The AML1/CBFβ transcriptional complex is essential for the formation of definitive hematopoietic stem cells (HSCs). Moreover, development of the hematopoietic system is exquisitely sensitive to the level of this complex. To investigate the effect of AML1 dosage on adult hematopoiesis, we compared the hematopoietic systems of AML1+/– and AML1+/+ mice. Surprisingly, loss of a single AML1 allele resulted in a 50% reduction in long-term repopulating hematopoietic stem cells (LTR-HSCs). This decrease did not, however, extend to the next level of hematopoietic differentiation. Instead, AML1+/– mice had an increase in multilineage progenitors, an expansion that resulted in enhanced engraftment following transplantation. The expanded pool of AML1+/– progenitors remained responsive to homeostatic mechanisms and thus the number of mature cells in most lineages remained within normal limits. Two notable exceptions were a decrease in CD4+ T cells, leading to an inversion of the CD4+ to CD8+ T-cell ratio and a decrease in circulating platelets. These data demonstrate a dosage-dependent role for AML1/CBFβ in regulating the quantity of HSCs and their downstream committed progenitors, as well as a more restricted role in T cells and platelets. The latter defect mimics one of the key abnormalities in human patients with the familial platelet disorder resulting from AML1 haploinsufficiency.
Introduction
The cloning of leukemia-associated translocations has led to the identification of a series of transcription factors that have critical functions in normal hematopoiesis. A prominent target of these translocations are the genes encoding the AML1/CBFβ transcription factor complex.1-4 AML1 (also known as RUNX1) is a member of a family of DNA-binding proteins that includes AML2 (RUNX3) and AML3 (RUNX2). These proteins bind to the core enhancer DNA sequence TGT/cGGT with a second subunit CBFβ, and both DNA binding and heterodimerization are mediated through a central domain in the AML family of proteins termed the runt-homology domain, or RHD. CBFβ does not itself contact DNA but instead increases the DNA-binding affinity of the AML family of proteins and protects them from ubiquitin-proteasome–mediated degradation.5-8
Homozygous deletion of either AML1 or CBFβ results in embryonic lethality, with embryos dying at embryonic day 12.5 (E12.5) from an absence of definitive hematopoiesis and central nervous system hemorrhages.9-12 Importantly, intra-aortic hematopoietic stem cells (HSC) clusters are absent in AML1-deficient embryos,13,14 and recent experiments have demonstrated that all embryonic HSCs express AML1.15 These data suggest that AML1/CBFβ is essential for the generation of definitive HSCs from mesodermal progenitors.
How AML1 carries out this critical developmental function remains unknown. Experimental data from a variety of systems have demonstrated that AML1/CBFβ regulates the expression of many hematopoietic-specific genes.1-4 However, despite the identification of a large number of AML1/CBFβ target genes, none appear to be essential for the generation of definitive HSCs. Thus, the critical targets involved in the formation of HSCs remain to be defined.
Importantly, the biologic functions of AML1 appear to be exquisitely sensitive to its intracellular levels. The best characterized example of this is the effect of AML1 haploinsufficiency on the temporal and spatial generation of definitive HSCs during embryogenesis.14 Loss of a single AML1 allele results in a temporal shift in the emergence of HSCs, resulting in an increase in HSCs at E10.5 in both the aorta-gonad-mesonephros (AGM) and yolk sac, followed 1 day later by a reduction in the ability of the AGM to support HSC maintenance and/or expansion.14-16 A possible explanation for the observed early increase in the number of HSCs in AML1+/– embryos was suggested by experiments geared toward assessing the in vitro differentiation capacity of AML1+/– ES cells.17 In these latter experiments, it was demonstrated that ES cells that were haploinsufficient for AML1 had an accelerated rate of mesoderm to hematopoietic cell differentiation as compared to that seen with wild-type cells.
The most compelling evidence for an effect of dosage on the biologic function of AML1/CBFβ comes from the observation of germ-line AML1 deletions or mutations in patients with a familial platelet disorder (FPD) that is associated with a high predisposition to develop acute myeloid leukemia (AML).18,19 The FPD/AML syndrome is a rare autosomal dominant disorder characterized by mild to moderate thrombocytopenia, abnormal platelet structure, and decreased megakaryocyte colony formation.20,21 Patients demonstrate a high incidence of developing leukemia in their fourth to fifth decade of life, with an overall lifetime risk of approximately 30%.18,20 A subset of the identified AML1 mutations appear to result in haploinsufficiency. Thus, in humans, simple loss of a single allele of AML1 leads to both an alteration in the number and function of megakaryocytes and to an enhanced susceptibility of HSCs or committed progenitors to develop into acute leukemia. Further supporting the latter conclusion is the identification of somatically acquired point mutations in AML1 in about 3% to 5% of sporadic cases of AML,22-24 with the highest frequency observed in leukemias that have an undifferentiated morphology (French-American-British [FAB]-M0).22,23,25 Interestingly, heterozygous point mutations of another family member AML3 have been identified in patients with cleidocranial dysplasia (CCD), a skeletal disorder with autosomal dominant inheritance.26-29 Most of the point mutations identified in AML1 and AML3 occur in the RHD and result in a loss of either DNA- or CBFβ-binding or encode truncated proteins that lack the C-terminal transactivation domain.18,19,22-31
These observations suggest that a threshold level of AML1/CBFβ is required for the normal function of HSCs and their more lineage-committed progeny. Although the functional consequence of AML1 haploinsufficiency on murine embryonic hematopoiesis has been defined, no detailed characterization has been performed on the hematopoietic system of AML1+/– adult mice. We now present the results of such an analysis.
Materials and methods
Mice
AML1 haploinsufficient mice (AML1+/–) were generated as previously described.9 For transplantation studies, recipient mice were 6- to 8-week-old female C57BL/6.SJL (Ly5.1+) (Jackson Laboratory, Bar Harbor, ME). Mice were maintained in compliance with the Standards for Humane Care and Use of Laboratory Animals by Public Health Research.
Hematologic analysis
Peripheral blood was withdrawn from mice via tail vein and collected in tubes containing EDTA (ethylenediaminetetraacetic acid), and complete blood count (CBC) analysis was performed using a Hemavet 3700 analyzer (Drew Scientific, Dallas, TX).
Bone marrow cell preparation and bone marrow transplantation
Bone marrow (BM) cells were harvested from 2- to 4-month-old mice. Red blood cells (RBCs) were lysed in RBC lysis solution (Puregene, Minneapolis, MN) and the white blood cells (WBCs) used for further experiments. Donor cells (AML1+/– or +/+) were from C57BL/6 (Ly5.2+) mice, and recipient were Ly5.1+. Three hours prior to transplantation, recipient mice were lethally irradiated with a split dose of 950 cGy. For in vivo limiting dilution assay, cells were cotransplanted with 2 to 5 × 104 irradiation protective cells obtained from BM of Ly5.1+ mice. At 2 and 4 months after transplantation the level of donor cell engraftment was assessed in blood by fluorescence-activated cell-sorter scanner (FACS). Recipient mice with a level of donor cell engraftment equal to 1% at 4 months in both the myeloid and lymphoid lineages were considered positive for calculations of the number of long-term repopulating hematopoietic stem cells (LTR-HSCs). Data from 2 independent experiments were combined, and the frequency of LTR-HSCs was estimated by Poisson statistics (L-calc software; StemCell Technologies, Vancouver, BC, Canada). For competitive repopulating assays, each lethally irradiated recipient mouse received 2 × 106 competitive cells (Ly5.1+) and 2 × 105 test cells (Ly5.2+) from AML1+/– or +/+ donor mice. For serial transplantation, recipients were killed 3 to 4 months following transplantation, and Ly5.2+ BM cells were isolated by FACS. The sorted Ly5.2+ cells (0.5 to 1.0 × 106) were then injected into new Ly5.1+ recipient mice and engraftment assessed by FACS of the BM at 12 to 16 weeks.
Flow cytometric analysis
All antibodies were purchased from Pharmingen (San Jose, CA) and are listed in the Supplemental Data on the Blood website; see the Supplemental Figures link at the top of the online article. Stained cells were analyzed using a FACSCalibur (Becton Dickinson, San Jose, CA), and cell sorting was performed using a FACSAria, as previously described.32
Hematopoietic colony assays
For methycellulose colony-forming units–culture (CFU-C) assays, BM cells were resuspended in Iscove modified Dulbecco medium (StemCell Technologies) supplemented with 15% heat-inactivated fetal bovine serum. Appropriate number of hematopoietic cells were then cultured in MethoCult GF M3434 or MethoCult M3234 (StemCell Technologies) supplemented with recombinant EPO (rEPO; Ortho Biotech, Bridgewater, NJ), recombinant murine IL-3 (rmIL-3), rhIL-6, and rmSCF (PeproTech, Rocky Hill, NJ), as previously described.32
In vivo homing assay
In vivo hematopoietic cell homing assay was performed as previously described.35,36 Briefly, BM cells were labeled with the fluorescent dye carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes, Eugene, OR), washed, and then injected into lethally irradiated recipient mice. Mice were killed 4 to 5 hours after the transplantation, and the percentage of CFSE+ cells in the BM and spleen were determined by FACS.
Cell cycle analysis
Propidium iodide (PI) and dual staining with pyronin Y (PY) and Hoechst 33342 were used for cell cycle analysis of BM-derived cells using previously published procedures.37 Details of the methods are provided in the Supplemental Data. Quantitative reverse transcriptase–polymerase chain reaction analysis was performed to assess the level of expression of the mRNAs encoding p15INK4b, p16INK4a, p21waf1/cip1, p27kip1, and p57kip2. Amplification and detection were performed with primers and probes from Applied Biosystems (Foster City, CA), using methods previously described.38
Results
Hematological analysis
To minimize the genetic background effects on the analysis of hematopoiesis, all of the mice used in this study were 2- to 4-month-old age-matched littermates back-crossed onto the C57BL/6 background for more than 10 generations. We first performed a CBC on AML1+/– and +/+ mice. This analysis revealed no significant differences in the peripheral blood hematocrit level, hemoglobin level, RBC number, or the number or differential of WBCs as a result of AML1 haploinsufficiency (Table 1). However, similar to patients with the FPD/AML syndrome, we identified an approximately 15% reduction in the number of platelets in the peripheral blood of AML1+/– mice (Table 1). We next examined the morphologic appearance of the hematopoietic organs, including BM, spleen, lymph nodes, and thymus. Although no significant morphologic abnormalities were identified in the AML1+/– mice, there was a trend toward an increase in the number of megakaryocytes within their BM. However, when the number of megakaryocytes per high-power field was quantified, the difference between AML1+/– versus AML1+/+ was not statistically significant (data not shown). In addition, no significant differences were noted in the overall BM cellularity between these 2 groups (Supplemental Data, Figure 1S).
Peripheral blood analysis of AML1+/+ and +/- mice
Hematologic parameter . | AMLI+/+ . | AML1+/- . |
|---|---|---|
| RBC count (× 106/uL) | 9.56 ± 0.83 | 9.49 ± 0.57 |
| HB level (g/dL) | 13.56 ± 0.7 | 13.62 ± 0.59 |
| HCT level (%) | 44.63 ± 3.73 | 43.63 ± 2.49 |
| WBC count (× 103/uL) | 9.94 ± 1.2 | 10.77 ± 0.98 |
| Differential of WBC | ||
| Lymphocyte (%) | 79.31 ± 3.6 | 77.25 ± 3.07 |
| Neutrophil (%) | 16.87 ± 2.97 | 18.80 ± 3.63 |
| Monocyte (%) | 3.41 ± 1.36 | 3.28 ± 1.19 |
| PLT (× 103/uL) | 787.75 ± 46.11 | 672.31 ± 54.45* |
Hematologic parameter . | AMLI+/+ . | AML1+/- . |
|---|---|---|
| RBC count (× 106/uL) | 9.56 ± 0.83 | 9.49 ± 0.57 |
| HB level (g/dL) | 13.56 ± 0.7 | 13.62 ± 0.59 |
| HCT level (%) | 44.63 ± 3.73 | 43.63 ± 2.49 |
| WBC count (× 103/uL) | 9.94 ± 1.2 | 10.77 ± 0.98 |
| Differential of WBC | ||
| Lymphocyte (%) | 79.31 ± 3.6 | 77.25 ± 3.07 |
| Neutrophil (%) | 16.87 ± 2.97 | 18.80 ± 3.63 |
| Monocyte (%) | 3.41 ± 1.36 | 3.28 ± 1.19 |
| PLT (× 103/uL) | 787.75 ± 46.11 | 672.31 ± 54.45* |
Analyses were performed on 12-week-old sex-matched littermates with 12 individual mice per genotype. Results are given as the average for the group ± standard error.
RBC indicates red blood cell count; HB, hemoglobin; HCT, hematocrit; WBC, white blood cell count; and PLT, platelet counts.
P < .0001, exact Wilcoxon test.
To complement this morphologic analysis, we also determined the percentage of cells of different hematopoietic lineages in the BM and spleen by FACS. In agreement with the results from the peripheral blood analysis, no significant differences were detected in the number of myeloid, erythroid, or lymphoid cells in the BM or spleen of AML1+/– versus +/+ mice (Figure 1A). However, as previously reported by others,32,39,40 there was an inversion in the CD4+ to CD8+ T-cell ratio in the spleens of AML1+/– mice when compared to wild-type littermates (Figure 1B). Thus, haploinsufficiency of AML1 resulted in minimal effects on the steady-state levels of hematopoietic cells in the young adult mouse, with significant changes limited to platelets and T cells.
Percentage of lineage-positive cells in the BM and spleen of AML1+/+ and +/– mice. (A) BM cells were isolated from either wild-type (left) or AML1+/– (right) mice and stained for Mac1 and GR-1, CD3 and B220, or Ter119, and the relative frequency of the individual hematopoietic lineages was determined by FACS. The percentage of cells positive for the indicated cell surface markers is denoted in each panel. (B) Spleen cells were isolated from wild-type (left) or AML1+/– (right) mice and stained for CD4 and CD8, and the frequency of cells staining with these antibodies was determined by FACS. The percentage of CD4+ and CD8+ cells, along with the CD4+ to CD8+ T-cell ratio, is indicated. The profile shown is from a single pair of age- and sex-matched littermates. Identical results were obtained from 2 other pairs of mice.
Percentage of lineage-positive cells in the BM and spleen of AML1+/+ and +/– mice. (A) BM cells were isolated from either wild-type (left) or AML1+/– (right) mice and stained for Mac1 and GR-1, CD3 and B220, or Ter119, and the relative frequency of the individual hematopoietic lineages was determined by FACS. The percentage of cells positive for the indicated cell surface markers is denoted in each panel. (B) Spleen cells were isolated from wild-type (left) or AML1+/– (right) mice and stained for CD4 and CD8, and the frequency of cells staining with these antibodies was determined by FACS. The percentage of CD4+ and CD8+ cells, along with the CD4+ to CD8+ T-cell ratio, is indicated. The profile shown is from a single pair of age- and sex-matched littermates. Identical results were obtained from 2 other pairs of mice.
AML1 haploinsufficiency results in an increase in multilineage and lineage-restricted progenitors
We next directly assessed the functional consequences of a reduced AML1 gene dosage on lineage-committed hematopoietic progenitors. We first used the methycellulose CFU-C assay to compare the number of myeloid-committed progenitors in the BM of AML1+/– and +/+ mice. Surprisingly, the total number of myeloid colonies (erythroid, granulocyte, macrophage, granulocyte/macrophage, and mixed) detected in the BM of AML1+/– mice were 50% greater than the number detected in marrow from wild-type littermates (79.8 ± 13.6/1 × 104 total BM cells versus 52.2 ± 18.2/1 × 104 cells, respectively, P < .05, Figure 2A). This increase was primarily due to an increase in the frequency of progenitors that give rise to granulocyte (G)– and/or macrophage (M)–restricted colonies (CFU-G/M), (63.2 ± 10.3 versus 39.2 ± 11.4, respectively, P < .005) or to more primitive mixed colonies CFU-granulocyte erythrocyte macrophage megakaryocyte (CFU-GEMM)9 (6 ± 0.8 versus 4.2 ± 1.8, P < .005). No significant differences were observed in the number of erythroid colonies (BFU-E, 11.26 ± 4.1 versus 7.4 ± 3.5, respectively, P = .08).
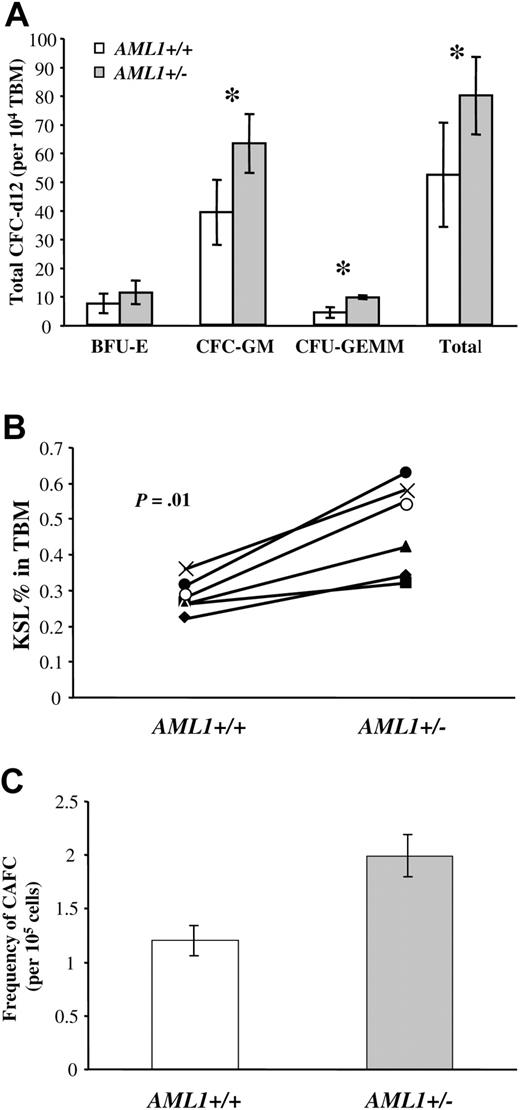
Analysis of multilineage and lineage-committed progenitors in the BM of AML1+/+ and +/– mice. (A) Total bone marrow (TBM) cells (1 × 104) from AML1+/+ or +/– mice were plated in duplicate in methylcellulose cultures containing IL-3, IL-6, SCF, and EPO, and the number of hematopoietic colonies was morphologically determined after 12 days of culture. Results are the average ± standard deviation from 5 mice for each genotype. The groups with statistically significant difference are marked by an asterisk. (B) TBM cells from AML1+/+ and +/– mice (6 mice per genotype) were analyzed by FACS to determine the number of c-Kit+Sca1+Linlow/– (KSL). The percentage of KSL cells in total BM is indicated for each pair of age- and sex-matched AML1+/+ and +/– mice. The average percentage of KSL cells in AML1+ /+ versus +/– mice was 0.28 ± 0.02 and 0.47 ± 0.05, respectively (P = .01). (C) TBM cells from AML1+/+ and +/– mice were serial diluted and then plated in long-term stromal-supported hematopoietic cultures using a 96-well format. Twenty replicate wells were seeded at each dilution, and wells containing clusters of 6 or more cobblestone cells after 28 days of culture were scored as positive. The combined data from 3 independent experiments were used to estimate the number of CAFC day 28 by Poisson statistics. This analysis indicated an increase in the frequency of CAFC-d28 in AML1+/– versus +/+ mice (1 in 50 263 ± 5 871 versus 1 in 82 953 ± 9298, respectively; P < .001).
Analysis of multilineage and lineage-committed progenitors in the BM of AML1+/+ and +/– mice. (A) Total bone marrow (TBM) cells (1 × 104) from AML1+/+ or +/– mice were plated in duplicate in methylcellulose cultures containing IL-3, IL-6, SCF, and EPO, and the number of hematopoietic colonies was morphologically determined after 12 days of culture. Results are the average ± standard deviation from 5 mice for each genotype. The groups with statistically significant difference are marked by an asterisk. (B) TBM cells from AML1+/+ and +/– mice (6 mice per genotype) were analyzed by FACS to determine the number of c-Kit+Sca1+Linlow/– (KSL). The percentage of KSL cells in total BM is indicated for each pair of age- and sex-matched AML1+/+ and +/– mice. The average percentage of KSL cells in AML1+ /+ versus +/– mice was 0.28 ± 0.02 and 0.47 ± 0.05, respectively (P = .01). (C) TBM cells from AML1+/+ and +/– mice were serial diluted and then plated in long-term stromal-supported hematopoietic cultures using a 96-well format. Twenty replicate wells were seeded at each dilution, and wells containing clusters of 6 or more cobblestone cells after 28 days of culture were scored as positive. The combined data from 3 independent experiments were used to estimate the number of CAFC day 28 by Poisson statistics. This analysis indicated an increase in the frequency of CAFC-d28 in AML1+/– versus +/+ mice (1 in 50 263 ± 5 871 versus 1 in 82 953 ± 9298, respectively; P < .001).
To quantitatively assess the number of more immature multilineage progenitors, we next determined the number of c-Kit+Sca-++Linlow/– (KSL) cells in the BM by FACS. This population of cells typically constitutes less than 1% of the BM and contains nearly all LTR-HSCs. However, most KSL cells represent more restricted short-term repopulating HSCs, multilineage progenitors, or lineage-committed progenitors.41-44 Because of the experimental variability inherent in the flow analysis of such a rare population of cells, we analyzed 6 independent pairs of age- and sex-matched AML1+/– and +/+ littermates. Although no differences were noted in the total BM cellularity between mouse strains in this pair-wise analysis (Figure 1S), a significant increase was detected in the number of KSL cells in the BM of AML1+/– versus AML1+/+ mice (0.47% ± 0.05% versus 0.28% ± 0.02%, respectively, P = .01, Figure 2B).
To further verify the increase in hematopoietic progenitors in AML1+/– BM, we used an in vitro stromal long-term BM culture system to quantitate the number of primitive hematopoietic cells. In this culture BM cells are grown over a layer of irradiated stromal cells for a period of up to 4 weeks.34 Progenitors migrate beneath the stromal layer and grow as clusters of cobblestone-appearing cells that are easily visualized using phase contrast microscopy. The cobblestone area–forming cells (CAFCs) that appear early (day 10) correspond to colony forming unit spleen day 12 (CFU-S12), whereas the CAFCs that appear after 28 days represent more primitive progenitors.34 Using limiting dilution analysis, the frequency of CAFC can be calculated by Poisson statistics. When this approach was used to assess the number of day-28 CAFCs in AML1+/– and +/+, we observed a small but statistically significant increase in the frequency of CAFCs in the BM of the AML1+/– mice (P < .001, Figure 2C). Taken together, these data demonstrate an increase in multilineage and lineage-committed progenitors in AML1+/– mice.
AML1+/– mice have a decrease in the number of LTR-HSCs
To directly assess the number of LTR-HSCs in BM of AML1+/– and +/+ mice, we performed an in vivo limiting dilution BM transplantation assay. Serial dilutions of BM cells from “test” mice (AML1+/– or +/+, Ly5.2+) were transplanted into lethally irradiated Ly5.1+ syngeneic recipients (Supplemental Data, Figure 2S). Engraftment was assessed at 4 months after transplantation by defining the number of Ly5.2+ cells in the peripheral blood of recipients by FACS.45 Transplantations were done using 4 to 5 donors and 5 to 16 recipients per dilution, and recipients with more than or equal to 1% donor cell engraftment were considered positive for calculations of the number of HSCs using Poisson statistics.45,46 There was no statistical difference in the level of short-term repopulating hematopoietic stem cells (STR-HSCs) between AML1+/– and AML1+/+ mice at 2 months after transplantation. Surprisingly, however, we observed a nearly 50% reduction in the frequency of LTR-HSCs at 4 months after transplant in AML1+/– versus AML1+/+ mice (P < .01, Table 2). Importantly, recipient mice from both AML1+/– and wild-type donors showed both myeloid and lymphoid engraftment (data not shown), suggesting that no difference existed in the ability of the LTR-HSCs to differentiate along various hematopoietic lineages.
AML1 haploinsufficiency results in a decrease in LTR-HSCs
Cell number* . | AML1+/+ mice + for donor engraftment/total mice . | AML1+/- mice + for donor engraftment/total mice . |
|---|---|---|
| 200 000 | 5/5† | 8/8 |
| 100 000 | 5/5 | 5/5 |
| 32 000 | 10/10 | 14/16 |
| 16 000 | 11/12 | 8/15 |
| 8 000 | 3/12 | 1/15 |
| 4 000 | 5/16 | 1/6 |
| 2 000 | 0/6 | 0/6 |
| LTR-HSCs | 1 in 11 171 ± 2509‡ | 1 in 22 878 ± 5102 |
Cell number* . | AML1+/+ mice + for donor engraftment/total mice . | AML1+/- mice + for donor engraftment/total mice . |
|---|---|---|
| 200 000 | 5/5† | 8/8 |
| 100 000 | 5/5 | 5/5 |
| 32 000 | 10/10 | 14/16 |
| 16 000 | 11/12 | 8/15 |
| 8 000 | 3/12 | 1/15 |
| 4 000 | 5/16 | 1/6 |
| 2 000 | 0/6 | 0/6 |
| LTR-HSCs | 1 in 11 171 ± 2509‡ | 1 in 22 878 ± 5102 |
Results represent 2 independent experiments, with a mixture of BM cells from 2 to 3 donor mice used in each experiment.
Number of “test” BM cells from AML1+/+ or +/- mice injected into each recipient.
Mice positive for donor engraftment were defined as those with more than 1% Ly5.2+ cells in the peripheral blood by FACS in both the lymphoid and myeloid gates as assessed by forward and side scatter.
The LTR-HSC frequency was calculated by Poisson statistics. The difference between AML1+/+ and +/- was statistically significant (P = .006).
The observed decrease in LTR-HSCs in the AML1+/– mice could be due to a reduction in the absolute number of LTR-HSCs or, alternatively, could reflect a decrease in the ability of AML1+/– LTR-HSCs to home into hematopoietic organs. To rule out the latter possibility, we assessed the ability of hematopoietic cells from AML1+/– and wild-type mice to home into the hematopoietic organs of recipient mice following transplantation. Using an in vivo homing assay,35,36 we failed to detect any significant differences in the homing capacity of AML1+/– versus AML1+/+ cells (Supplemental Data, Figure 3S). Therefore, the reduction in the number of LTR-HSCs in AML1+/– does not appear to be the result of a homing defect but instead appears to reflect an absolute decrease in their numbers in AML1+/– mice.
LTR-HSCs from AML1 haploinsufficient mice show enhanced engraftment levels
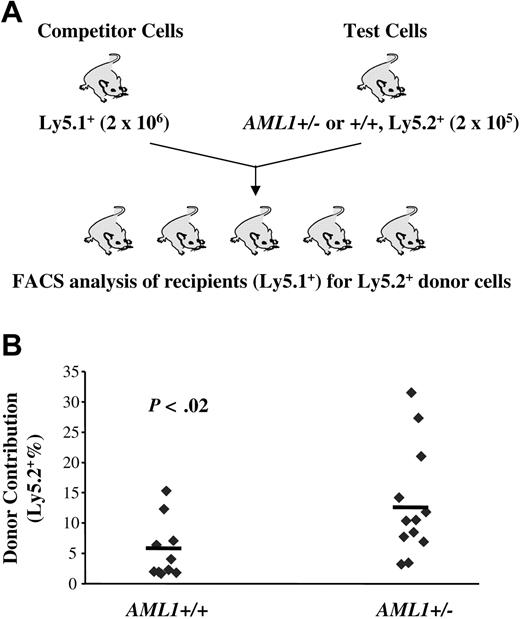
The data presented so far reveal a reduction in LTR-HSCs in AML1 haploinsufficient mice and a paradoxical increase in lineage-restricted progenitors. These data suggested that loss of one AML1 allele alters the balance between HSCs and their progeny, favoring the expansion of more mature progenitors. If this hypothesis is correct, HSCs from AML1+/– mice should, in a competitive long-term repopulating assay, yield a higher percentage of mature cells than wild-type HSCs. To test this hypothesis, we mixed “test” cells (Ly5.2+) from either AML1+/– or +/+ mice with “competitor” cells (Ly5.1+) in a 1:10 ratio and transplanted this mixed cell population into lethally irradiated Ly5.1+ recipients. The percentage of Ly5.2+ cells in the peripheral blood of recipient mice was then monitored monthly. In agreement with our hypothesis, we consistently observed an elevated percentage of Ly5.2+ cells in the peripheral blood of recipient mice transplanted with cells from AML1+/– donors from 4 weeks through 24 weeks after BM transplantation (Figure 4; Table 3). A similar level of enhanced engraftment was seen when a ratio of competitor-to-test cells of 1:1 was used (data not shown). Importantly, although AML1+/– cells demonstrated only an approximately 2-fold increase in level of engraftment, this was based on the use of BM samples that had only one half the number of LTR-HSCs found in wild-type mice. Thus, the competitive growth advantage of AML1+/– cells was at least 4-fold greater than wild-type cells. A higher percentage engraftment also was seen in AML1+/– secondary transplant recipients, further supporting a better repopulating capacity (Supplemental Data, Figure 4S). These data suggest that although haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs, those that are generated yield an increased number of multilineage and lineage-committed progenitors, as well as their mature progeny.
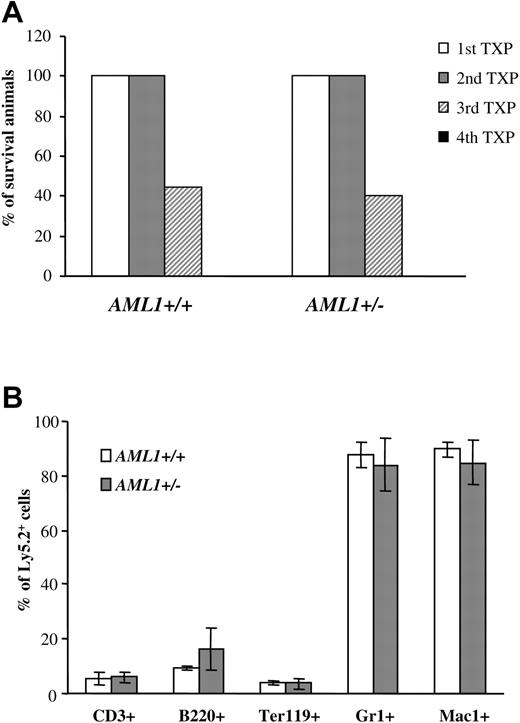
Similar self-renewal capacity seen for AML1+/– and +/+ LTR-HSCs. (A) 1 × 106 BM cells (Ly5.2+) from AML1+/+ or +/– mice were injected into lethally irradiated recipients. Three to four months after transplantation, BM cells were harvested from animals that had received transplants, donor-derived Ly5.2+ cells were sorted by FACS, and 0.5 to 1 × 106 Ly5.2+ cells were injected into the secondary recipients. This procedure was then repeated sequentially 3 to 4 months after each transplantation. The percentage of mice surviving from 4 sequential cycles is shown (n = 9-10 mice/group). (B) BM cells from secondary recipients were analyzed for multilineage engraftment by FACS. The percentage of Ly5.2+ T cells (CD3+), B cells (B220+), erythroid (Ter119+), and GR1+ and Mac1+ myeloid cells is shown. (n = 4 mice/group). Error bars represent standard deviation.
Similar self-renewal capacity seen for AML1+/– and +/+ LTR-HSCs. (A) 1 × 106 BM cells (Ly5.2+) from AML1+/+ or +/– mice were injected into lethally irradiated recipients. Three to four months after transplantation, BM cells were harvested from animals that had received transplants, donor-derived Ly5.2+ cells were sorted by FACS, and 0.5 to 1 × 106 Ly5.2+ cells were injected into the secondary recipients. This procedure was then repeated sequentially 3 to 4 months after each transplantation. The percentage of mice surviving from 4 sequential cycles is shown (n = 9-10 mice/group). (B) BM cells from secondary recipients were analyzed for multilineage engraftment by FACS. The percentage of Ly5.2+ T cells (CD3+), B cells (B220+), erythroid (Ter119+), and GR1+ and Mac1+ myeloid cells is shown. (n = 4 mice/group). Error bars represent standard deviation.
AML1+/- HSCs show enhanced donor cell engraftment under conditions of competitive repopulation
. | Mean level of engraftment (%) . | . | . | |
|---|---|---|---|---|
| Time points . | AML1+/+ . | AML1+/- . | P . | |
| Week 4 | 5.372* | 8.212 | .0169† | |
| Week 8 | 5.608 | 9.314 | .0692 | |
| Week 12 | 4.855 | 10.713 | .0112 | |
| Week 16 | 4.419 | 10.706 | .0206 | |
| Week 20 | 4.913 | 11.481 | .0426 | |
| Week 24 | 4.589 | 11.796 | .0169 | |
. | Mean level of engraftment (%) . | . | . | |
|---|---|---|---|---|
| Time points . | AML1+/+ . | AML1+/- . | P . | |
| Week 4 | 5.372* | 8.212 | .0169† | |
| Week 8 | 5.608 | 9.314 | .0692 | |
| Week 12 | 4.855 | 10.713 | .0112 | |
| Week 16 | 4.419 | 10.706 | .0206 | |
| Week 20 | 4.913 | 11.481 | .0426 | |
| Week 24 | 4.589 | 11.796 | .0169 | |
The results are the average of 2 independent experiments. For each experiment, 5 to 6 mice per genotype received transplants using a ratio of 10:1 competitors to test cells. The level of engraftment in the peripheral blood (percentage of Ly5.2+ cells) was determined by FACS.
The statistical significance of the differences between the genotypes at each time point was assessed using an exact Wilcoxon test.
LTR-HSCs could generate an increase in committed progenitors by either shifting cell divisions away from self-renewal and toward differentiation and/or by generating lineage-committed progenitors that have an enhanced ability to expand in response to cytokines. Serial BM transplantations were used to assess directly the self-renewal capacity of LTR-HSCs. 1 × 106 BM cells (Ly5.2+) from each genotype were injected into lethally irradiated Ly5.1 primary recipients. Three to four months after transplantation, 0.5 to 1 × 106 Ly5.2+ BM cells from the animals that received transplants were used as donor cells for secondary recipients (Ly5.1+), and the procedure was then repeated sequentially. By the third cycle, fewer than 50% of the recipients showed engraftment, and by the fourth cycle, no engraftment was observed and all recipient mice died (Figure 3A). Importantly, however, no significant differences were seen in the engraftment potential during serial transplantation between AML1+/– and +/+ (Figure 4A). Moreover, recipient mice from both AML1+/– and +/+ donors showed full multilineage engraftment (Figure 4B), indicating that no defect existed in the differentiation of AML1+/– LTR-HSCs. Taken together, these data suggested that no significant differences were observed between the self-renewal capacity of LTR-HSCs from AML+/– or +/+ mice.
AML1+/– cells produce a higher engraftment level in a competitive long-term repopulating assay. (A) Schematic diagram of the in vivo competitive repopulating assay. 2 × 105 test BM cells from 2 to 3 AML1+/– or +/+ mice (Ly5.2+) were mixed together with a 10-fold higher number (2 × 106) of competitor cells (Ly5.1+) and then transplanted into lethally irradiated Ly5.1+ mice (n = 10-12 mice/group). The percentage of Ly5.2+ cells in the peripheral blood was determined by FACS every 4 week after transplantation. (B) Percentage of Ly5.2+ cells in the blood at week 24 after transplantation.
AML1+/– cells produce a higher engraftment level in a competitive long-term repopulating assay. (A) Schematic diagram of the in vivo competitive repopulating assay. 2 × 105 test BM cells from 2 to 3 AML1+/– or +/+ mice (Ly5.2+) were mixed together with a 10-fold higher number (2 × 106) of competitor cells (Ly5.1+) and then transplanted into lethally irradiated Ly5.1+ mice (n = 10-12 mice/group). The percentage of Ly5.2+ cells in the peripheral blood was determined by FACS every 4 week after transplantation. (B) Percentage of Ly5.2+ cells in the blood at week 24 after transplantation.
To examine the proliferative properties of lineage-committed progenitors, we assessed the CFU-C capacity of BM cells from AML1+/– and +/+ mice in response to varying concentrations of cytokines. Total BM cells were plated in duplicate in methycellulose containing cultures supplemented with cytokine cocktails of interleukin 3 (IL-3), IL-6, stem cell factor (SCF), and erythropoietin (EPO) at 5 different concentration that varied over a 2-log range (from the standard concentration of IL-3, 10 ng/mL; IL-6, 10 ng/mL; SCF, 50 ng/mL; and EPO, 3 U/mL, to a lowest concentration of IL-3, 0.1 ng/mL; IL-6, 0.1 ng/mL; SCF, 0.5 ng/mL; and EPO, 0.05 U/mL). At all tested concentrations, we observed a slight increase in the number of CFU-G/M, CFU-GEMM, and total colonies in the AML1+/– BM; however, no differences were noted in the size or number of cells per colony at any of the tested concentrations of cytokines (negative data not shown). Similarly, no differences were detected in the number of colonies or the number of cells per colony when KSL cells were used in these in vitro hematopoietic colony assays. These data suggested that no significant differences can be detected in the proliferative capacity between AML1+/– and +/+ progenitors using this in vitro assay.
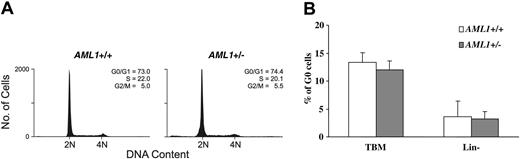
To further examine the cell cycle status of hematopoietic progenitors, lineage-negative BM cells were stained with PI and the distribution of cells in the different phases of the cell cycle assessed by FACS. As shown in Figure 5A, no significant difference was seen in the percentage of cells in the different cell cycle phases between the 2 genotypes. The cell cycle status was further analyzed by dual staining with the RNA dye PY and DNA dye Hoechst 33342. Again, no significant differences were noted in the percentage of cells in G0 (PY low, Hoechst low) in either total BM cells or in the lineage-negative subpopulation (Figure 5B). Consistent with these observations, we saw no difference in the level of mRNA expression of the key cell cycle regulators p15INK4b, p16INK4a, p21waf1/cip1, p27kip1, and p57kip2 between KSL cells from AML1+/– and wild-type animals (negative data not shown, n = 3). Thus, although an increase in lineage-committed progenitors is consistently seen in the AML1+/– mice, the underlying mechanism responsible for this expansion remains to be identified.
Cell cycle analysis of AML1+/– and +/+ BM cells. (A) Lineage-negative BM cells were labeled with PI and subjected to FACS analysis. Shown are representative PI FACS profiles of wild-type and AML1+/– cells. (B) Total BM cells (TBM) and lineage-negative BM cells (Lin–) were dual-stained with the RNA dye PY and DNA dye Hoechst 33342, and the percentage of G0 cells (PY low, Hoechst low) was determined by FACS. The values shown represent the mean and standard deviation of 6 independent experiments.
Cell cycle analysis of AML1+/– and +/+ BM cells. (A) Lineage-negative BM cells were labeled with PI and subjected to FACS analysis. Shown are representative PI FACS profiles of wild-type and AML1+/– cells. (B) Total BM cells (TBM) and lineage-negative BM cells (Lin–) were dual-stained with the RNA dye PY and DNA dye Hoechst 33342, and the percentage of G0 cells (PY low, Hoechst low) was determined by FACS. The values shown represent the mean and standard deviation of 6 independent experiments.
Discussion
A detailed analysis of the hematopoietic system of mice that are haploinsufficient for the DNA-binding subunit of the AML1/CBFβ transcription factor complex revealed a number of subtle but significant alterations at the level of HSCs and progenitors. At the top of the hematopoietic hierarchy, loss of a single allele of AML1 resulted in a dramatic 50% reduction in the total number of LTR-HSCs as measured by transplantation. Paradoxically, this decrease was not reflected at the next level of hematopoietic differentiation and lineage commitment. Instead, AML1+/– mice were found to have an increase in multilineage progenitors as reflected by an increase in total BM KSL cells, CAFCs, and myeloid and mixed CFU-Cs. The expansion of progenitors remained evident following transplantation of AML1+/– LTR-HSCs, with recipient mice showing an enhanced level of engraftment when measured under conditions of competitive repopulation. The expanded pool of committed progenitors, however, remained responsive to the homeostatic mechanisms that control the number of mature cells, and thus no significant increases were noted in mature hematopoietic cell numbers in AML1+/– mice. In fact, careful analysis of the number of cells of specific lineages revealed that haploinsufficiency of AML1 resulted in a decrease in the number of CD4+ T cells leading to an inversion of the CD4+ to CD8+ T-cell ratio, as previously reported,32,39,40 and a decrease in the number of circulating platelets. Taken together, these data demonstrate an important role for AML1 in regulating the quantity of HSCs and their downstream lineage-committed progenitors, as wells as a more restricted role in T cells and megakaryocytes.
In previous studies from our laboratory,14 it was demonstrated that haploinsufficiency of AML1 resulted in a quantitative early increase in LTR-HSCs in the developing embryo at E10.5, followed by a rapid decrease in the ability of the AGM to support the further generation or maintenance of these cells. By E11.5 the total number and size of hematopoietic clusters emerging from the aorta, vitelline, and umbilical arteries was decreased in AML1+/– embryos. This decrease was further reflected by a decrease in the number of CFU-S11 within the yolk sac, AGM, and fetal liver at E11.5 to 12.5. Our data now demonstrate that the early decrease in the number of LTR-HSCs persists into adulthood. Thus, the 50% reduction in the number of LTR-HSCs in adult AML1+/– mice is likely to be a direct result of a decrease in their generation and/or maintenance in the developing embryo. The LTR-HSCs that developed in AML1+/– mice, however, showed full differentiation potential, contributing to both myeloid and lymphoid lineages following transplantation. Understanding how AML1 haploinsufficiency leads to the early appearance of LTR-HSCs followed by a decrease in their total number will require defining the components of the AML1/CBFβ transcriptional cascade that are involved in the generation, expansion, and maintenance of LTR-HSCs.
In contrast to the effects on LTR-HSCs, AML1 haploinsufficiency leads to an increase in more committed progenitors, suggesting that AML1 may negatively regulate the number of hematopoietic progenitors. This effect was recently shown to be even more dramatic following the complete loss of AML1 in adult mice using a conditional Runx1 null allele.47 Although the number and engraftment potential of LTR-HSCs were maintained following the complete loss of AML1, a significant increase was observed in the number of KSL cells, CFU-Cs, and megakaryocyte progenitors. The relative increase in the number of progenitors following the complete loss of AML1 was greater than that observed following AML1 haploinsufficiency, suggesting an AML1 dose-response effect.
It is formally possible that the increase in progenitors in AML1+/– mice is the result of a shift in the proliferation of LTR-HSCs away from self-renewal divisions and more toward differentiation. This possibility, however, appears unlikely in that LTR-HSCs from AML1+/– mice showed similar self-renewal capacity to wild-type stem cells in a serial transplantation assay. Alternatively, the expansion in committed progenitors could result from a partial block in myeloid terminal differentiation caused by haploinsufficiency. However, similar multilineage engraftment was observed for both AML1+/+ and +/– donors, even after secondary transplantations, and no evidence of a block in myeloid differentiation was observed. A third explanation for the expansion in committed progenitors is that AML1+/– multilineage and lineage-restricted progenitors might have an enhanced proliferation and/or self-renewal capacity, resulting in an absolute expansion in their numbers under steady-state conditions. We observed no evidence for an enhanced proliferative capacity in AML1+/– progenitors in response to cytokine stimulation nor a significant alteration in the percentage of progenitors actively within the cell cycle between AML1+/– and +/+ mice. These data suggest that a simple alteration in proliferative capacity is unlikely to be responsible for the observed expansion in progenitors.
An alteration in the self-renewal capacity between AML1+/– and +/+ progenitors is an interesting possibility in light of the finding that expression of the t(8;21)-encoded AML1-ETO fusion protein leads to an increase in the self-renewal capacity of murine48 and human hematopoietic progenitors,49 including lineage-restricted progenitors. AML1 haploinsufficiency could similarly alter the self-renewal capacity of these cells, leading to their expansion. However, when we directly assessed this possibility, we failed to detect any significant difference in the self-renewal capacity between AML1+/– and +/+ progenitors using a serial CFU-S assay (W.S. and J.R.D., unpublished data, May 2004). By contrast, following the complete loss of AML1,47 a subtle increase was observed in the self-renewal capacity of progenitors, although the magnitude of this increase was significantly less than that induced by the expression of the AML1-ETO.48-50 Taken together, these data suggest that the self-renewal capacity of hematopoietic progenitors is influenced by the intracellular level of AML1. However, if AML1 haploinsufficiency induces a change in this biologic process, it is below our ability to detect it using the assays employed. Thus, defining the underlying mechanism responsible for the enhanced number of hematopoietic progenitors that results from a decrease in AML1 dosage will require genetic approaches to elucidate the components of the AML1/CBFβ-induced transcriptional cascade that are important for regulating this biological process.
The reduction in CD4+ T cells in peripheral lymphoid organs and the resultant decrease in the CD4+ to CD8+ T-cell ratio that results from AML1 haploinsufficiency have been previously reported.39,40,51 Importantly, AML1 and its related family member, AML2 (RUNX3), have been demonstrated to play distinct roles at several stages of T-cell development.51 Specifically, using a conditional AML1 knock-out allele, it was recently demonstrated that AML1 is required for the repression of CD4 in immature double-negative (DN) thymocytes, for T-cell receptor β (TCRβ) selection during the transition of DN to double-positive (DP) thymocytes, and for up-regulation of CD8 in DP and single-positive thymocytes. Moreover, expression of AML1 appears to be necessary for the normal expansion or survival of CD4+ T cells in peripheral lymphoid organs. By contrast, AML2 is essential for the repression of CD4 in mature CD8+ T cells and for their antigen-specific responsiveness. The observed decrease in CD4+ T cells in lymphoid organs in adult AML1+/– mice suggests a unique sensitivity of these mature cells to the dosage of AML1. Interestingly, CD4+ T cells express an approximately 2-fold higher level of AML1 than CD8+ T cells.32 It remains to be determined how a decrease in the intracellular level of AML1 leads to a reduction in the number of these cells in the periphery.
The reduction in the number of circulating platelets in AML1+/– mice, although not previously reported, was predicted based on the decrease in platelet numbers seen in humans with germ-line AML1 mutations.18,19 Importantly, the decrease in platelet numbers resulting from AML1 haploinsufficiency in the mouse was less dramatic than that observed following the complete loss of AML1, again suggesting a dose-response effect on megakaryocyte development/differentiation.47 Determining where AML1 functions in the developmental pathway from the formation of megakaryoctes through to the production of mature platelets remains to be defined; however, the complete loss of AML1 leads to a partial arrest in the differentiation of megakaryocytres. A similar but less severe block in megakaryocyte polyploidization and differentiation was seen in the megakaryocytes of AML1+/– mice (W.S. and J.R.D., unpublished data, May 2004). In addition, the expression of AML1 in biopotential megakaryocyte-erythroid progenitors and its down-regulation in more mature erythroid cells and up-regulation in megakaryocytes suggest that AML1/CBFβ might play an important role in the commitment to the megakaryocytic lineage.32,52
Although mice with AML1 haploinsufficiency replicate some of the phenotypic features of patients with the FPD/AML syndrome, one important difference is the absence of the development of AML in these mice. The AML1+/– mice have a similar life span as AML1+/+ animals and fail to spontaneously develop hematopoietic malignancies. Moreover, when these mice were treated with the chemical carcinogen N-ethyl-N-nitrosourea (ENU) at a dose previously demonstrated to cooperate with the expression of AML1-ETO to induce AML,50 no AML developed during an 18-month period of observation (Z. Cai and J.R.D., unpublished data, December 2002). The absence of leukemia development in these mice compared to humans with AML1 haploinsufficiency could reflect a difference in the length of time available for the acquisition of cooperating mutations. In humans with FPD/AML, leukemia typically developed sometime in the fourth to fifth decade. Therefore, the short life span of mice may preclude the acquisition of a sufficient number of cooperating mutations to result in the generation of overt leukemia. Alternatively, the possibility exists that the human disease may result not from simple haploinsufficiency of AML1 but instead may more commonly be the result of either the expression of dominant-negative mutants of AML1 or biallelic loss of this gene.
In summary, our results suggest that a threshold level of AML1/CBFβ is required for the normal function of HSCs and progenitors. Genetic alterations that decrease the activity of this complex below this level directly alter the normal growth and differentiation of these cells. The modest expansion in progenitors that occurs as a result of AML1 haploinsufficiency may increase the pool of cells susceptible to the acquisition of secondary mutations and may thus contribute to the increased incidence of leukemia in patients with germ-line mutations in AML1. Further studies on the functional consequences of the expression of mutant forms of AML1 that have been identified in patients with FPD/AML should provide further insights into the mechanism by which alterations in the functional level of AML1/CBFβ lead to an enhanced susceptibility to leukemia.
Prepublished online as Blood First Edition Paper, August 5, 2004; DOI 10.1182/blood-2003-12-4349.
Supported in part by National Cancer Institute grants P01 CA71907-06 (J.R.D.), CA-21765 (Cancer Center CORE grant to St Jude Children's Research Hospital [SJCRH]), and by the American Lebanese and Syrian Associated Charities (ALSAC) of SJCRH.
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank the members of the Downing laboratory including Zhongling Cai, Shouli Yang, Parasakthy Kumarevelu, PhD, and Noel Lenny, PhD, for their assistance with the analysis of mice; Richard Cross, PhD, Richard Ashmun, PhD, and Ann Marie Hamilton-Easton, PhD, for assistance with FACS analysis; Dr Martine Roussel for providing plasmids encoding murine cell cycle regulators p15INK4b, p16INK4a, p21waf1/cip1, p27kip1, and p57kip2; and Cheng Cheng, PhD, and Deqing Pei from the Department of Biostatistics for assistance in the analysis of the data.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal