Contemporary treatment of pediatric acute myeloid leukemia (AML) requires the assignment of patients to specific risk groups. To explore whether expression profiling of leukemic blasts could accurately distinguish between the known risk groups of AML, we analyzed 130 pediatric and 20 adult AML diagnostic bone marrow or peripheral blood samples using the Affymetrix U133A microarray. Class discriminating genes were identified for each of the major prognostic subtypes of pediatric AML, including t(15;17)[PML-RARα], t(8;21)[AML1-ETO], inv16 [CBFβ-MYH11], MLL chimeric fusion genes, and cases classified as FAB-M7. When subsets of these genes were used in supervised learning algorithms, an overall classification accuracy of more than 93% was achieved. Moreover, we were able to use the expression signatures generated from the pediatric samples to accurately classify adult de novo AMLs with the same genetic lesions. The class discriminating genes also provided novel insights into the molecular pathobiology of these leukemias. Finally, using a combined pediatric data set of 130 AMLs and 137 acute lymphoblastic leukemias, we identified an expression signature for cases with MLL chimeric fusion genes irrespective of lineage. Surprisingly, AMLs containing partial tandem duplications of MLL failed to cluster with MLL chimeric fusion gene cases, suggesting a significant difference in their underlying mechanism of transformation.
Introduction
Acute myeloid leukemia (AML) is a relatively rare malignancy in the pediatric population, comprising only 15% to 20% of the acute leukemias diagnosed in this age group.1 Nevertheless, it remains a challenging disease with an inferior treatment outcome compared with pediatric acute lymphoblastic leukemia (ALL). Despite the introduction of new drugs, the aggressive use of allogeneic and autologous bone marrow transplantation, and improvements in supportive care, overall cure rates of AML in most contemporary treatment protocols remain below 60%.2-5 Further improvements in cure rates are likely to come from a better understanding of both the molecular abnormalities responsible for the formation and growth of the leukemic cells, and the mechanisms underlying drug resistance.
Increasingly, contemporary treatment protocols are incorporating methods for both accurate diagnosis and subsequent risk stratification. To achieve this requires not only distinguishing myeloblasts from lymphoblasts, but also assessing the extent of lineage commitment and differentiation, as well as the presence of specific molecular lesions or chromosomal abnormalities. Efforts over the last several decades have revealed AML to be a heterogeneous disease, with marked differences in cure rates between various genetic subtypes.6-9 Acute promyelocytic leukemia was the first clear example of a clinically distinct AML subtype, being characterized by FAB-M3 morphology and expression of the t(15;17)-encoded promyelocytic leukemia–retinoic acid receptor alpha (PML-RARα) fusion protein.10-14 Treatment with all-trans retinoic acid, which targets the PML-RARα fusion protein, induces differentiation and significantly improves cure rates when combined with chemotherapy.15-19 More recent work has resulted in the classification of AMLs into one of 3 prognostic or risk groups: favorable, including t(15;17)[PML-RARα], t(8;21)[AML1-ETO], inv16[CBFβ-MYH11], and, in the pediatric population, t(9;11); intermediate, including other MLL chimeric fusion genes or normal karyotypes; and unfavorable, including -5/del(5q),-7/del(7q), inv3/t(3;3), +8, and complex karyotypes.7,8,20,21 An additional poor risk subtype of AML that is seen primarily in the pediatric population is acute megakaryocytic leukemia (FAB-M7).22,23
High throughput parallel expression analysis using DNA-based microarrays has recently been applied to the diagnosis of acute leukemias and to the exploration of their underlying molecular pathology.24-26 Work from a number of different laboratories have identified unique expression signatures for the 3 major subtypes of favorable risk adult AML -t(15;17), t(8;21), and inv16,27-30 and for several rare genetic subtypes of AML.24,31 Whether these expression profiles will allow the accurate diagnosis of these specific subtypes of AML in the pediatric population remains to be determined. Similarly, it remains to be determined whether distinct expression signatures exist for some of the standard and high risk forms of pediatric AML.
To address these issues, we utilized oligonucleotide microarrays to analyze the expression of more than 22 000 genes in diagnostic leukemic blasts from 130 pediatric AML patient samples using the Affymetrix U133A oligonucleotide microarray. Our data demonstrate that expression profiling is not only a robust approach for the accurate identification of known lineage and molecular subtypes of pediatric AML, but also provides new insights into their underlying biology. In addition, only minimal differences were identified between the expression profiles of pediatric and adult AML cases that contained the same genetic lesions, suggesting that these de novo leukemia subtypes are the same diseases in different age groups. Lastly, a common expression signature was identified for acute leukemias that contain MLL chimeric fusion genes, irrespective of their lineage. This signature provides novel insights into the altered transcriptional program induced by the presence of an MLL fusion gene. Importantly, cases of AML that contain a partial tandem duplication of the MLL gene failed to express this transcriptional program, suggesting that these leukemias have a mechanism of transformation that differs from that of cases with MLL chimeric fusion genes.
Materials and methods
Samples
Bone marrow (BM) aspirates or peripheral blood (PB) samples were obtained at the time of diagnosis from pediatric (n=130) or adult (n=20) patients with de novo AML. Informed consent for the use of the leukemic cells for research was obtained from parents, guardians, or patients (as age-appropriate) in accordance with the Declaration of Helsinki, and study approval was obtained from the SJCRH institutional review board (IRB).
Mononuclear cells were purified from the diagnostic BM or PB samples by density gradient centrifugation and cryopreserved in liquid nitrogen. Samples included BM aspirates (n = 139), PB (n = 10), or therapeutic apheresis sample (n = 1). The majority (122 of 130) of the pediatric samples were from patients who were subsequently treated on St Jude Children's Research Hospital (SJCRH) AML protocols: AML83, AML87, AML91, or AML97. Results of these protocols have been published elsewhere.32-35 After 1992, children with t(15;17)-positive acute promyelocytic leukemia were not treated on AML studies but instead were treated either on POG9710 or by best clinical management. To increase the number of cases with PML-RARα and AML1-ETO, diagnostic samples (3 PML-RARα and 5 AML1-ETO) were obtained from pediatric patients in El Salvador who were treated through the SJCRH International Outreach Program.
All pediatric AML samples used in this study with the exception of one had a blast percentage equal to or greater than 65% after ficoll purification (Table S1). The average blast percentage for the samples included in this study was 86.9%. Only 10 samples had blast counts of less than 75% and only 2 samples had a blast count of less than 70%. All adult AML samples evaluated in this study had a blast count of more than 75% after ficoll purification.
The diagnosis and classification of AML were based on morphologic, cytochemical, and immunophenotypic criteria according to the revised French-American-British (FAB) classification.36-38 The diagnostic samples were also characterized by conventional cytogenetics, reverse transcriptase–polymerase chain reaction (RT-PCR) assays for PML-RARα, AML1-ETO, CBFβ-MYH11, and for the presence of MLL chimeric fusion genes by at least 2 of the following methods: cytogenetics, 11q23 fluorescence in situ hybridization (FISH), or RT-PCR for t(9;11)[MLL-AF9], t(11;19)[MLL-ENL], t(11;19)[MLL-ELL], t(10;11)[MLL-AF10], or t(4;11)[MLL-AF4]. Samples that lacked evidence of any of these chromosomal rearrangements were evaluated for the presence of internal tandem duplications of the MLL gene using a previously described RT-PCR–based assay (Table S6).39
Gene expression profiling
Detailed protocols for RNA extraction, assessment of integrity, and generation of labeled cRNA have been previously described, and can be obtained at http://www.stjuderesearch.org/data/ALL1 and http://www.stjuderesearch.org/data/ALL3. Arrays were scanned using a laser confocal scanner (Agilent, Palo Alto, CA) and then analyzed with Affymetrix Microarray Suite 5.0 (MAS 5.0). Detection values (present, marginal, or absent) were determined by default parameters, and signal values were scaled by global methods to a target value of 500. Minimal quality control parameters for inclusion in the study included more than 10% present calls and a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) 3′-to-5′ ratio of less than or equal to 3. Microarrays included in the study had an average present call of 38.75% (range, 20.2-50.4). Primary data are available at our web site at http://www.stjuderesearch.org/data/AML1.
Statistical analysis
Analysis was performed using 2-dimensional hierarchical clustering, principal component analysis (PCA), and discriminant analysis with variance (DAV) (GeneMaths software version 2.01; Applied Maths, Austin, TX). For class prediction, the pediatric data set was split into training (100 samples) and test (30 samples) sets, which were stratified with regard to AML1-ETO, CBFβ-MYH11, MLL chimeric fusion genes, PML-RARα, and FAB-M7 (Table 1). Prior to analysis, a variation filter was applied that removed any probe sets absent in all samples, had a maximum signal value within the data set less than or equal to 100, or had a maximum to minimum signal value of less than or equal to l00 (Table S5). Significance analysis of microarray (SAM) was performed exclusively using cases in the training set to select class discriminating genes.40 The discriminating genes were then used in an artificial neural network (ANN), supervised learning algorithm, and class assignment accuracies were initially assessed by 3-fold cross validation on the randomly selected stratified training set. The true accuracy was then determined on a blinded test group consisting of the remaining one-fourth of the samples. Details of the supervised learning algorithms and their use have been previously described.26,41
Pediatric AML subgroup distribution
Genetic subgroup . | Training, no. . | Test, no. . |
|---|---|---|
| PML-RARα | 11 | 4 |
| AML1-ETO | 16 | 5 |
| CBFβ-MYH11 | 10 | 4 |
| MLL chimeric fusion genes | 18 | 5 |
| FAB-M7 | 7 | 3 |
| Other | 38 | 9 |
| Total | 100 | 30 |
Genetic subgroup . | Training, no. . | Test, no. . |
|---|---|---|
| PML-RARα | 11 | 4 |
| AML1-ETO | 16 | 5 |
| CBFβ-MYH11 | 10 | 4 |
| MLL chimeric fusion genes | 18 | 5 |
| FAB-M7 | 7 | 3 |
| Other | 38 | 9 |
| Total | 100 | 30 |
Cases were randomly distributed into training and test sets, stratifying for genetic subgroups and FAB-M7.
Results
Expression profile of genetic risk groups of pediatric AML
Expression profiles were obtained from diagnostic samples of leukemic blasts from 130 pediatric patients with AML using the Affymetrix HG-U133A microarrays. The leukemia samples had an average blast percentage close to 90%. Cases were selected to provide a representation of the known morphologic, genetic, and prognostic subtypes of pediatric AML, and included cases with t(15;17)[PML-RARα], t(8;21)[AML1-ETO], inv16[CBFβ-MYH11], MLL chimeric fusion genes, acute megakaryocytic morphology (FAB-M7), or lacking any of these features (Table 1, Table S1, Table S2). The primary data are available at http://www.stjuderesearch.org/data/AML1.
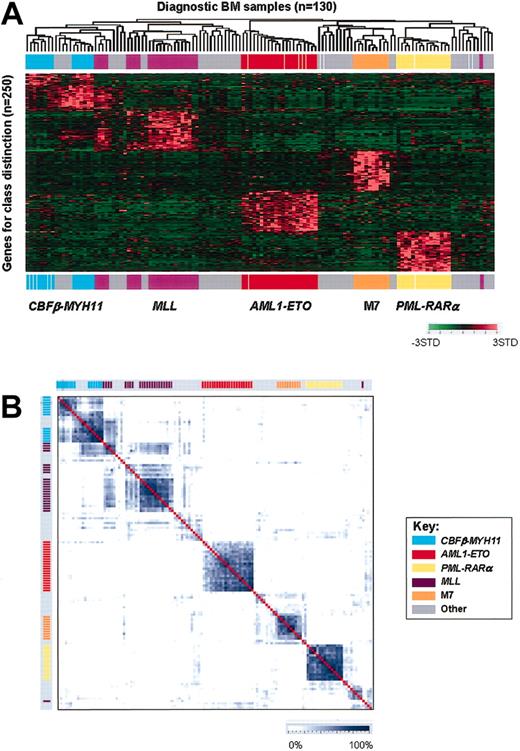
To reduce the complexity of the data, a variation filter was applied to remove probe sets that showed minimal variation across the data set (“Materials and methods”). In an initial analysis of the filtered data using an unsupervised 2-dimensional hierarchical clustering algorithm, relatively good clusters were observed for several of the genetic and morphologic AML subtypes, although the tightness of clustering was less than that seen in pediatric ALL (Figure 1, Figure S1, and Yeoh et al26 and Ross et al41 ). As shown in Figure 1, relatively tight grouping was observed for the genetic subgroups AML1-ETO, PML-RARα, and MLL chimeric fusion genes, and for the morphologic subgroups FAB-M3, M7, and M4/M5. Unexpectedly, however, AMLs that expressed the inv16-encoded CBFβ-MYH11 failed to cluster using a variety of different unsupervised clustering algorithms. The failure of this subgroup to tightly cluster indicates significant heterogeneity within the gene expression profile of these cases.
Unsupervised cluster analysis of pediatric AMLs. Expression profiles of the diagnostic leukemic blasts from 130 cases of pediatric AML were obtained using the U133A Affymetrix microarray. The expression data were then filtered to remove any probe sets that failed to show significant variation in expression across the data set. The remaining 17 051 probe sets were then used in an unsupervised 2-dimensional hierarchical clustering algorithm, and the resultant dendrogram is shown. Indicated below the dendrogram are the genetic subtype and FAB morphology for each case according to the indicated color codes.
Unsupervised cluster analysis of pediatric AMLs. Expression profiles of the diagnostic leukemic blasts from 130 cases of pediatric AML were obtained using the U133A Affymetrix microarray. The expression data were then filtered to remove any probe sets that failed to show significant variation in expression across the data set. The remaining 17 051 probe sets were then used in an unsupervised 2-dimensional hierarchical clustering algorithm, and the resultant dendrogram is shown. Indicated below the dendrogram are the genetic subtype and FAB morphology for each case according to the indicated color codes.
We next set out to identify expression signatures for each of the known prognostically important AML subtypes including AML1-ETO, PML-RARα, CBFβ-MYH11, MLL chimeric fusion genes, and FAB M7. Discriminating genes were selected using SAM on a training set of pediatric AML cases. The numbers of discriminating probe sets per leukemia subtype at a 5% false discovery rate (FDR) were: AML1-ETO, 764; PML-RARα, 2521; CBFβ-MYH11, 63; MLL chimeric fusion genes, 2218; and FAB M7, 1242. Consistent with the observed heterogeneity noted above in the expression profile of CBFβ-MYH11 leukemias, this subtype also had the smallest number of class discriminating genes.
The expression profiles obtained using the top 50–ranked genes for the 5 prognostically important subgroups are illustrated in Figure 2A using a 2-dimensional hierarchical clustering algorithm (see also Figure S2 and Table S7-S11). As shown, using the unique class-specific expression signatures, we were able to obtain relatively tight clustering of cases within each of the 5 leukemia subtypes, including those expressing CBFβ-MYH11. Thus, distinct expression signatures can be identified for each of the known prognostically important AML leukemia subtypes.
Expression profiles of pediatric AMLs. (A) Hierarchical clustering of 130 diagnostic pediatric AML samples (columns) versus 250 class discriminating genes (rows). The genes used in this analysis are the top 50–ranked genes per group as selected by SAM. For genes that had more than one probe set selected as a class discriminator, the highest-ranked probe set was used for this figure. Probe set signal values were normalized to the mean across the entire data set and the relative value for each case is represented by a color, with red representing high expression and green representing low expression (scale shown in the lower right). The genetic subtype of each case is indicated by colored bars across the top and bottom of the panel. (B) Similarity plot of 130 pediatric AML diagnostic samples using the top 50–ranked genes (1 probe set per gene) for each subgroup as selected by SAM. Similarities are plotted using a scale that is based on Pearson correlation coefficients calculated for pairwise comparisons using the expression data. The degree of similarity between cases is displayed using the blue color scale at the bottom of the figure. Genetic groups are indicated by the color bars along the top and side of the similarity plot and are arranged identically as in panel A.
Expression profiles of pediatric AMLs. (A) Hierarchical clustering of 130 diagnostic pediatric AML samples (columns) versus 250 class discriminating genes (rows). The genes used in this analysis are the top 50–ranked genes per group as selected by SAM. For genes that had more than one probe set selected as a class discriminator, the highest-ranked probe set was used for this figure. Probe set signal values were normalized to the mean across the entire data set and the relative value for each case is represented by a color, with red representing high expression and green representing low expression (scale shown in the lower right). The genetic subtype of each case is indicated by colored bars across the top and bottom of the panel. (B) Similarity plot of 130 pediatric AML diagnostic samples using the top 50–ranked genes (1 probe set per gene) for each subgroup as selected by SAM. Similarities are plotted using a scale that is based on Pearson correlation coefficients calculated for pairwise comparisons using the expression data. The degree of similarity between cases is displayed using the blue color scale at the bottom of the figure. Genetic groups are indicated by the color bars along the top and side of the similarity plot and are arranged identically as in panel A.
Despite the ability to identify class-specific expression signatures, significant heterogeneity in the signatures continued to be observed among cases in both the CBFβ-MYH11 and MLL chimeric fusion gene subgroups (Figure 2A). To further assess the relationship both within and between leukemia subtypes, we next analyzed the expression data in pair-wise comparisons to assess the degree of relatedness between cases. The data are displayed using a 2-dimensional plot in which similarities are plotted using a scale that is based on Pearson correlation coefficients calculated for pair-wise comparisons using the expression data for the 250 class discriminating genes. As shown in Figure 2B, the similarities of cases within a leukemia subtype were very high for AML1-ETO, PML-RARα, and FAB-M7. By contrast, more heterogeneity was observed among cases within the CBFβ-MYH11 and MLL chimeric fusion gene subgroups.
Close examination of the expression profiles and similarity plots for cases with either MLL chimeric fusion genes or CBFβ-MYH11 suggests the existence of distinct subgroups. However, the observed variation could not be completely explained by differences in the structure of chromosomal rearrangements, extent of differentiation, or presence of specific secondary mutations (Figure S2 and associated description). Thus, the underlying reason for the observed heterogeneity remains unknown.
Biologic insights from the class defining genes
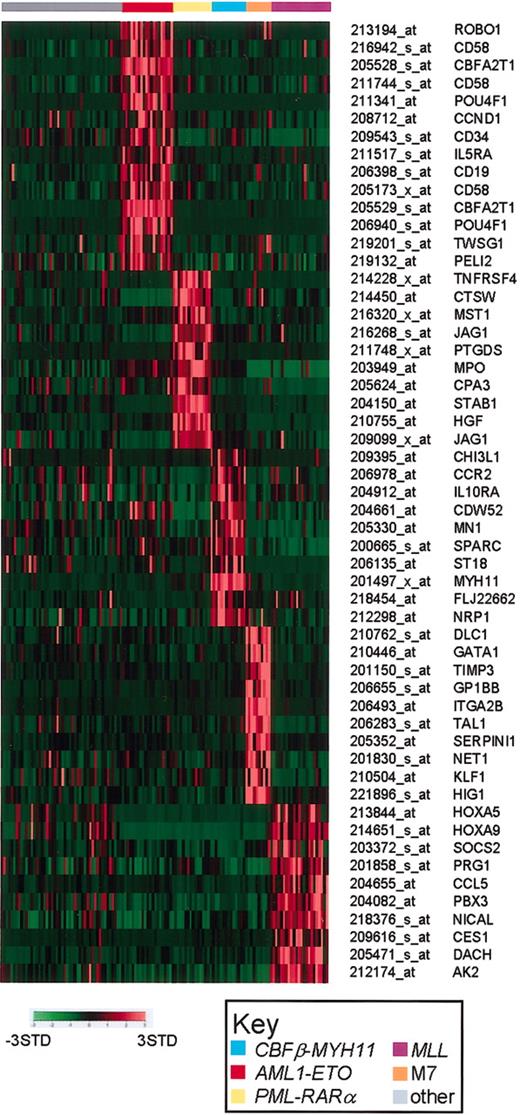
The identified class discriminating genes should provide unique insights into the underlying biology of the different leukemia subtypes and have the potential to serve as unique class-specific diagnostic or therapeutic targets. The patterns of expression of a select subset of class discriminating genes are shown in Figure 3. For each leukemia subtype, the class specific genes shown include examples of previously defined class-specific markers, as well as subtype-specific genes defined exclusively by our analysis.
AML subtype-specific class discriminating genes. Shown are representative genes that are highly correlated with the individual genetic subtypes of AML. Probe set signal values are normalized to the mean for the data set and the expression for each case is then represented by color, with red representing deviation above the mean and green representing deviation below the mean. The leukemia subtype is indicated at the top of the figure, and the Affymetrix probe set number and gene symbol are listed on the right side of the figure.
AML subtype-specific class discriminating genes. Shown are representative genes that are highly correlated with the individual genetic subtypes of AML. Probe set signal values are normalized to the mean for the data set and the expression for each case is then represented by color, with red representing deviation above the mean and green representing deviation below the mean. The leukemia subtype is indicated at the top of the figure, and the Affymetrix probe set number and gene symbol are listed on the right side of the figure.
The aberrant coexpression of the hematopoietic progenitor marker CD34 and the B-cell antigen CD19 are a hallmark of AML1-ETO–expressing leukemic blasts.42 Similarly, the unique high-level expression of ETO (CBFA2T1) is expected. More surprising is the number of AML1-ETO class discriminating genes whose homologs in Drosophila have been shown to be involved in developmental processes. These include roundabout (ROBO1), twisted (TWSG1), and pellino homolog 2 (PEGI2). Although no direct functional relationship has been established between the Drosophila AML1 homolog (runt, a pair-rule gene) and these other genes, their specific high expression in this leukemia subtype warrants further exploration. Lastly, the POU4F1 transcription factor, a proposed modulator of p53 transcription, is overexpressed more than 30-fold, and is the second-highest-ranking AML1-ETO class discriminating gene (Table S7).43
The class discriminating genes for PML-RARα, CBFβ-MYH11, and FAB-M7 each include genes that encode proteins characteristic of the specific myeloid differentiation stage or lineage. For example, PML-RARα includes hepatocyte growth factor (HGF), myeloperoxidase (MPO), and carboxypeptidase A3 (CPA3); CBFβ-MYH11 includes CDW52 and chitinase 3-like (CHI3L1); and FAB-M7 includes the megakaryocytic lineage markers glycoprotein Ib and IIb (GP1BB and ITGA2B). Also prominent among the list of class discriminating genes are growth factors, growth factor receptors, and somewhat surprising, putative tumor suppressors. The latter includes meningioma (disrupted in balanced translocation) 1 (MN1), and suppressor of tumorgenicity 18 (ST18) in CBFβ-MYH11, and deleted in liver cancer 1 (DLC1) in FAB-M7. Direct sequence analysis of these genes will be required to determine if they encode wild-type or mutant proteins. The genes included in the MLL chimeric fusion gene signature are described in more detail in “The significance of MLL gene rearrangement in pediatric ALL and AML.”
CBFβ-MYH11 and AML1-ETO target the genes that encode the AML1/CBFβ transcription factor complex and constitute 2 of the major subtypes of “so-called” core-binding factor leukemias. To better assess the relationship between these leukemia subtypes, we determined the number of genes whose expression significantly differed between subtypes. In this analysis the smaller the number of differentially expressed genes between 2 leukemia subtypes the closer their relationship. As shown in Table 2, CBFβ-MYH11 leukemias were most closely related to cases having either AML1-ETO or MLL gene rearrangements. By contrast, AML1-ETO leukemias showed significantly more similarity to CBFβ-MYH11 than to MLL chimeric fusion gene cases. Thus, as expected, a high degree of similarity is identified between the 2 major subtypes of core-binding factor leukemias; however, a surprising degree of similarity is also seen between CBFβ-MYH11 and MLL chimeric fusion gene cases, and may be reflective of similarities in their extent of monocytic differentiation.
Pair-wise comparisons showing the number of probe sets that differ between group 1 and group 2 by SAM
Group 1/Group 2 . | CBFβ-MYH11 . | AML1-ETO . | MLL . | PML-RARα . | M7 . |
|---|---|---|---|---|---|
| CBFβ-MYH11 | - | 637 | 547 | 2081 | 1519 |
| AML1-ETO | 637 | - | 2140 | 1533 | 1599 |
| MLL | 547 | 2140 | - | 2742 | 1610 |
| PML-RARα | 2081 | 1533 | 2742 | - | 1992 |
| FAB-M7 | 1599 | 1599 | 1610 | 1992 | - |
Group 1/Group 2 . | CBFβ-MYH11 . | AML1-ETO . | MLL . | PML-RARα . | M7 . |
|---|---|---|---|---|---|
| CBFβ-MYH11 | - | 637 | 547 | 2081 | 1519 |
| AML1-ETO | 637 | - | 2140 | 1533 | 1599 |
| MLL | 547 | 2140 | - | 2742 | 1610 |
| PML-RARα | 2081 | 1533 | 2742 | - | 1992 |
| FAB-M7 | 1599 | 1599 | 1610 | 1992 | - |
Values determined using SAM at the 90th percentile and a 1% false discovery rate.
indicates identity
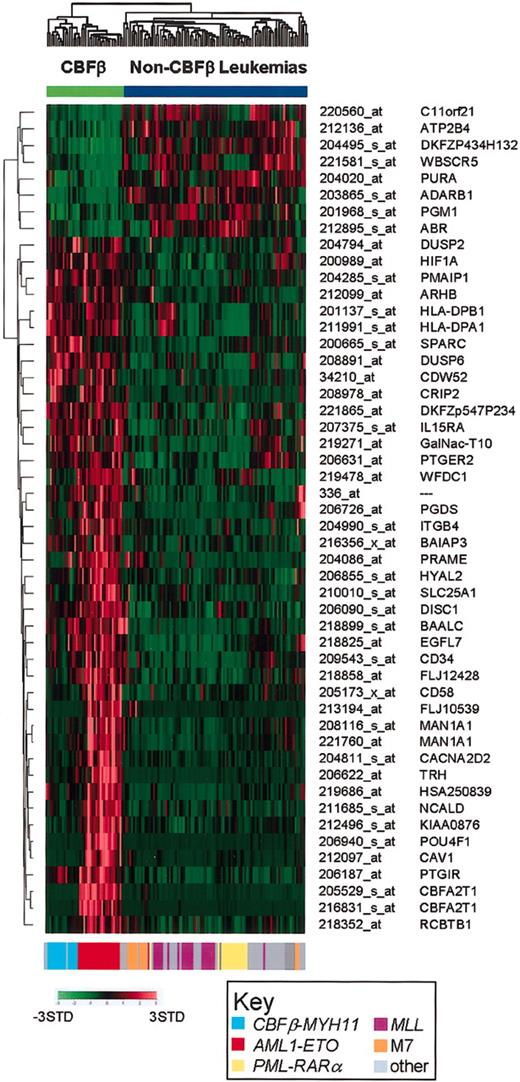
Based on the known relationship between CBFβ-MYH11 and AML1-ETO leukemias, and the observed similarities outlined above, we also selected genes that could discriminate these 2 subtypes of core-binding factor leukemias from all other leukemia subtypes. The top 50 discriminating genes are shown in Figure 4 and listed in Table S12. As illustrated, a subset of the discriminating genes are either over- or underexpressed in both CBFβ-MYH11 and AML1-ETO subgroups, whereas the expression of other discriminating genes appears to be primarily expressed in AML1-ETO cases. Most of the latter genes were also selected as AML1-ETO only class discriminating genes, as shown in Figure 2A.
Expression signature of core-binding factor AMLs. Two-dimensional hierarchical clustering of the 130 AML cases using the top 50–ranked discriminating probe sets for the core-binding factor (CBF) leukemias (AML1-ETO and CBFβ-MYH11 cases). The genetic subtype of each case is presented by a color-coded bar at the bottom of the figure, using the same color scheme used in Figures 1, 2, 3. The probe set number and gene symbol for the discriminating genes are listed on the right. The normalized expression level for each gene is represented by a color using the scale shown in the lower left corner. Cases were clustered using a cosine function.
Expression signature of core-binding factor AMLs. Two-dimensional hierarchical clustering of the 130 AML cases using the top 50–ranked discriminating probe sets for the core-binding factor (CBF) leukemias (AML1-ETO and CBFβ-MYH11 cases). The genetic subtype of each case is presented by a color-coded bar at the bottom of the figure, using the same color scheme used in Figures 1, 2, 3. The probe set number and gene symbol for the discriminating genes are listed on the right. The normalized expression level for each gene is represented by a color using the scale shown in the lower left corner. Cases were clustered using a cosine function.
Expression profiling as a diagnostic tool
A major goal of this study was to assess the ability of gene expression profiling to accurately diagnose the prognostically important AML subtypes. To examine this, class discriminating genes identified using SAM were used in an ANN-based supervised learning algorithm to classify cases into PML-RARα, AML1-ETO, CBFβ-MYH11, MLL chimeric fusion gene, or FAB-M7. The assignment to one of these leukemia subtypes required that the ANN generated node value for classification exceeded a 95% confidence level (view Supplemental Data on the Blood web site; see the Supplemental Data link at the top of the online article). Any case that was not classified with high confidence into one of these 5 subgroups was labeled as “other.” Using the top 20– to top 50–ranked discriminating genes for each subgroup, very high prediction accuracies were achieved on a randomly selected training set that consisted of three-fourths of the total cases. When this classification model was then applied to a blinded test set consisting of the remaining 30 samples, 100% diagnostic accuracies were obtained for PML-RARα, AML1-ETO, CBFβ-MYH11, and FAB-M7, and 93% accuracy for cases with MLL chimeric fusion gene, for an overall accuracy of 93% (95% confidence interval [CI], 79%-99%; Table 3). Optimal class assignment was achieved with as few as 5 genes (the smallest number tested) for PML-RARα, AML1-ETO, CBFβ-MYH11, and FAB-M7, and 35 genes for cases with MLL gene rearrangements (data not shown).
Diagnostic accuracies for pediatric AML
. | Training, n = 100 . | Test, n = 30 . |
|---|---|---|
| PML-RARα | 100 | 100 |
| AML1-ETO | 100 | 100 |
| CBFβ-MYH11 | 100 | 100 |
| MLL | 98 | 93 |
| FAB-M7 | 100 | 100 |
. | Training, n = 100 . | Test, n = 30 . |
|---|---|---|
| PML-RARα | 100 | 100 |
| AML1-ETO | 100 | 100 |
| CBFβ-MYH11 | 100 | 100 |
| MLL | 98 | 93 |
| FAB-M7 | 100 | 100 |
Values determined using 25 genes selected by SAM at 90th percentile with 5% FDR rate for PML-RARα, AML1-ETO,CBFβ-MYH11, and FAB-M7 and 50 genes for MLL.
Since the incidence of AML is significantly higher in adults than in pediatric patients, we next assessed whether the expression profiles identified using pediatric patients could be used to accurately classify these specific subtypes of de novo AML in adults. For this analysis, a selected set of 20 adult de novo AML diagnostic samples were used (Table S3). This sample data set contained examples of the specific leukemia subtypes under study, with the exception of FAB-M7, which is exceedingly rare in adult patients. We first used SAM to calculate the number of genes at a 1% FDR that differed between like subgroups of pediatric and adult de novo AML. For this analysis, a sufficient number of adult cases were available only for PML-RARα, AML1-ETO, and MLL chimeric fusion gene subgroups. As shown in Table 4, minimal or no significant differences existed between adult and pediatric cases within these specific subtypes of AML. Next we used the adult cases as a second blinded test set and applied the discriminating genes and supervised learning algorithms developed using pediatric AML cases to see if we could accurately classify the adult cases. As shown in Table 5, using ANN we were able to obtain very high diagnostic accuracies for each of the genetic and morphologic subtypes, with an overall accuracy of 90% (95% CI, 68%-98%). Although no FAB-M7 cases were included in this data set, no samples were misclassified as FAB-M7. These data demonstrate that there are minimal differences between pediatric and adult cases of these specific subtypes of de novo AML. Moreover, the class discriminating genes selected using the pediatric cases can be used to accurately diagnose adult cases.
Number of probe sets selected to differentiate between pediatric and adult AML subtypes
. | 1% FDR . | 5% FDR . |
|---|---|---|
| PML-RARα | 0 | 0 |
| AML1-ETO | 0 | 172 |
| MLL | 0 | 17 |
. | 1% FDR . | 5% FDR . |
|---|---|---|
| PML-RARα | 0 | 0 |
| AML1-ETO | 0 | 172 |
| MLL | 0 | 17 |
Genes selected with SAM at 90th percentile with the indicated false discovery rate (FDR).
Adult AML case distribution and classification accuracy
. | No. of cases . | Accuracy, % . |
|---|---|---|
| PML-RARα | 4 | 100 |
| AML1-ETO | 6 | 100 |
| CBFβ-MYH11 | 2 | 95† |
| MLL | 6 | 95 |
| Other | 2 | NA |
| Overall | 20 | 90 |
. | No. of cases . | Accuracy, % . |
|---|---|---|
| PML-RARα | 4 | 100 |
| AML1-ETO | 6 | 100 |
| CBFβ-MYH11 | 2 | 95† |
| MLL | 6 | 95 |
| Other | 2 | NA |
| Overall | 20 | 90 |
There were no FAB-M7 cases in the adult data set. No samples were mistakenly predicted as FAB-M7.
NA indicates not applicable.
The CBFβ-MYH11 data set contains one case that expresses both CBFβ-MYH11 and BCR-ABL, a combination described once before.45 The ANN supervised learning algorithm failed to classify this case as CBFβ-MYH11
Although knowing the specific subtypes of AML can influence therapeutic decisions, additional prognostic markers are needed to more accurately predict whether a patient can be cured by a specific therapeutic approach. Identifying a gene expression–based outcome predictor that could provide additional prognostic information, either independent of or within a genetic subtype, would be a significant advance. We examined the association of expression with outcome in pediatric AML. For this analysis, we used a cohort of 98 patients treated on the AML 87, AML 91, and AML 97 protocols, excluding those patients with t(15;17). Genes were selected using a generalized Mantel statistic that examined the association of expression with time to relapse (Supplemental Data, section VI).44 This procedure selected 3 probe sets whose significance at the α equals .001 level was robust against the exclusion of any one patient from analysis. Two of the probe sets were also significant in a multivariable Cox proportional hazards regression analysis applied to the training cohort. The training cohort data were then used to develop a prognostic score function based on the expression of the 2 probe sets. In the validation cohort, time to relapse or progression became significantly shorter (P = .0442) as the value of this score increased. To further explore the prognostic significance of these genes, we applied the score function for the 2 identified genes to the adult cases included in our study (see Supplemental Data, section V). Although the same trend was observed in this small adult cohort, the association was not statistically significant at the traditional α equals 0.05 level (P = .0898). Thus, the value of these genes as predictors of prognosis independent of the genetic subtypes of de novo AML will require assessing their performance in larger cohorts of pediatric and adult patients.
A recently published paper reported the identification of 35 genes that could serve together as an outcome predictor in pediatric AML.29 When tested on our data set, the expression level of these genes did not show correlation with outcome (Supplemental Data, section V).
The significance of MLL gene rearrangements in pediatric ALL and AML
Translocations targeting the MLL gene are seen across the spectrum of acute leukemias including both B- and T-lineage ALLs and AMLs. Cases of B-precursor ALL with MLL chimeric fusion genes, like several other genetic subtypes of ALL, can be easily identified as a unique biologic subtype by their expression profile.25,26,41 As noted above, we can also identify an expression signature that can accurately identify cases of AML with MLL chimeric fusion genes. Defining expression signatures associated with MLL chimeric fusion genes irrespective of the lineage of the acute leukemia should provide valuable insights into common downstream pathways that are required for MLL-mediated transformation. To explore this possibility, we combined the pediatric AML data set from this study with a data set of 132 pediatric ALL cases,41 and 5 additional pediatric cases of T-ALL with MLL translocations (Table S4). These 267 pediatric acute leukemias include 48 cases with MLL chimeric fusion genes (20 B-lineage ALLs, 5 T-lineage ALLs, and 23 AMLs), and 219 acute leukemias that lack this genetic lesion (98 B-lineage ALLs, 14 T-lineage ALLs, and 107 AMLs). No examples of therapy-induced AMLs are included in this data set.
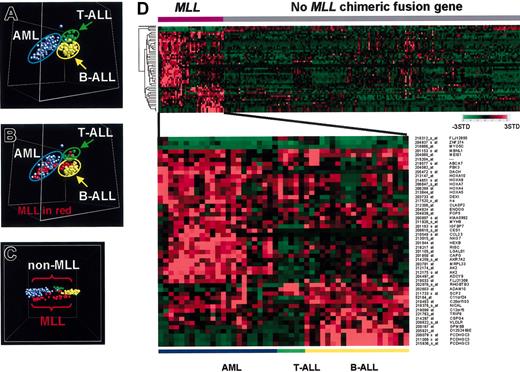
We initially analyzed this data set using the unsupervised clustering algorithm PCA to assess the major grouping of the cases based solely on their gene expression profiles. Using all genes that passed a variation filter, 3 major subgroups were identified and shown to correspond to B-lineage ALL (B-ALL), T-lineage ALL (T-ALL), and AML (Figure 5A). Importantly, the cases with MLL chimeric fusion genes segregated according to their lineage (Figure 5B). Thus in this analysis, cases with MLL chimeric fusion genes did not cluster as a unique subgroup, but instead clustered according to their lineage of origin.
Gene expression profiles of pediatric acute leukemia with MLL chimeric fusion genes. (A) Multidimensional scaling plot generated using unsupervised principle components analysis with a combined data set containing 130 AML cases, 132 ALL cases,41 and 5 additional T-lineage ALL (T-ALL) cases that contain MLL chimeric fusion genes. A variation filter was applied to remove any probe sets that showed minimal variation in expression across this data set, and the analysis was performed with the remaining 17 944 probe sets. Each case is represented by a colored sphere, with AML cases indicated by blue, B-progenitor lineage ALL (B-ALL) by yellow, and T-ALL by green. Acute leukemia cases cluster based on lineage. (B) The same PCA analysis as shown in panel A, except that cases that contain an MLL chimeric fusion gene are indicated in red. The cases containing the MLL chimeric fusion gene continue to cluster according to lineage. (C) Multidimensional scaling plot generated using the supervised learning algorithm, discriminants analysis with variance (DAV) with the expression data from the 267 acute leukemia samples generated using the 17 944 probe sets that passed the variation filter. Cases are color coded as described for panel B. Cases with an MLL chimeric fusion gene (in red) can be separated in gene space from the leukemias that lack this genetic lesion. (D) Expression profile of the top 50–ranked MLL discriminating genes. The probe set number and gene symbol for the discriminating genes are listed on the right. The normalized expression level for each gene is represented by color using the scale shown.
Gene expression profiles of pediatric acute leukemia with MLL chimeric fusion genes. (A) Multidimensional scaling plot generated using unsupervised principle components analysis with a combined data set containing 130 AML cases, 132 ALL cases,41 and 5 additional T-lineage ALL (T-ALL) cases that contain MLL chimeric fusion genes. A variation filter was applied to remove any probe sets that showed minimal variation in expression across this data set, and the analysis was performed with the remaining 17 944 probe sets. Each case is represented by a colored sphere, with AML cases indicated by blue, B-progenitor lineage ALL (B-ALL) by yellow, and T-ALL by green. Acute leukemia cases cluster based on lineage. (B) The same PCA analysis as shown in panel A, except that cases that contain an MLL chimeric fusion gene are indicated in red. The cases containing the MLL chimeric fusion gene continue to cluster according to lineage. (C) Multidimensional scaling plot generated using the supervised learning algorithm, discriminants analysis with variance (DAV) with the expression data from the 267 acute leukemia samples generated using the 17 944 probe sets that passed the variation filter. Cases are color coded as described for panel B. Cases with an MLL chimeric fusion gene (in red) can be separated in gene space from the leukemias that lack this genetic lesion. (D) Expression profile of the top 50–ranked MLL discriminating genes. The probe set number and gene symbol for the discriminating genes are listed on the right. The normalized expression level for each gene is represented by color using the scale shown.
To see if a unique expression signature could be defined for the MLL chimeric fusion gene containing leukemias irrespective of their lineage, we next analyzed the data using the supervised learning algorithm DAV. As shown in Figure 5C, although this algorithm continued to demonstrate a strong correlation between lineage and gene expression profile, we could now also appreciate a separation in gene space of MLL versus non-MLL chimeric fusion gene cases. Thus, this analysis suggested the existence of a shared gene expression signature among cases with MLL chimeric fusion genes.
To identify the genes that contributed to this signature, SAM was used to identify non–lineage-restricted MLL class discriminating genes. This analysis identified 1059 genes whose expression patterns were statistically associated with the presence of an MLL chimeric fusion gene at a 1% FDR (the top 100 genes are listed in Table S13). The expression patterns of the top 50–ranked non–lineage-restricted MLL class discriminating genes are illustrated in the 2-dimensional dendogram shown in Figure 5D. In this analysis, all cases with MLL chimeric fusion genes were grouped on the left by lineage. As shown, the majority of the MLL class discriminating genes are overexpressed in this leukemia subtype. Among these are genes that are expressed in the majority of MLL chimeric fusion gene–containing cases, including MBNL1, MEIS1, HOXA4, HOXA5, HOXA9 HOXA10, and MYH9 (Table S14), as well as genes that show a more lineage-restricted pattern of expression, being preferentially expressed in either AMLs or T- and B-lineage ALLs. Use of the MLL-specific discriminating genes in an ANN supervised learning algorithm yielded an overall diagnostic accuracy of 96% (95% CI, 90%-99%) when tested on a blinded test set of 100 cases.
In addition to chromosomal translocation, the MLL gene can also be altered by an internal partial tandem duplication (PTD).46 MLL-PTDs are typically found in AML cases that have either normal cytogenetics or a trisomy of chromosome 11.47,48 The 47 cases of pediatric AML that lacked evidence of PML-RARα, AML-ETO, CBFβ-MYH11, MLL chimeric fusion genes, or FAB-M7 morphology were analyzed for evidence of MLL-PTD using a RT-PCR–based assay.39 Thirteen cases (28% of the analyzed samples) contained MLL-PTD. Quite surprisingly, these cases failed to cluster with the other MLL chimeric fusion gene AML cases when all 130 AML cases were analyzed using a 2-dimensional hierarchical clustering algorithm (Figure S1). Moreover, many fewer discriminating genes were identified for the combined group of MLL-PTD and MLL chimeric fusion gene cases, than for a group consisting exclusively of cases with MLL chimeric fusion genes. In addition, use of these discriminating genes in an ANN supervised learning algorithm yielded a very low accuracy of class prediction (data not shown). Lastly, when MLL-PTD was considered as a single leukemia subgroup, no class discriminating genes could be identified at a 5% FDR. Thus, taken together, these data suggest that MLL-PTDs are heterogeneous at a molecular level and are distinct from AMLs that contain chromosomal translocation of MLL that result in the formation of chimeric fusion genes.
Discussion
Gene expression profiling using microarray-based methodologies has provided new insights into the biology of a variety of hematopoietic malignancies, and has shown promise as a tool to aid in the accurate diagnosis and risk-stratification of patients.24-26,41,49-51 Recent applications to the acute leukemias have revealed distinct expression signatures for the individual lineages of the leukemic blasts,49 as well as for many of the known prognostic subtypes of pediatric ALL and adult AML.25-27 These gene signatures have proven to be robust discriminators of the specific subtypes of leukemia, showing diagnostic accuracies that, in many cases, exceed those achieved using routine diagnostic approaches. We now extend these studies by reporting the results from the expression analysis of diagnostic leukemic blasts from 130 pediatric and 20 adult patients with de novo AML. Our results demonstrate distinct expression signatures for each of the known prognostic subtypes of pediatric AML, including t(8;21)[AML1-ETO], inv16[CBFβ-MYH11], t(15;17)[PML-RARα], MLL chimeric fusion genes, and FAB-M7. Moreover, using the identified expression signatures in an ANN-based supervised learning algorithm, we achieved an overall diagnostic accuracy of 93%. More importantly, the pediatric AML subtype-specific expression signatures were present essentially unchanged in adult AML cases containing the identical genetic lesions. Thus, the identified class discriminating expression signatures should prove valuable in the development of custom AML diagnostic microarrays for use in the clinical setting. In addition, we identified a limited set of genes whose high expression correlated with a poor outcome. However, because of the relative small size of our data set, the true prognostic significance of these genes will require validation in larger cohorts of pediatric and adult patients. Lastly, by combining the described AML data set with a previously published pediatric ALL data set,41 we identified an expression signature that was specific for the presence of an MLL chimeric fusion gene, irrespective of the lineage of the leukemic blasts. This signature provides novel insights into the downstream transcriptional cascade resulting from the expression of an MLL chimeric fusion gene.
The diagnostic expression signatures identified in this paper, as well as those presented in several other recently published studies,27-29,52,53 represent only a first step in moving this methodology in the clinical setting. Importantly, the signatures developed to date allow the identification of a limited subset of the known prognostically important AML subtypes. It remains to be determined whether expression signatures can be identified for some of the other known AML subtypes, including -5/del(5q), -7/del(7q), and inv3/t(3;3). Moreover, between 25% and 45% of adult and pediatric cases of AML are reported to lack evidence of a clonal chromosomal abnormality.9 A variety of genetic lesions including mutations of AML1, N-RAS, K-RAS, C/EBPα, and the FLT-3 receptor have been identified in varying proportions of these cases.6,54-60 Whether unique expression signatures can be defined for some of these lesions remains unknown. Although only a minority of the cases in our study were analyzed for these lesions, we nevertheless were able to detect a suggestion of clustering for a small subset of the cases containing FLT-3 activating mutations (data not shown). The examination of a large number of well-characterized cases will be required to determine if unique expression signatures can be defined for these genetic lesions, and if so, whether they are robust enough to accurately diagnose the presence of the lesions in blinded clinical samples. Two recent studies have demonstrated that for some genetic lesions this may be possible.52,53
The identified expression signature for each of the different leukemia subtypes provides a unique insight into their underlying pathobiology. Although many testable hypotheses can be generated from the list of class discriminating genes, they remain speculative at best. Direct experimentation will be required to determine which of the identified genes play a mechanistic role in the growth of the leukemic cells. Moreover, defining the genes that are aberrantly expressed within the leukemic cells as compared with normal bone marrow–derived hematopoietic stem cells and lineage-committed progenitors will provide important insights into the altered biology of the leukemic cells. The generated data set provides an invaluable resource for the latter kinds of analyses.
The presented FAB-M7 expression signature represents the first detailed analysis of this specific subtype of AML. FAB-M7 is known to be a heterogeneous leukemia subtype with a minority of patients having Down syndrome, and an independent subset having leukemic blasts that contain the t(1;22).61 In the examined cohort, only 2 patients had Down syndrome and no examples of the t(1;22) were included. Thus, the data provide a view of only a limited subset of FAB-M7 leukemias. In this cohort, most of the identified genes have previously been shown to be expressed in cells of the megakaryocyte lineage.62-64 These include glycoproteins IIb/IIIa (GP1BB and ITGA2B), GATA1, and MRPS12. In addition, we identified a number of novel genes not previously associated with this leukemia subtype. Included in this latter list are the BMP-2 inducible nuclear serine/threonine kinase (BMP2K) and the putative tumor suppressor deleted in liver cancer 1 (DLC1). Exploring the functional role of the identified genes in the pathogenesis of this leukemia subtype should provide insights that could lead to improvements in our ability to treat this poor-risk AML subtype.
The identified expression signature of MLL chimeric fusion gene cases provides an important view into the downstream targets of these mutant transcriptional regulatory proteins. It is likely that within the identified set of genes are targets whose altered expression is essential for the development and growth of the leukemic clone. Among the MLL class discriminating genes we identified a subset of 21 genes that showed a relatively uniform level of expression in all MLL chimeric fusion gene cases, irrespective of lineage (Table S14). Many of the genes in this list have previously been implicated in MLL chimeric fusion protein-mediated transformation, including MEIS1 and the HOX genes (HOXA4, HOXA5, HOXA7, HOXA9, and HOXA10).65-69 Others, however, are unique to this analysis. A comparison of the 21 relative expression levels of the identified genes to their level of expression in normal BM-derived CD34+ hematopoietic progenitors revealed that approximately half were expressed at levels nearly identical to that seen in normal hematopoietic progenitors (Table S14). By contrast, other genes in this list were significantly overexpressed, including HOXA9, HOXA10, CAPG, NICAL, ABCA7, MYH9, VLDLR, ARPC2, PTPRC, and RAC2. Several of these genes, including CAPG, ABCA7 and MYH9, are normally highly expressed on cells of the monocyte/macrophage lineage, and thus likely reflect a degree of myeloid/monocytic gene expression, or more correctly, inappropriate expression in the B- and T-lineage cases. Others genes encode products that suggest that their inappropriate expression may be functionally important. Rac2 in particular has recently been shown to play an important role in intergrin-mediated stem cell signaling, and to inhibit cell death under certain growth conditions.70 Deciphering which genes are simply reflective of the stage of differentiation, and which are mechanistically important, will require direct experiments to assess the functional effects that result from the loss of expression.
One of the most surprising results from our analysis was the lack of a distinct expression signature for AMLs with MLL-PTD and the inability to define a clear relationship of these cases to other AMLs with MLL chimeric fusion genes. These data suggest that MLL-PTD induces the altered growth of hematopoietic progenitors through a mechanism that shows minimal, if any, relationship to the altered transcriptional pathway that results from the expression of MLL chimeric fusion genes. Comparing and contrasting the mechanisms of transformation induced by these 2 distinct classes of MLL mutations should provide valuable insights that will eventually lead to better ways to treat these AML subtypes.
In summary, the data presented suggest that expression profiling could provide a robust platform for the accurate diagnosis and classification of the major prognostic subtypes of AML in both pediatric and adult patients. Moreover, the database generated through this study coupled with those generated through our past efforts should provide valuable resources for the investigation of the altered biology that underlies the various genetic subtypes of acute leukemia.
Supported in part by National Cancer Institute grants P01 CA71907-06 (J.R.D.), CA-21765 (Cancer Center CORE grant to St Jude Children's Research Hospital), T32-CA70089 and St Jude Physician Scientist Training Program (M.E.R.), NHRI-EX92-9011SL (L.Y.S.), and by the American Lebanese and Syrian Associated Charities (ALSAC) of St Jude Children's Research Hospital. C.-H.P. is the recipient of the American Cancer Society F.M. Kirby Clinical Professorship.
M.E.R. and R.M. contributed equally to this work and both should be considered first authors.
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Prepublished online as Blood First Edition Paper, June 29, 2004; DOI 10.1182/blood-2004-03-1154.
The authors thank the staff of the Molecular Pathology laboratory and the Hartwell Center for Bioinformatics and Biotechnology at St Jude Children's Research Hospital (SJCRH) for technical support. We also thank the SJCRH Tumor Bank and Michael Jaynes for assistance in obtaining cryopreserved samples.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal