In humans, epithelial Langerhans cells (LCs) and monocyte-derived/interstitial dendritic cells (DCs) constitute 2 myeloid DC sublineages. Molecular mechanisms involved in their development from common myeloid progenitors remain poorly defined. Here we demonstrate that the nuclear factor-κB (NF-κB) transcription factor RelB regulates the generation of monocytic CD14+CD11b+ precursors of interstitial DCs from human hematopoietic progenitors. RelB overexpression promoted, whereas endogenous RelB inhibition (by p100ΔN) blocked, precursor cell development along this DC subset pathway. RelB inhibition specifically arrested precursor progression from CD14loCD11b- to CD14+CD11b+ stages. Precursors were still capable of LC and granulocyte differentiation but were defective in macrophage–colony-stimulating factor (M-CSF)–dependent monocyte/macrophage differentiation. RelB inhibition markedly differed from classical NF-κB signaling inhibition because IκBα superrepressor (IκBα-SR), but not p100ΔN, impaired LC/DC differentiation, DC adhesion, and progenitor cell proliferation. Although RelB up-regulation and nuclear translocation are regarded as hallmarks of human myeloid DC maturation, ectopic RelB overexpression failed to promote DC maturation. Our results suggest that RelB regulates human monopoiesis and monocyte-derived DC subset development.
Introduction
Dendritic cells (DCs) originate from hematopoietic precursors and comprise specialized DC subtypes that differ in phenotype, function, and localization. In humans, epidermal Langerhans cells (LCs), interstitial DCs, and plasmacytoid DCs (pDCs) are recognized as separate DC lineages.1-3 CD34+ progenitors from umbilical cord blood represent well-characterized model cells for studying human DC subset development and maturation in vitro.4 The 2 myeloid-related human DC subtypes, LCs and interstitial DCs, codevelop from CD34+ cells in response to granulocyte macrophage–colony-stimulating factor (GM-CSF) plus tumor necrosis factor-α (TNFα) stimulation,5,6 and adding transforming growth factor-β1 (TGF-β1) polarizes precursors toward the LC pathway.7,8 Despite considerable knowledge of cytokine requirements and the identification of putative shared precursors of LC/DC and monocytes,9 transcriptional processes underlying their development remain poorly defined. Retroviral gene transduction experiments revealed that CCAAT/enhancer binding protein (C/EBP) and PU.1 transcription factors reciprocally control LC differentiation from CD34+ cells,10 and the Id proteins Id2 and Id3 inhibit human pDC but not myeloid DC development from CD34+ progenitors.11 However, little is known about processes that control human myeloid DC subset differentiation from putative shared precursors.
RelB is a member of the NF-κB transcription factor family, which comprises 5 members in mammals. RelA (p65), c-Rel, RelB, NF-κB1 (encoding the precursor molecule p100 and the processed form p52), and NF-κB2 (encoding the precursor molecule p105 and the processed form p50). The different members can form a variety of homodimers and heterodimers and are associated with inhibitory proteins (the IκBs), which retain the dimers in the cytoplasm. Like the IκBs, the precursor molecules p100 and p105 can sequester NF-κB molecules in the cytoplasm through their ankyrin-rich C-terminal domains.12 Recent studies have shown that 2 independent NF-κB activation cascades differentially control the nuclear translocation of NF-κB transcription factors.13,14 The classical NF-κB signaling cascade regulates dimers consisting of p65, c-Rel, and p50 and induces the phosphorylation and ubiquitin-dependent degradation of the IκBs (IκBα, IκBβ, and IκBϵ). In contrast to p65, c-Rel, and p50, RelB is not bound to the IκBs but is retained in the cytoplasm by p100.15,16 Processing of p100 to p52, which results in the translocation of p52/RelB dimers into the nucleus, is induced by the nonclassical NF-κB signaling cascade.
RelB is associated with mature DCs17-19 and is up-regulated and translocated into the nuclei of DCs in response to various stimuli that induce their maturation.20-24 However, RelB function has not been described in human DC development or maturation. In mice, RelB is essential for generating a specific DC subset (CD11c+/CD11b+/CD8α-/Dec205-) from hematopoietic stem cells.25 Furthermore, RelB levels are elevated in DCs of autoimmune diabetic mice,26 and RelB regulates murine DC antigen-presentation function in vivo.27 Here, we asked whether retroviral overexpression or RelB inhibition in primary human progenitor cells might regulate DC subset generation or DC maturation. We addressed this question by using an in vitro model of human DC generation from CD34+ cord blood progenitors that allowed us to analyze DC lineage development and DC maturation under well-defined culture conditions. We demonstrated in this study that RelB regulates the development of human DC subsets by promoting the differentiation of monocytic intermediates. In addition, we observed that ectopic RelB is insufficient to promote DC maturation.
Materials and methods
Isolation of cord blood CD34+ cells
Cord blood samples from healthy donors were collected during healthy full-term deliveries. Approval was obtained from the Medical University of Vienna institutional review board for these studies. Informed consent was provided according to the Declaration of Helsinki. Mononuclear cells were obtained by density gradient centrifugation over Lymphoprep (Axis-Shield PoC AS, Oslo, Norway). CD34+ cells were isolated from mononuclear cord blood cells using MACS Direct CD34 Progenitor Cell Isolation Kit (Miltenyi Biotech, Bergisch Gladbach, Germany) according to the instructions of the manufacturer. The purity of the isolated CD34+ cells analyzed by flow cytometry was greater than 95%.
CD34+ cell expansion and DC, monocyte, and granulocyte generation
To generate DCs, CD34+ cells were plated at a density of 1 to 2 × 104/mL in 24-well plates (Nunc, Rochester, NY; Costar, Cambridge, MA) in the presence of cytokines, as described.8 Briefly, serum-free medium X-VIVO 15 (BioWhittaker, Walkersville, MD) was supplemented with l-glutamine (2.5 mM), penicillin (125 U/mL), and streptomycin (125 U/mL). Cultures contained the following cytokine cocktail: GM-CSF (200 ng/mL), stem cell factor (SCF) (20 ng/mL), FLT3 ligand (FLT3L) (50 ng/mL), TNF-α (2.5 ng/mL), and TGF-β1 (0.1 or 0.5 ng/mL). Cultures were grown for the time periods indicated in the text, and DC maturation was induced by adding 500 ng/mL CD40 ligand, (CD40L trimer), kindly provided by Immunex (Seattle, WA), for 48 hours. Progenitor cell expansion cultures contained FLT3L, SCF, and thrombopoietin (TPO), each at 50 ng/mL in the described serum-free culture system. Monocyte generation cultures included M-CSF (100 ng/mL), interleukin-6 (IL-6) (20 ng/mL), FLT3L (50 ng/mL), and SCF (20 ng/mL); granulocyte generation cultures contained G-CSF (100 ng/mL), SCF (50 ng/mL), and FLT3L (50 ng/mL). SCF, TPO, M-CSF, G-CSF, and TNF-α were purchased from PeproTech (London, United Kingdom); TGF-β1 was purchased from RD Systems GmbH (Wiesbaden, Germany); FLT3L was obtained from Amgen (Seattle, WA); and GM-CSF and IL-6 were kindly provided by Novartis Research Institute (Vienna, Austria).
Retroviral constructs and gene transduction
The cDNAs of RelB (human; full length, obtained from R. Thomas, Brisbane, Australia), p100ΔN (amino acid [aa] 407-900 of the coding cDNA of p100, obtained from C.V. Paya, Rochester, MN), and a superrepressor form of IκBα (S32/36A, obtained from R. de Martin, Vienna, Austria) were subcloned into the multiple cloning site of the Moloney murine leukemia virus (MMLV)–based retroviral vector pBMN (obtained from G. P. Nolan, Stanford, CA), upstream of an internal ribosome entry site (IRES), followed by either enhanced green fluorescence protein (GFP) or murine CD8α (mCD8α). To produce recombinant amphotropic retrovirus, vectors were transiently transfected into the packaging cell line Phoenix-GP (Gag-Pol) using a calcium-phosphate protocol (https://www.stanford.edu/group/nolan/protocols/pro_helper_dep.html). Phoenix-GP was cotransfected with an expression plasmid encoding gibbon ape leukemia virus (GALV) envelope (gift from D. B. Kohn, Los Angeles, CA). Before gene transduction, fresh or thawed CD34+ cells were stimulated overnight in X-VIVO 15 medium supplemented with the cytokines SCF (50 ng/mL), FLT3L (50 ng/mL), and TPO (50 ng/mL). Afterward, 1 mL retroviral supernatant (harvested 36-48 hours after transfection of packaging cells) was added to 5 × 104 CD34+ cells in the presence of platebound RetroNectin (Takara Bio, Shiga, Japan) using nontissue culture–treated 24-well plates (Cellstar; Greiner Bio-One GmbH, Kremsmuenster, Austria) following the instructions of the manufacturer. Infections were repeated 2 to 3 times at intervals of 12 to 24 hours using fresh virus supernatants and in the presence of the DC cytokine cocktail or the cytokines SCF, FLT3L, and TPO, depending on the experimental settings. Within 60 hours of the first transduction cycle, cells were harvested and recultured in lineage-specific growth media.
Cell lines
HL60 cells expressing the ecotropic MMLV receptor (HL60e) were kindly provided by B. Fletcher (Miami, FL). U937T cells28 were obtained from G. Grosveld (Memphis, TN). To generate U937Te cells, U937T cells were transduced with a retroviral vector encoding the MMLV ecotropic receptor (obtained from R. de Martin) upstream of IRES-mCD8α (retroviral pBMN–based backbone; obtained from G. P. Nolan). Afterward, cells were repeatedly sorted using fluorescence-activated cell sorting (FACS) for mCD8αbright cells to obtain a highly infectable subclone (ie, U937Te cells). HL60e or U937Te cells were maintained in RPMI plus 10% fetal calf serum (FCS) medium. TF-1 cells were purchased from ATCC (Manassas, VA) and were transduced with the above-described pBMN-MMLV ecoreceptor construct, and subclones were generated as described. TF-1e cells were maintained in RPMI plus 10% FCS medium supplemented with 20 ng/mL recombinant human GM-CSF (rhGM-CSF). CD11b and CD14 expression on U937Te cells was induced with 25 ng/mL 1α,25-dihydroxyvitamin D3 (Sigma, Vienna, Austria) for 72 hours. U937 NF-κB reporter cells were generated by transducing U937 cells with a self-inactivating retroviral vector backbone (obtained from G. P. Nolan) in which we inserted a 5 × NF-κB–GFP reporter cassette (obtained from R. de Martin), followed by single-cell cloning.
Immunofluorescence and flow cytometry
Immunofluorescence stainings were performed as previously described.29 Murine monoclonal antibodies (mAbs) of the following specificities were used: phycoerythrin (PE)–conjugated mAbs specific for CD86, human leukocyte antigen (HLA)–DR, CD54 (PharMingen, Heidelberg, Germany); CD80, CD11b, Langerin (BD Biosciences, Palo Alto, CA), CD83 (Immunotech, Marseille, France); and biotinylated mAbs against CD1a (clone VIT 6) CD40 (clone G28-5), CD14 (clone MEM 18), and CD11b (LM-2; obtained from O. Majdic Vienna, Austria). As second-step reagents, we used streptavidin (SA)–peridinin chlorophyll (PerCP) (BD Biosciences). Allophycocyanin (APC)–conjugated mAbs specific for CD1a and CD14 were purchased from BD Biosciences. Anti-E–cadherin mAb (clone HECD-1) was obtained from RD Systems. PerCP-conjugated rat mAb against murine CD8α (lyt2, clone 53-6.7) was obtained from PharMingen; isotype control mAbs were provided by O. Majdic. For analysis, a FACSCalibur flow cytometer and CellQuest software were used (Becton Dickinson, Mountain View, CA). Cell sorting was performed using a FACSVantage (Becton Dickinson).
For intracellular staining of RelB, cells were first attached to adhesion slides (Marienfeld, Germany) according to the manufacturer's instructions. Cells were then fixed and permeabilized with the reagent combination Fix & Perm (Caltag Laboratories, Burlingame, CA; An der Grub, Kaumberg, Austria), as recommended. Afterward cells were incubated with a rabbit antimouse RelB antibody (Santa Cruz Biotechnology, CA) diluted 1:1000 (in phosphate-buffered saline [PBS]/1% bovine serum albumin [BSA]) overnight at 4°C, followed by an Alexa 568–conjugated goat antirabbit antibody, diluted 1:1000 (Molecular Probes, Eugene, OR). Microscopy was performed using the confocal laser scanning microscope LSM-510 (Zeiss, Jena, Germany) with the LSM Image Examiner V3.1 acquisition software.
AnnexinV-PE (BD Biosciences PharMingen) versus 7-amino-actinomycin D (7-AAD; Sigma-Aldrich, St Louis, MO) stainings were performed according to the manufacturer's protocol. For in vitro cell proliferation studies, the PKH26 red fluorescence cell linker kit (Sigma-Aldrich) was used according to the manufacturer's instructions.
Mixed-leukocyte reaction
Gene-transduced GFP+CD1a+ cells underwent FACS and were γ-irradiated (25 Gy). Graded numbers of these stimulator cells were cocultured with a constant number of 5 × 104 to 1 × 105 highly purified (greater than 98%) allogeneic T cells in RPMI medium containing 10% FCS using round-bottom, 96-well tissue culture plates (Nalge Europe, Brussels, Belgium). Cultures were pulsed at culture day 5 for 18 hours with 1 μCi [0.037 MBq]/well methyl-3H-thymidine (Amersham, Buckinghamshire, United Kingdom). Incorporated radioactivity was measured using a 1450 microbeta plate reader (Wallac-Trilux Instrument; Life Science, Vienna, Austria). Results are presented as the mean ± SD obtained from triplicate cultures.
Western blot analysis
Whole cell lysates of primary cells were prepared as follows: cells were pelleted, washed once with ice-cold PBS, and resuspended in an appropriate volume of lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 2.5 mM EDTA [ethylenediaminetetraacetic acid], 1% Triton X-100) supplemented with protease inhibitors (protease inhibitor cocktail set III; Calbiochem, San Diego, CA). After 15-minute incubation on ice, extracts were centrifuged at 14 000 rpm (16.100 g) for 10 minutes at 4°C. U937 cells were transduced with the above-described retroviral constructs, and GFP+ cells were sorted and expanded. Nuclear and cytoplasmic extracts were prepared using the nuclear extract kit from Active Motif (Carlsbad, CA) according to the manufacturer's instructions with an additional washing step of the isolated nuclei. The protein concentration of the extracts was determined using a Bradford-based protein assay from Bio-Rad (Hercules, CA). For Western blot analysis, 10 to 15 μg protein of the prepared extracts were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (8%-12%) and were transferred to a polyvinylidene difluoride (PVDF) membrane. Immunoblotting was performed using the following rabbit polyclonal antibodies: anti-RelB sc-226, anti-p65 sc-372, anti-cRel sc-272, anti-p52 sc-298, and anti-IκBα sc-371. Goat polyclonal antiactin sc-10806, rabbit polyclonal antihistone H1 sc-1617, or rabbit anti-SP1 sc-59 was used to control loaded protein amounts (Santa Cruz Biotechnology). Detection was performed using horseradish peroxide (HRP)–conjugated antirabbit immunoglobulin G (IgG; Pierce Biotechnology, Rockford, IL) or HRP-conjugated antigoat IgG (Santa Cruz Biotechnology) in combination with the chemiluminescent substrate SuperSignal (Pierce Biotechnology). For detection of p100ΔN, rabbit serum against the C-terminal domain of p100 was used (N. Rice, Frederick, MD).
Statistics
Data are presented as mean ± SD. For comparisons, a 2-sided paired student t test was used. P values less than .05 were considered statistically significant.
Results
RelB is required for the generation of human CD11b+ DCs
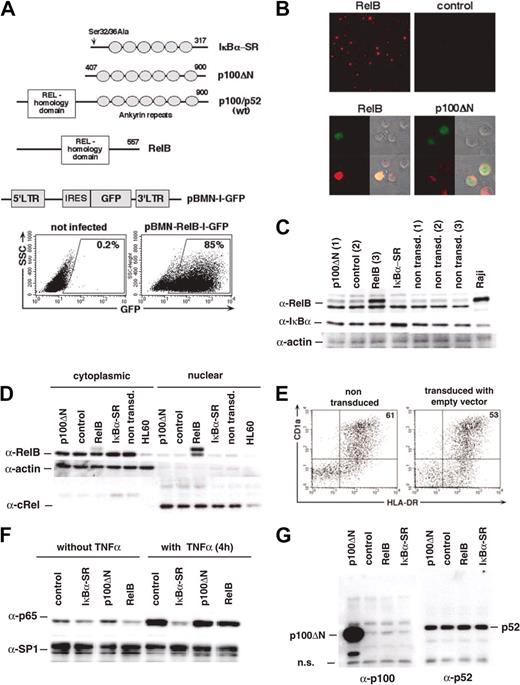
CD34+ cord blood progenitor/stem cells can give rise to precursors of 2 separate human CD1a+ DC sublineages.2,6 LCs develop from CD1a+CD11b- intermediates,9 whereas interstitial DCs develop from CD14+CD11b+ intermediates. These latter intermediates share many features with blood monocytes.6 To functionally analyze RelB in human DC development, we inserted human RelB or a truncated version of p100 (p100ΔN15), which contains only the RelB inhibitory domain (Figure 1A) upstream of IRES-GFP into a retroviral vector. It has been shown that nuclear translocation of RelB is efficiently blocked by this p100ΔN molecule in HeLa cells.15 For comparison, we created a third retroviral construct encoding the superrepressor mutant of IκBα,30 IκBα-SR, upstream of IRES-GFP. We delivered p100ΔN, empty vector (control), RelB, or IκBα-SR into CD34+ progenitor cells and induced them to differentiate into DCs, as described.8 Overexpression of RelB in these primary cells was confirmed using immunofluorescence microscopy (Figure 1B) and by Western blot analysis of FACS-sorted GFP+ gene–transduced cells (Figure 1C). Additionally, we transduced U937 cells with RelB-IRES-GFP or control constructs. Nuclear and cytoplasmic extracts of GFP+ cells were analyzed (Figure 1D). As expected, high RelB levels could be detected in the nuclear extracts of RelB-transduced cells. In contrast, p100ΔN–transduced primary cells showed a pronounced cytoplasmic RelB localization (Figure 1B), consistent with its capacity to capture RelB in the cytoplasm.15 Retroviral expression of p100ΔN was confirmed by Western blot analysis of cytoplasmic extracts from U937 cells (Figure 1G). Approximately 2-fold IκBα overexpression levels were observed in GFP+ IκBα-SR–transduced primary cells (Figure 1C). As expected, IκBα-SR inhibited TNF-α–induced nuclear translocation of p65 in U937 cells (Figure 1F).
Retroviral gene transduction. CD34+ cells or U937 cells were transduced with retroviral vectors encoding p100ΔN, empty control, RelB, or IκBα-SR upstream of IRES-GFP. (A) Schematic representation of the IκBα-SR, p100ΔN compared with wild-type (wt) p100/p52 and RelB as well as the retroviral backbone pBMN-IRES-GFP used in our study. Representative FACS diagrams show gated GFP+ gene–transduced cells. (B) Representative immunofluorescence analysis of RelB expression in gene-transduced DCs. Numbers indicate the percentages of the gated cells. (Upper panels) DCs transduced with RelB or empty vector. (lower panels) Subcellular localization of RelB-in RelB or p100ΔN-transduced DCs. RelB (red), GFP (green), phase-contrast and merged images. (C) Overexpression of RelB and IκBα-SR in DCs. CD34+ cells were cultured for 8 days and then sorted for GFP+ and GFP- fractions. Lanes 1 to 4 represent total cell extracts of GFP+ cells transduced with p100ΔN (1), control (2), RelB (3), and IκBα-SR, as indicated. Lanes 5 to 7 represent GFP- cells p100ΔN (1), control (2), and RelB (3). Raji cells (lane 8) were included as a RelB-positive control. (D) Western blot analysis of cytoplasmic (lanes 1-5) and nuclear (lanes 7-11) extracts prepared from FACS-sorted U937 cells transduced with the indicated constructs. Nontransduced HL60 cells (lanes 6 and 12) are compared. (E) DCs derived in vitro from CD34+ cells. FACS diagrams show representative nontransduced versus control-transduced cells. (F) IκBα-SR inhibits TNF-α–induced nuclear translocation of p65. Gene-transduced U937 cells were stimulated for 4 hours with 5 ng/mL TNF-α. Nuclear extracts from these cells were probed with anti-p65 and anti-SP1 antibodies (loading control). (G) Overexpression of p100ΔN was confirmed by immunoblotting of cytoplasmic U937 cell extracts with rabbit anti-p100 serum (left blot). The same extracts were analyzed in parallel for endogenous p52 levels (right blot).
Retroviral gene transduction. CD34+ cells or U937 cells were transduced with retroviral vectors encoding p100ΔN, empty control, RelB, or IκBα-SR upstream of IRES-GFP. (A) Schematic representation of the IκBα-SR, p100ΔN compared with wild-type (wt) p100/p52 and RelB as well as the retroviral backbone pBMN-IRES-GFP used in our study. Representative FACS diagrams show gated GFP+ gene–transduced cells. (B) Representative immunofluorescence analysis of RelB expression in gene-transduced DCs. Numbers indicate the percentages of the gated cells. (Upper panels) DCs transduced with RelB or empty vector. (lower panels) Subcellular localization of RelB-in RelB or p100ΔN-transduced DCs. RelB (red), GFP (green), phase-contrast and merged images. (C) Overexpression of RelB and IκBα-SR in DCs. CD34+ cells were cultured for 8 days and then sorted for GFP+ and GFP- fractions. Lanes 1 to 4 represent total cell extracts of GFP+ cells transduced with p100ΔN (1), control (2), RelB (3), and IκBα-SR, as indicated. Lanes 5 to 7 represent GFP- cells p100ΔN (1), control (2), and RelB (3). Raji cells (lane 8) were included as a RelB-positive control. (D) Western blot analysis of cytoplasmic (lanes 1-5) and nuclear (lanes 7-11) extracts prepared from FACS-sorted U937 cells transduced with the indicated constructs. Nontransduced HL60 cells (lanes 6 and 12) are compared. (E) DCs derived in vitro from CD34+ cells. FACS diagrams show representative nontransduced versus control-transduced cells. (F) IκBα-SR inhibits TNF-α–induced nuclear translocation of p65. Gene-transduced U937 cells were stimulated for 4 hours with 5 ng/mL TNF-α. Nuclear extracts from these cells were probed with anti-p65 and anti-SP1 antibodies (loading control). (G) Overexpression of p100ΔN was confirmed by immunoblotting of cytoplasmic U937 cell extracts with rabbit anti-p100 serum (left blot). The same extracts were analyzed in parallel for endogenous p52 levels (right blot).
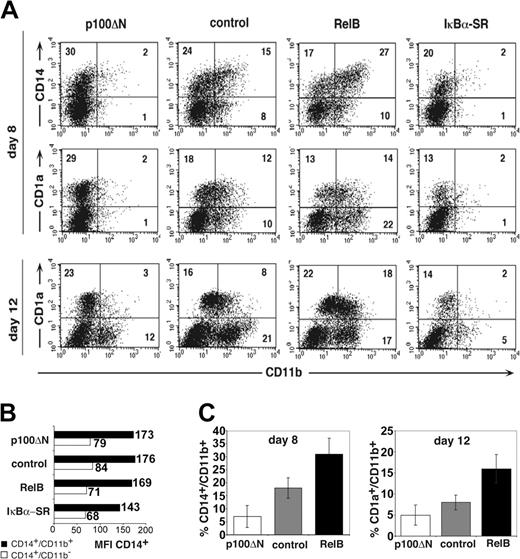
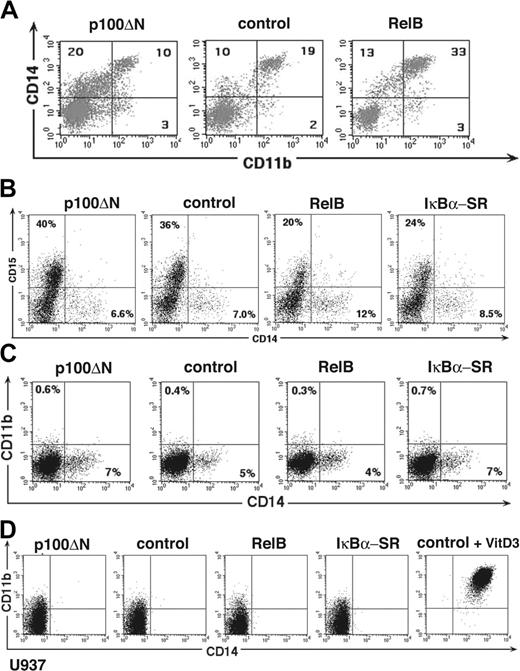
For phenotypic analysis, gene-transduced primary cells were identified as GFP+ cells (according to gate settings in Figure 1A). These GFP+ cells were analyzed for expression of CD1a, CD14, and CD11b at several time points using flow cytometry (after 4-12 days; Figure 2A). Nontransduced and empty vector–transduced cells were phenotypically comparable (Figure 1E). RelB overexpression significantly increased percentages of CD11b+CD14+ cells compared with control-transduced cells (Figure 2A, upper panel; Figure 2C, left). The inverse effect was observed for p100ΔN–mediated inhibition of RelB. Ectopic p100ΔN significantly inhibited the generation of CD11b+CD14+ cells (Figure 2C, right) and, in turn, increased percentages of cells that were CD14dim (CD14lo) in the absence of CD11b (CD14loCD11b- cells) (Figure 2A, upper left). Analysis of the CD14 mean fluorescence intensities (MFIs; Figure 2B) confirmed that CD14+CD11b- cells (Figure 2A, upper left quadrants) expressed on average lower amounts of CD14 than CD11b+CD14+ cells (upper right quadrants). Similar MFI values were observed for p100ΔN-, control-, and RelB-transduced cells (Figure 2B), supporting observations that p100ΔN and RelB alter the relative frequencies of these 2 phenotypically defined subsets. Later during culture (day 12), significantly higher percentages of RelB-transduced cells coexpressed CD11b and CD1a than control-transduced cells. Conversely, p100ΔN significantly inhibited the generation of these CD11b+CD1a+ cells (Figure 2A and 2C, right diagram). Total percentages of CD1a+ cells were, on average, equivalent among p100ΔN compared with control-transduced cells, and RelB or p100ΔN did not influence cell proliferation (Figure 3E). These data suggest that RelB plays a pivotal role for the generation of CD11b+CD1a+ DC and their CD11b+CD14+ monocytic intermediates.
Reciprocal effects of RelB and p100ΔN on the differentiation of CD11b+ DCs. CD34+ cells were transduced with retroviral vectors encoding p100ΔN, empty control, RelB, or IκBα-SR upstream of IRES-GFP and were induced to develop into DCs. (A) FACS diagrams show transduced cells (gated on GFP+ cells, as shown in Figure 1A) analyzed for CD11b versus CD14 (day 8) or CD11b versus CD1a (days 8, 12). Markers were set according to negative control stainings. Data are representative of 4 independent experiments. Numbers indicate percentages of cells within the quadrants. (B) CD14 MFI (numbers to the right of each bar) of cells from FACS diagrams shown in panel A. ▪ indicates CD14+CD11b+ cells (top right quadrants). ▦ indicates CD14+CD11b- cells (top left quadrants). (C) Bar diagrams represent mean ± SD of CD14+CD11b+ (day 8; left) or CD1a+CD11b+ (day 12; right) cells in 4 independent experiments. Paired Student t test statistical analysis: percentage CD14+CD11b+ (left bar diagrams; p100ΔN vs control, P = .002; RelB vs control, P = .004); percentage CD1a+CD11b+ (right bar diagrams; p100ΔN vs control, P = .046; RelB vs control, P = .003).
Reciprocal effects of RelB and p100ΔN on the differentiation of CD11b+ DCs. CD34+ cells were transduced with retroviral vectors encoding p100ΔN, empty control, RelB, or IκBα-SR upstream of IRES-GFP and were induced to develop into DCs. (A) FACS diagrams show transduced cells (gated on GFP+ cells, as shown in Figure 1A) analyzed for CD11b versus CD14 (day 8) or CD11b versus CD1a (days 8, 12). Markers were set according to negative control stainings. Data are representative of 4 independent experiments. Numbers indicate percentages of cells within the quadrants. (B) CD14 MFI (numbers to the right of each bar) of cells from FACS diagrams shown in panel A. ▪ indicates CD14+CD11b+ cells (top right quadrants). ▦ indicates CD14+CD11b- cells (top left quadrants). (C) Bar diagrams represent mean ± SD of CD14+CD11b+ (day 8; left) or CD1a+CD11b+ (day 12; right) cells in 4 independent experiments. Paired Student t test statistical analysis: percentage CD14+CD11b+ (left bar diagrams; p100ΔN vs control, P = .002; RelB vs control, P = .004); percentage CD1a+CD11b+ (right bar diagrams; p100ΔN vs control, P = .046; RelB vs control, P = .003).
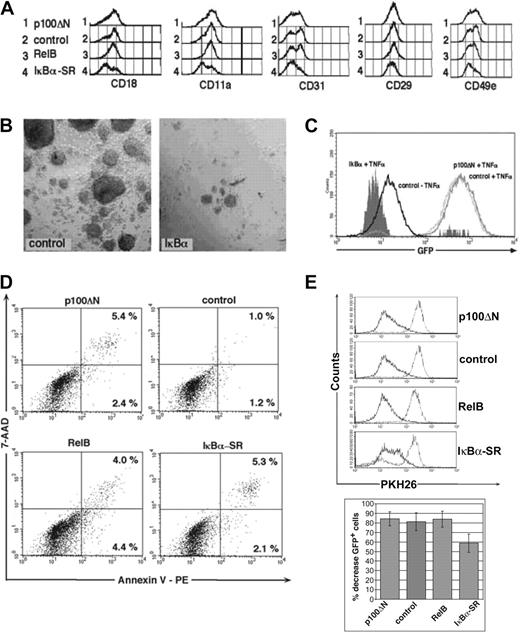
Comparison of p100ΔN and IκBα-SR. CD34+ cells were transduced with retroviral vectors encoding the indicated molecules and were induced to develop into DCs. (A) Histograms represent expression patterns of the indicated cytoadhesion molecules by gated GFP+ cells. (B) Microphotographs show culture morphology of cells after 30-minute 1g sedimentation over 7.5% HAS. (left) Control (empty vector). (right) IκBα-SR. (C) U937 NF-κB–GFP reporter cells were infected with IκBα-SR–IRES-mCD8α, p100ΔN–IRES-mCD8α, or empty vector (control). Two days after infection, cells were stimulated with TNF-α (25 ng/mL) for 48 hours and were analyzed for GFP induction by FACS. Overlay histograms show control-transduced cells with or without TNF-α stimulation, and IκBα-SR or p100ΔN-transduced cells stimulated with TNF-α. (D) Annexin versus 7-AAD stainings of representative day 8–generated GFP+-gated cells. Numbers indicate percentages of gated cells within the quadrants. (E) Bar diagrams show consistent time-linked decreases in average percentages of GFP+ cells in IκBα-SR compared with p100ΔN-, control-, or RelB-transduced cultures. Bars represent mean ± SD percentage decreases of GFP+ cells determined at days 7 to 9 after gene transduction over values observed at 48 to 72 hours after gene transduction; 48- to 72-hour values represent 100%. FACS data in panels D and E are representative of 3 independent experiments. Bar histograms represent the mean ± SD of 4 independent experiments. P = .009 for IκBα-SR compared with control.
Comparison of p100ΔN and IκBα-SR. CD34+ cells were transduced with retroviral vectors encoding the indicated molecules and were induced to develop into DCs. (A) Histograms represent expression patterns of the indicated cytoadhesion molecules by gated GFP+ cells. (B) Microphotographs show culture morphology of cells after 30-minute 1g sedimentation over 7.5% HAS. (left) Control (empty vector). (right) IκBα-SR. (C) U937 NF-κB–GFP reporter cells were infected with IκBα-SR–IRES-mCD8α, p100ΔN–IRES-mCD8α, or empty vector (control). Two days after infection, cells were stimulated with TNF-α (25 ng/mL) for 48 hours and were analyzed for GFP induction by FACS. Overlay histograms show control-transduced cells with or without TNF-α stimulation, and IκBα-SR or p100ΔN-transduced cells stimulated with TNF-α. (D) Annexin versus 7-AAD stainings of representative day 8–generated GFP+-gated cells. Numbers indicate percentages of gated cells within the quadrants. (E) Bar diagrams show consistent time-linked decreases in average percentages of GFP+ cells in IκBα-SR compared with p100ΔN-, control-, or RelB-transduced cultures. Bars represent mean ± SD percentage decreases of GFP+ cells determined at days 7 to 9 after gene transduction over values observed at 48 to 72 hours after gene transduction; 48- to 72-hour values represent 100%. FACS data in panels D and E are representative of 3 independent experiments. Bar histograms represent the mean ± SD of 4 independent experiments. P = .009 for IκBα-SR compared with control.
Inhibition of RelB does not impair LC differentiation
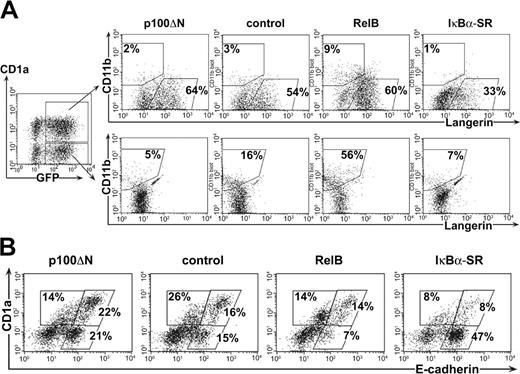
We found that RelB inhibition by p100ΔN impairs the generation of CD11b+ DCs. Therefore, we analyzed whether inhibiting RelB similarly influences LC development. To predominantly generate LCs among CD1a+ cells, we increased the amounts of exogenous TGF-β1 from 0.1 to 0.5 ng/mL.7 LCs were identified by the coexpression of E-cadherin, Langerin (CD207), and high levels of CD1a in the absence of CD11b.9,31,32 To analyze the generation of LCs and monocytes in our culture system, GFP+CD1a+ and GFP+CD1a- cells were separately analyzed for Langerin or CD11b expression (Figure 4A). Among control-transduced cells, most CD1a+ cells were identified as Langerin+CD11b- LCs, whereas a subset of CD1a- cells showed Langerin-CD11b+ monocytic characteristics. This confirmed the coemergence of LCs and monocytes in these cultures. We performed a comparative analysis of p100ΔN, RelB, and IκBα-SR on the generation of these 2 cell populations (Figure 4A). Inhibiting RelB by p100ΔN did not impair the percentages of Langerin+CD11b- LCs but did slightly increase them. On average, slightly higher percentages of Langerin+ cells were observed among p100ΔN-than among control-transduced cells (Table 1). Consistent with this, slightly higher percentages of E-cadherin+CD1a+ cells were found among p100ΔN-transduced cells than among control-transduced cells (Figure 4B; experiment 1, 19% vs 15%; experiment 2, 48% vs 39%). In line with the above data, p100ΔN inhibited the generation of CD1a-Langerin-CD11b+ monocytes. Conversely, ectopic RelB substantially increased the percentages of CD1a-Langerin-CD11b+ monocytes, whereas percentages of LCs remained on average unchanged. Therefore, RelB influences the generation of monocytes but not of LCs.
RelB inhibition impairs the generation of monocytes but not of LCs. Gene-transduced progenitor cells were stimulated in the presence of 0.5 ng/mL TGF-β1 to generate LCs. (A) CD1a+GFP+ and CD1a-GFP+ cells were gated as shown (left diagram) and analyzed for Langerin versus CD11b expression. (B) Gated GFP+ cells analyzed for E-cadherin versus CD1a expression. Region settings were performed according to control stainings. Data are representative of at least 3 independent experiments. Percentages on the plots are the frequency of gated cells lying within the indicated regions.
RelB inhibition impairs the generation of monocytes but not of LCs. Gene-transduced progenitor cells were stimulated in the presence of 0.5 ng/mL TGF-β1 to generate LCs. (A) CD1a+GFP+ and CD1a-GFP+ cells were gated as shown (left diagram) and analyzed for Langerin versus CD11b expression. (B) Gated GFP+ cells analyzed for E-cadherin versus CD1a expression. Region settings were performed according to control stainings. Data are representative of at least 3 independent experiments. Percentages on the plots are the frequency of gated cells lying within the indicated regions.
Lack of inhibitory effects of p100ΔN on the in vitro generation of Langerin (CD207)+ cells from CD34+ cord blood progenitors
Experiment . | p100ΔN . | Control . | RelB . | IκBα-SR . |
|---|---|---|---|---|
| 1 | 28* | 23 | 24 | 17 |
| 2 | 33 | 26 | 26 | 12 |
| 3 | 28 | 23 | 21 | NT |
| 4 | 37 | 25 | 27 | NT |
| 5 | 27 | 21 | 21 | NT |
| 6 | 32 | 27 | 25 | NT |
| 7 | 40 | 43 | 31 | 13 |
| 8 | 40 | 28 | 19 | NT |
| 9 | 18 | 14 | 20 | 9 |
| Mean | 31† | 26† | 24 | ND |
| SD | 7 | 8 | 4 | ND |
Experiment . | p100ΔN . | Control . | RelB . | IκBα-SR . |
|---|---|---|---|---|
| 1 | 28* | 23 | 24 | 17 |
| 2 | 33 | 26 | 26 | 12 |
| 3 | 28 | 23 | 21 | NT |
| 4 | 37 | 25 | 27 | NT |
| 5 | 27 | 21 | 21 | NT |
| 6 | 32 | 27 | 25 | NT |
| 7 | 40 | 43 | 31 | 13 |
| 8 | 40 | 28 | 19 | NT |
| 9 | 18 | 14 | 20 | 9 |
| Mean | 31† | 26† | 24 | ND |
| SD | 7 | 8 | 4 | ND |
In all experiments, CD34+ cells were cultured in DC generation cultures in the presence of cytokines and were gene transduced with retroviral vectors, as described in “Materials and methods.” Nine independent cord blood donor samples were analyzed.
NT indicates not tested; ND, not determined.
GFP+ cells were gated and analyzed for Langerin expression. Values represent percentages of Langerin- cells among GFP+ cells
Significantly higher percentages of Langerin+ cells among p100ΔN-transduced cells than among control-transduced cells. P = .0043; paired Student t test analysis
IκBα-SR and p100ΔN markedly differ in inhibitory effects on DCs
We also included the classical NF-κB pathway inhibitor IκBα-SR in our experiments. Percentages of IκBα-SR–IRES-GFP–transduced cells gradually decreased during culture, whereas no such effect was observed for p100ΔN-IRES-GFP–transduced cells (Figure 3E, bar diagrams). This effect of IκBα-SR was mediated by decreased cell proliferation rather than induction of apoptosis, as evidenced from PKH26 dye labeling (Figure 3E) and Annexin-V staining experiments (Figure 3D). Phenotypic analysis showed decreased percentages of CD1a+ cells and monocytes (Figure 2) and decreased percentages of Langerin+CD1a+ cells (Figure 4) among IκBα-SR–transduced cells compared with control-transduced cells. Interestingly, IκBα-SR–transduced cultures contained elevated percentages of cells showing immature LC characteristics (E-cadherindimCD1a-; Figure 4B, lower right diagram) compared with cells transduced with empty control vector or p100ΔN. In addition, IκBα-SR, but not p100ΔN, reduced the expression of several cytoadhesion molecules (Figure 3A), including integrin beta chains CD18 (β2) and CD29 (β1), integrin α chains CD11a and CD49e, and CD31 platelet/endothelial cell adhesion molecule-1 (PECAM-1). As seen from the histograms in Figure 3A, changes for CD29 and CD49e were minor (in 2 experiments, MFI values for control versus IκBα-SR–transduced cells were CD29 MFI, 52 versus 38 and 59 versus 45; CD49e MFI, 36 versus 24 and 31 versus 22). Additionally, as expected from the diminished expression of these molecules,33 IκBα-SR–transduced cultures showed less tight cell cluster formation. This is evident from the observation that purification of IκBα-SR–transduced clusters over a 1g human serum albumin gradient34 failed to recover typical LC clusters (Figure 3B). In contrast, p100ΔN (or RelB) did not influence homotypic LC clustering (data not shown). Therefore, the efficient transition of LC precursors to LCs and homotypic LC adhesion requires classical NF-κB signaling, whereas RelB-dependent signaling seems dispensable. Diminished DC differentiation by IκBα-SR is likely mediated by the inhibition of TNF-α signaling. We confirmed this using a NF-κB reporter assay. We transduced the myelomonocyte line U937 with an NF-κB-GFP reporter cassette and generated a TNF-α–inducible subclone. We observed that IκBα, but not p100ΔN, suppressed NF-κB activity in these cells in response to 48-hour TNF-α stimulation (Figure 3C). Together, these experiments show that p100ΔN and IκBα-SR substantially differ in the regulation of myeloid DC development.
Ectopic RelB is insufficient to induce DC maturation
Enhancement of RelB expression and its nuclear translocation are hallmarks of human DC maturation in response to various stimuli.23 However, it is unknown whether RelB is functionally involved in human DC maturation. The culture model used in our study yields high percentages of immature DCs, which can be induced in a controlled manner to undergo DC maturation (CD40L for 48 hours29 ). Therefore, this system is well suited for examining whether RelB overexpression might promote human DC maturation. Both DC subsets generated in serum-free media showed immature DC characteristics unless they were further stimulated with ligands that induce their maturation. We found that RelB-transduced DCs resembled control cells in the expression of DC maturation–associated marker molecules, including CD86, CD80, HLA-DR, CD40, CD54, and CD83 (Figure 5A). Furthermore, RelB did not promote CD40L-induced DC maturation, as exemplified by comparable CD1a and CD83 expression (Figure 5B; n = 14 independent experiments). Additionally, these CD40L-stimulated cells did not differ phenotypically at earlier time points (24 hours; data not shown). In line with these observations, the capacity of RelB-transduced and control-transduced CD1a+ cells to stimulate allogeneic T cells was found to be equivalent (n = 4; Figure 5C). We further observed that RelB inhibition by p100ΔN does not inhibit DC maturation, suggesting that RelB inhibition does not prevent human DC maturation (data not shown). Together, these experiments showed that enforced RelB expression in human CD34+-derived DCs is insufficient to promote DC maturation.
Effects of ectopic RelB on DC maturation. Immature or CD40L stimulated DCs transduced with RelB or control vector were analyzed for DC maturation-associated molecules. (A) Overlay histograms of gated GFP+ cells analyzed for the indicated marker molecules. Filled histograms represent RelB-transduced cells and solid black histograms represent empty control vector transduced cells. (B) FACS diagrams of gated GFP+ cells analyzed for CD83 versus CD1a expression. Parallel cultures were harvested and analyzed at day 10 (top panel) or were further cultured in the presence of CD40L and then harvested 48 hours later (bottom panel). Percentages of CD1a+CD83+ cells are indicated on the plots. (C) Cells were transduced with empty vector or RelB-IRES-GFP. Generated cells were FACS-sorted to obtain CD1a+GFP+ or CD1a-GFP+ cell fractions, and cells were subsequently stimulated with CD40L for 24 hours. Cells were then analyzed for their capacity to stimulate allogeneic T cells. (A-C) Data are representative of at least 4 experiments.
Effects of ectopic RelB on DC maturation. Immature or CD40L stimulated DCs transduced with RelB or control vector were analyzed for DC maturation-associated molecules. (A) Overlay histograms of gated GFP+ cells analyzed for the indicated marker molecules. Filled histograms represent RelB-transduced cells and solid black histograms represent empty control vector transduced cells. (B) FACS diagrams of gated GFP+ cells analyzed for CD83 versus CD1a expression. Parallel cultures were harvested and analyzed at day 10 (top panel) or were further cultured in the presence of CD40L and then harvested 48 hours later (bottom panel). Percentages of CD1a+CD83+ cells are indicated on the plots. (C) Cells were transduced with empty vector or RelB-IRES-GFP. Generated cells were FACS-sorted to obtain CD1a+GFP+ or CD1a-GFP+ cell fractions, and cells were subsequently stimulated with CD40L for 24 hours. Cells were then analyzed for their capacity to stimulate allogeneic T cells. (A-C) Data are representative of at least 4 experiments.
RelB augments monopoiesis but not granulopoiesis
Our data show that RelB regulates the generation of CD11b+CD14+ monocytic DC intermediates (Figures 2 and 4). We analyzed whether this effect is specific to our DC culture system or whether RelB might generally affect human monopoiesis. Therefore, RelB and p100ΔN-transduced CD34+ progenitors were stimulated with M-CSF plus IL-6, which join in inducing human monopoiesis.35 We found that RelB overexpression increased whereas p100ΔN reduced the percentages of CD11b+CD14+ cells compared with control-transduced cultures (Figure 6A). In line with these data (Figure 2), higher percentages of CD11b-CD14lo cells were observed among p100ΔN-transduced cells (Figure 6A). When analyzing vitamin D3–induced monocyte differentiation of primary CD34+ cells, we observed similar RelB-dependent regulation of CD11b+CD14+ cell generation (data not shown). For comparison, we stimulated cells with a cytokine combination that promotes granulocyte development (SCF, FLT3L, and G-CSF). RelB failed to enhance, but instead diminished, CD15+CD14- granulocyte differentiation (Figure 6B). Furthermore, granulocyte differentiation remained unimpaired by p100ΔN-mediated RelB inhibition. Therefore, RelB selectively promotes human monocyte development but not granulopoiesis.
RelB promotes human monopoiesis but not granulopoiesis. CD34+ cells or U937 cells were transduced with retroviral vectors encoding p100ΔN, empty control, or RelB 5′ of IRES-GFP. Diagrams show representative gated GFP+ cells (A) CD34+ cells stimulated in the presence of M-CSF plus IL-6, FL. and SCF for 10 days analyzed for CD11b versus CD14 expression. (B) CD34+ cells stimulated in the presence of G-CSF, SCF, and FLT3L for 6 days and analyzed for CD14 versus CD15 expression. (C) U937 cells analyzed for CD14 versus CD11b expression (top panel). Right diagram show a positive control (cells 72 hours after vitamin D3 stimulation). (D) CD34+ cells stimulated in the presence of expansion mix (SCF, FLT3L, TPO) for 7 days analyzed for CD14 versus CD11b expression. Markers were set according to negative control stainings. (A-D) Data are representative of at least 3 experiments. Numbers on the plots represent percentages of gated cells within the indicated quadrants.
RelB promotes human monopoiesis but not granulopoiesis. CD34+ cells or U937 cells were transduced with retroviral vectors encoding p100ΔN, empty control, or RelB 5′ of IRES-GFP. Diagrams show representative gated GFP+ cells (A) CD34+ cells stimulated in the presence of M-CSF plus IL-6, FL. and SCF for 10 days analyzed for CD11b versus CD14 expression. (B) CD34+ cells stimulated in the presence of G-CSF, SCF, and FLT3L for 6 days and analyzed for CD14 versus CD15 expression. (C) U937 cells analyzed for CD14 versus CD11b expression (top panel). Right diagram show a positive control (cells 72 hours after vitamin D3 stimulation). (D) CD34+ cells stimulated in the presence of expansion mix (SCF, FLT3L, TPO) for 7 days analyzed for CD14 versus CD11b expression. Markers were set according to negative control stainings. (A-D) Data are representative of at least 3 experiments. Numbers on the plots represent percentages of gated cells within the indicated quadrants.
To assess the conditions under which RelB is able to induce monopoiesis, we delivered our constructs into myelomonocyte lines (U937, TF1, and HL60) and primary CD34+ cells stimulated in a progenitor cell expansion mix (SCF, FLT3L, and TPO). We observed that ectopic expression of RelB failed to induce or enhance CD11b or CD14 expression in these experiments (Figure 6C; data not shown). These experiments show that RelB enhances human in vitro monopoiesis from CD34+ cells, and this effect requires cytokine conditions that promote monopoiesis.
Discussion
Two human myeloid–related DC subsets can be generated from CD34+ hematopoietic cord blood progenitors.1,2,6 Studies of their development previously led to the identification of putative shared LC/DC/monocyte precursors.9 Although it is known that lineage differentiation of these precursors is influenced by cytokines, underlying molecular mechanisms remain poorly characterized.
Here we demonstrated that the transcription factor RelB plays an important role in human DC subset development by promoting the generation of monocytic intermediates. We found that RelB overexpression promotes, whereas RelB inhibition impairs, the development of monocyte intermediates from putative shared LC/DC/monocytic precursors. RelB inhibition still allowed alternative myeloid differentiation pathways, as evidenced from undisturbed LC or granulocyte development. Furthermore, our data suggest that RelB generally promotes monopoiesis because RelB inhibition impaired the transition of CD11b-CD14lo precursor cells into CD11b+CD14+ monocytes, irrespective of the specific growth conditions used to induce human monopoiesis. Based on these results, we propose the following model (Figure 7): RelB regulates the generation of CD11b+CD14+ monocytes from common myeloid/DC progenitors. These cells are capable of differentiating into CD11b+CD1a+ monocyte–derived DCs. If this default pathway is blocked in response to RelB inhibition (by ectopic p100ΔN expression), cells accumulate at CD11b-CD14lo or CD11b-CD14- myeloid precursor cell stages. If stimulated appropriately, these precursors develop into LCs (with TGF-β1) or granulocytes (with G-CSF). Because CD11b-CD14+ cells rapidly undergo LC differentiation in response to TGF-β1,9 LC differentiation may transit through a CD14+ stage. Alternatively, it may transit through a more immature CD14- monopoietic stage (lysozyme+CD14-).7 Our data, therefore, support the concept that CD14loCD11b- precursors represent postcommitted shared human LC/DC/monocyte precursors.9 We propose that RelB is involved in the monocyte lineage fate decision of these precursors.
Model of RelB-mediated regulation of human DC subset generation. RelB promotes the differentiation CD14+CD11b+ monocytic intermediates from CD34+ hematopoietic progenitor cells. These cells can give rise to CD1a+CD11b+ interstitial-type DCs. CD34+ progenitors in which RelB is inhibited (by stable ectopic expression of the RelB inhibitory protein p100ΔN) are arrested at a CD11b- myeloid cell stage capable of differentiating into other myeloid lineages, including LCs. Our data suggest that CD14loCD11b- monocytes represent shared monocyte/LC/DC precursors that require RelB for differentiation into monocytic intermediates of interstitial DCs.
Model of RelB-mediated regulation of human DC subset generation. RelB promotes the differentiation CD14+CD11b+ monocytic intermediates from CD34+ hematopoietic progenitor cells. These cells can give rise to CD1a+CD11b+ interstitial-type DCs. CD34+ progenitors in which RelB is inhibited (by stable ectopic expression of the RelB inhibitory protein p100ΔN) are arrested at a CD11b- myeloid cell stage capable of differentiating into other myeloid lineages, including LCs. Our data suggest that CD14loCD11b- monocytes represent shared monocyte/LC/DC precursors that require RelB for differentiation into monocytic intermediates of interstitial DCs.
We showed that RelB inhibition impairs the development of monocyte-derived DCs, whereas LC differentiation and DC cluster adhesion remained unchanged. This contrasts the effects of IκBα-SR, which inhibits classical NF-κB signaling. We observed that LC/DC differentiation, DC adhesion, and myeloid progenitor cell proliferation are profoundly inhibited by IκBα-SR, as expected based on the findings of previous studies.36-41 Therefore, therapeutic interference with nonclassical NF-κB/RelB signaling might represent an attractive strategy to modulate the antigen-presenting cell system in vivo in a DC subset–specific manner without simultaneously affecting all human DC functions and subsets. Interestingly, IκBα-SR inhibited cell differentiation and proliferation without detectably enhancing apoptosis of cultured primary cells. This seems to be contrary to previous reports showing IκBα-SR–induced cell death in many cell types. Several possible explanations for these differences might be envisioned. Among them are lower gene copy numbers in our study compared with previous studies and cell type–specific differences. In line with this, primary CD34+ cells were previously reported to be more resistant to apoptosis induction by NF-κB inhibition compared with phenotypically identical myeloid leukemia stem cells.42
Human and murine DC subsets are difficult to compare because of their phenotypic differences (eg, CD8α, CD1a, or myeloid markers) and potential developmental differences (eg, possible lymphoid-related origin of LCs).2 Nevertheless, our data corroborate those obtained in the murine system. It has been shown that RelB-deficient murine hematopoietic cells fail to give rise to a specific DC subset (CD11c+/CD11b+/CD8α-/Dec205- splenic DCs).25 Furthermore, in line with our data demonstrating that RelB inhibition does not impair human granulopoiesis and LC development, murine RelB-deficient mice contain these lineages.25,43,44
RelB-dependent DCs in our study coexpressed CD1a and CD11b. Based on these characteristics they resembled monocyte-derived DCs generated in response to GM-CSF and IL-4,9,45 inflammatory dendritic epidermal cells (IDECs) of patients with atopic dermatitis,46,47 and LC histiocytosis cells.31,48 Conversely, several human DC subsets—including human LCs,31 pDCs,49,50 and peripheral blood DCs50 —lack CD11b. We speculate that RelB might promote a monocyte-derived human DC pathway in vivo. These cells might give rise to DCs, which are specifically recruited to inflammatory sites. Adding serum favors the monocytic over the LC pathway from progenitors.7,9 Given that RelB is rapidly induced in response to serum stimulation,51,52 factors contained in serum might promote monocyte-DC differentiation from progenitors through a RelB-dependent mechanism in vitro. Local stimuli in tissue microenvironments (interstitium or dermis) might replace serum factors under homeostatic conditions in vivo.
We observed that ectopic RelB fails to enhance DC maturation. Previous studies described enhanced expression and nuclear translocation of RelB as a hallmark of human myeloid DC maturation in response to various stimuli that cause DC maturation,23 leading to the speculation that RelB might functionally participate in myeloid DC maturation. Recently, it was shown that RelB is recruited to the nucleus with a markedly delayed kinetics compared with other NF-κB members during human DC maturation,24 resulting in an exchange of p65 and c-Rel by RelB-containing dimers over time.53 Furthermore, it was demonstrated that RelB binding to the IL-12p40 promoter correlates with a drop in RNApolII occupancy, suggesting that RelB contributes to the transcriptional down-regulation of this gene and potentially of other DC-related genes.53 Given this delayed kinetics, together with our observations that RelB overexpression fails to promote human DC maturation, it is unlikely that RelB is involved in initiating DC maturation. Instead, RelB might play a thus far unrecognized regulatory role in DCs that have already undergone maturation. Interestingly, RelB inhibition by p100ΔN did not impair DC maturation (data not shown), suggesting that RelB is not required for DC maturation. This is in line with the observation that monocyte-derived DCs, in which the nonclassical NF-κB pathway is inhibited by adenoviral expression of dominant negative NF-κB–inducing kinase (NIK), are still capable of effective T-cell activation in an allogeneic mixed-leukocyte reaction (MLR).54 It should be taken into account that RelB might influence antigen-presenting functions in vivo in mice because of perturbations in DC subset frequencies that differ in antigen-presentation functions.
The observed RelB-dependent effects seem to be restricted to certain differentiation stages, and it must be considered that cultures of CD34+ cells represent heterogeneous populations comprising cells at distinct stages of differentiation. For detailed biochemical studies, it will be critical to establish more homogenous primary cell systems that selectively mimic these transition steps. In addition, it must be considered that RelB-dependent enhancement of CD14+CD11b+ cell generation from CD34+ cells in our model required cytokine conditions that promote monopoiesis or DC differentiation (such as M-CSF, GM-CSF, and TNF-α) from progenitors. In comparison, CD34+ cells stimulated in expansion conditions (FLT3L, SCF, and TPO) or myelomonocytic model cell lines (U937, HL60, or TF1) did not show similar phenotypic changes in RelB or p100ΔN transduction experiments. Therefore, cytokines that promote monopoiesis or DC differentiation from primary cells may induce signaling processes that functionally cooperate with RelB (eg, nuclear translocation of RelB heterodimerization partners p50 or p52). In line with this possibility, the cotransfection of p50 and RelB exceeded RelB alone in its capacity to up-regulate antigen-presenting cell features in B-cell lines.55 Future mechanistic studies should address these questions in primary human cells.
We showed that inhibiting RelB impairs human monocyte-derived DC subset development, whereas precursors are still capable of differentiating along alternative myeloid pathways (LC and granulocytes). Together with our observations that RelB does not influence DC maturation, our in vitro data point to an important regulatory role of RelB on human DC subset regulation.
Prepublished online as Blood First Edition Paper, August 17, 2004; DOI 10.1182/blood-2004-02-0412.
Supported by START-Y156 grant from the Austrian Science Fund, Fond zur Förderung der wissenschaftlichen Forschung (FWF).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank all the collaborating nurses and doctors from the obstetric departments at Lainz Hospital and Kaiser Franz Josef Hospital, Vienna. Furthermore, we thank L. Heinz, M. Epstein, W. Ellmeier, A. Elbe-Burger, H. Stockinger, and W. Knapp for critical discussion.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal