF (B). (A) Increasing amounts of the GP Ibα Leu214 to Val229 peptide were incubated with immobilized VWF, VWF A1 domain, or AP1. Detection of peptide binding is described in “Materials and methods.” In the inhibition assay, free AP1 antibody and 60 μM peptide were added simultaneously. This figure is representative of 3 individual experiments. (B) Cells expressing wild-type GP Ibα were introduced into a parallel-platelet flow chamber in the presence of 250 μM peptide for 1 minute and allowed to settle on the surface of immobilized VWF (coated with 5 or 10 μg/mL VWF). The chamber was then perfused with TBS at flow rates that generated wall shear stresses of 20 dynes/cm2. The rolling velocities of cells in the absence of peptide were measured and compared with that in the presence of peptide. Error bars indicate SEM.
The GP Ibα AP1 peptide binds VWF, VWP A1, and AP1, but it does not inhibit GP Ib-mediated cell rolling on VWF. GP Ibα peptide encompassing Leu214 to Val229 binds immobilized VWF, VWF A1 domain, and AP1 (A) and affects the rolling of GP Ibα-expressing cells on immobilized VWF (B). (A) Increasing amounts of the GP Ibα Leu214 to Val229 peptide were incubated with immobilized VWF, VWF A1 domain, or AP1. Detection of peptide binding is described in “Materials and methods.” In the inhibition assay, free AP1 antibody and 60 μM peptide were added simultaneously. This figure is representative of 3 individual experiments. (B) Cells expressing wild-type GP Ibα were introduced into a parallel-platelet flow chamber in the presence of 250 μM peptide for 1 minute and allowed to settle on the surface of immobilized VWF (coated with 5 or 10 μg/mL VWF). The chamber was then perfused with TBS at flow rates that generated wall shear stresses of 20 dynes/cm2. The rolling velocities of cells in the absence of peptide were measured and compared with that in the presence of peptide. Error bars indicate SEM.
Adhesion of mutant CHOαβIX cells to immobilized VWF under flow
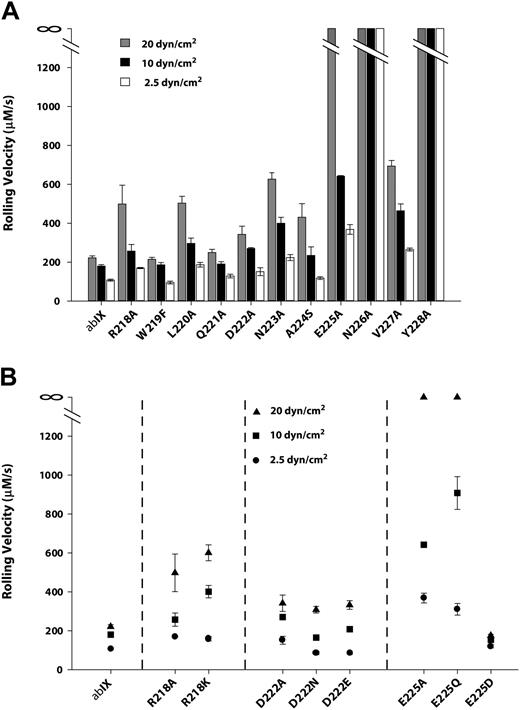
In the presence of hydrodynamic shear stress, CHOαβIX cells adhere to immobilized VWF in the absence of modulators, with the same molecular requirements as platelet adhesion.36,37 We therefore examined the binding of mutant-expressing cells to VWF under flow. Cells were allowed to settle on the VWF matrix and then subjected to shear stresses of 2.5, 10, or 20 dyn/cm2. The rolling velocity of the wild-type cells increased in response to increases in shear stress (Figure 7A), rolling at mean velocities of 107.4 μm/s, 161 μm/s, and 220.8 μm/s at 2.5, 10, and 20 dyn/cm2, respectively. The alanine mutants could be segregated into 2 groups based on the relative impairment in their interaction with the surface, as evidenced by increased rolling velocities. The mutants of residues from Arg218 to Asp222 as a group were markedly less impaired in their interaction with the VWF surface than the second group, comprising mutants of residues from Asn223 to Tyr228. In the first group, Trp219Phe, Gln221Ala, and Asp222Ala rolled at similar velocities as wild-type cells at each shear stress, whereas Arg218Ala and Leu220Ala rolled approximately twice as fast as wild-type cells at all shear stresses. In the second group, Asn226Ala and Tyr228Ala completely failed to bind the VWF surface, even at the lowest shear stress. At 20 dyn/cm2, Asn223Ala and Glu225Ala lost their ability to tether and roll on the VWF surface. Ala224Ser and Val227Ala also showed increased rolling velocities with increased shear stress, with both of the velocities significantly higher than those of all the mutants in the first group. In some cases, the rolling velocity may have underestimated the degree of impairment in the VWF interaction, because markedly fewer cells in mutants Asn223Ala, Ala224Ser, and Glu225Ala interacted with the surface (< 1 cell per view field). Cell lines expressing wild-type GP Ibα, by contrast, generally had more than 20 rolling cells per view field.
The interaction of cells expressing GP Ibα mutants with immobilized VWF under flow. Cells expressing either wild-type (A) or charge mutants of (B) GP Ibα were introduced into a parallel-platelet flow chamber and allowed to settle on the surface of immobilized VWF for 1 minute before the chamber was perfused with TBS at flow rates that generated wall shear stresses of 2.5, 10, or 20 dynes/cm2. The rolling velocities of the mutant-expressing cells were measured and compared with the rolling velocity of cells expressing wild-type GP Ibα.
The interaction of cells expressing GP Ibα mutants with immobilized VWF under flow. Cells expressing either wild-type (A) or charge mutants of (B) GP Ibα were introduced into a parallel-platelet flow chamber and allowed to settle on the surface of immobilized VWF for 1 minute before the chamber was perfused with TBS at flow rates that generated wall shear stresses of 2.5, 10, or 20 dynes/cm2. The rolling velocities of the mutant-expressing cells were measured and compared with the rolling velocity of cells expressing wild-type GP Ibα.
The results of flow experiments with mutants of the 3 charged residues—Arg218, Asp222, and Glu225—closely mirrored the results of ristocetin-induced binding (Figure 7B). Arg218Ala and Arg218Lys behaved similarly, with moderate increase in rolling velocities compared with wild-type cells, especially at the highest shear stress. Mutants Asp222Ala and Asp222Asn displayed a mild impairment in the surface interaction, which was not improved in the conservative Asp222Glu substitution. Mutations Glu225Ala and Glu225Gln again severely impaired the interaction, and Glu225Asp not only restored function but also seemed to induce a mild gain of function.
Discussion
In this study, we explored the functional role of the GP Ibα C-terminal flanking sequence in the interaction of the GP Ib-IX complex with VWF. We began by characterizing in detail the AP1 epitope. We created a series of GP Ibα mutants, either by generating human-canine chimeras, or by single amino acid mutations, which we then expressed as components of GP Ib-IX complexes in CHO cells. The location of the epitope was first narrowed to the sequence Arg218 to Tyr228, which comprises the α1 helix and β13 strand as defined by the crystal structure.3 Within this sequence, 3 charged amino acids, Arg218, Asp222, and Glu225, play an indispensable role in AP1 binding, as evinced by the following: (1) AP1 binding was lost when any 1 of these 3 amino acids was mutated to alanine or to other neutral amino acids, and (2) replacement of any of the amino acids with residues of similar charge either abolished or greatly diminished AP1 binding. In contrast, the binding of VM16d, another conformation-sensitive monoclonal antibody whose epitope lies within the C-terminal disulfide loops, was unaffected by these point mutations (not shown). These studies indicate not only that the binding sites of AP1 and VM16d are distinct, but also that the mutations do not change the overall conformation of the C-terminal flanking domain.
Further evidence for a role of this region in the binding of both VWF and AP1 came from the finding that a peptide spanning Leu214 to Val229 bound saturably to immobilized VWF, to the VWF A1 domain, and to AP1, with half-maximal binding of the peptide to the 3 ligands occurring at 2 μM of added peptide (Figure 6A). The binding to each ligand was completely abolished by free AP1.
We then investigated whether the mutations affected the interaction of GP Ibα with VWF in the presence of either botrocetin or ristocetin or under shear conditions. Extensive previous work has shown that a negatively charged cluster downstream of the C-terminal flank is important in botrocetin-induced VWF binding and that even minor perturbations of this site can alter binding drastically.38-40 Consistent with this, the mutants within the C-terminal flank affected botrocetin-induced VWF binding only slightly (Figure 4A). By contrast, each mutant was defective in ristocetin-induced binding (Figure 4B). Accordingly, based on their rolling velocities on a VWF matrix under flow, the alanine mutants could be segregated into 2 groups: the first mutant group, affecting amino acids Arg218 to Asp222, showed less variable rolling velocity than did the second group affecting residues between Asn223 and Tyr228. In addition, mutants of the charged residues in general showed the same functional profile in the flow studies as in the binding studies using ristocetin. These results are consistent with the fact that the binding of VWF induced by ristocetin more closely resembles the shear-induced interaction than does botrocetin-induced binding.11 The mechanisms underlying the different effects of those 2 modulators on GP Ibα-VWF interaction have not been clarified.
These data suggest that the sequence Arg218 to Tyr228 contains 2 regions with distinct roles in VWF binding: the α1 helix appears to have a regulatory role and the β turn and β13 strand seem to bind VWF directly. In favor of the α1 helix serving a regulatory role is the finding that none of the mutations from Arg218 to Asp222 completely abrogated VWF binding or rolling on immobilized VWF. This is in marked contrast to the effect of mutations within the leucine-rich repeat region of GP Ibα described in Bernard-Soulier syndrome, which can completely abolish VWF binding while still allowing expression of the polypeptide on the cell surface.17 The regulatory region may function by forming intramolecular bonds that correctly orient the β-switch.3 In favor of the interpretation that the β turn and β13 strand bind VWF directly is the finding that mutations within the region Asn223 to Tyr228 abolish the GP Ibα-VWF interaction without inducing gross conformational changes detectable by the conformation-sensitive antibodies AK2, AN51, and VM16d (not shown). This interpretation is also favored by the fact that a peptide based on the sequence Leu214 to Val229 binds saturably to intact VWF and the isolated A1 domain (Figure 6A).
Recently, 3 crystallographic structures of human GP Ibα were independently determined.3,4,35 The first was a structure of a native GP Ibα fragment corresponding to residues 1 to 288. The investigators in this study used computer modeling to fit the preexisting VWF A1 structure into a complex with GP Ibα.4 The second structure was of a complex between the GP Ibα N-terminus and the VWF A1 domain, which was made possible by the introduction of gain-of-function mutations into both GP Ibα and VWF A1.3 The third structure reported was of a complex between wild-type GP Ibα and wild-type VWF A1.35 In the 3 structures, key structural elements of GP Ibα have been described in terms of the palm, thumb, and wrist of a human hand4 corresponding, respectively, to the N-terminal β-hairpin and 8 tandem LRRs, the regulatory disulfide-loop (“R-loop”) of the C-terminal flank, and the anionic/sulfated “wrist” (269-282) forming a flexible hinge linking the ligand-binding domain to the membrane-proximal sialomucin domain.3 The 3 models all revealed the involvement of N-terminal (β-hairpin) and C-terminal (β-switch) flanking sequences of GP Ibα in binding VWF A1. Of interest, the modeled structure4 of the complex was different from the 2 structures in which the A1 domain and GP Ibα were crystallized together, with the VWF A1 domain being docked in opposite orientation. Based on a molecular model, Uff et al4 suggested that negatively charged binding surfaces on the concave face of the LRRs and the anionic region mediated 2-step binding to the top of VWF A1, which can be regulated by an unmasking mechanism involving conformational changes of the R-loop. Huizinga et al,3 on the other hand, suggested that long-range electrostatic interactions involving the region encompassed by residues 59 to 128 of GP Ibα and a positively charged patch on the bottom of the VWF A1 domain position the receptor and ligand for subsequent bidentate binding. Dumas et al35 provided a structure similar to that of Huizinga et al,3 and suggested a potential mechanism by which the GP Ibα M239V mutation associated with platelet-type von Willebrand disease increases the affinity for A1. Our results are consistent with the interaction demonstrated in the crystal structure of the VWF A1-GP Ibα complex,3 which shows that residues Glu225, Asn226, and Tyr228 of the β13 strand directly contact Arg632, Glu596, and Lys599 in the α3 helix of VWF A1, forming the first contact site within the C-terminal flank through hydrogen bonds or salt bridges. Another contact site involves the R-loop of GP Ibα, within the sequence Val227 to Asn242, which directly interacts with the β3 region of A1. It seems reasonable to speculate that perturbation of the first contact site in the C-terminal flank either by elimination of the binding sites or by a conformational change would attenuate the affinity for VWF. The crystal structure of the complex between wild-type VWF A1 and wild-type GP Ibα N-terminus supports this idea by providing evidence that remote amino acid substitutions can modulate the affinity of the GP Ibα-A1 interaction by allosterically altering the binding sites, supporting an allosteric mechanism for enhanced VWF binding to platelet in platelet-type von Willebrand disease. Our results indicate that perturbations within the sequence Asn223 and Tyr228 completely abolish the GP Ibα-VWF interaction induced by ristocetin or elevated shear stress. Nevertheless, the synthetic peptide spanning Leu214 to Val229 did not completely block the GP Ibα-VWF interaction, but did increase slightly the rolling velocity of GP Ibα-expressing CHO cells on VWF (Figure 6B). One possible explanation for the failure of the peptide to block binding is that the contacts between VWF and residues that comprise the AP1 epitope play an ancillary role in the overall interaction, supporting the interaction but not being absolutely required for it. Although the mutations may disrupt this particular contact site, the resulting alteration of the conformation may destabilize the interaction between downstream sequences (after Val227) and the β3 region of A1. In support of this, Vicente et al41 previously demonstrated that a peptide encompassing Lys231 to Ser245 inhibited VWF binding to platelets by more than 50%, whereas a peptide spanning Gln221 to Asp235 inhibited only by approximately 10%.
On the other hand, the α1 helix—which has been shown to lie parallel to the β sheet on the concave face of the “palm” made up of leucine-rich repeats4 —was not shown to bind directly to the A1 domain.3 Consistent with that, our results showed that mutations within this region have, at best, mild effects on the GP Ibα-VWF interaction. Based on our results, we speculate that the GP Ibα monoclonal antibody AP1 inhibits VWF binding either by steric hindrance to prevent direct binding or by stabilizing the protruding position of the R-loop, preventing it from undergoing the conformational change that unmasks the binding site.
Recently, 2 crystal structures of human GP Ibα in complex with thrombin were determined.25,26 Both reports claim 2 distinct interacting interfaces between thrombin and GP Ibα. In one, Asp222, Asn223, Glu225, and Tyr228 form extensive direct and water-mediated contacts with residues Lys21, Tyr71, and Lys106 in thrombin exosite I25 ; in the other, Arg218, Asp222, and Asn223, which form part of the AP1 epitope, were reported to participate in hydrogen bonding or salt-bridge interactions with thrombin exosite I.26 Consistent with this, we found that our GP Ibα peptide inhibited thrombin clotting activity in a dose-dependent manner (Y.P. and J.A.L., unpublished data, September 2004). As opposed to its role in VWF binding, the AP1 epitope of GP Ibα binds directly to thrombin exosite I.
In summary, we have mapped the AP1 epitope in GP Ibα to a region within the sequence Leu214 to Val229 and show that mutations within this region disrupt VWF binding. This disruption is probably direct and indirect, as the purified A1 domain binds specifically and saturably to a peptide based on the epitope, but the peptide itself has only a modest effect on GP Ibα-mediated cell adhesion to immobilized VWF. This work therefore puts into perspective the relative contributions in VWF binding of different regions in the C-terminal flank of GP Ibα. In addition, the AP1 epitope is of interest because AP1 blocks the binding not only of VWF to GP Ibα, but also of the important ligands thrombin and Mac-1.
Prepublished online as Blood First Edition Paper, August 19, 2004; DOI 10.1182/blood-2004-07-2544.
Supported by grants HL64796 and P50 HL65967 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal