Abstract
Immunoglobulin production is impaired in Hodgkin and Reed-Sternberg (HRS) cells of classical Hodgkin lymphoma (cHL) in spite of functional clonal rearrangements. The presence of “crippling” mutations in coding and regulatory regions, as well as down-regulation of B-cell-specific transcription factors, has been suggested as a potential reason for the lack of immunoglobulin (Ig) chain gene transcription. We have investigated the impact of epigenetic silencing in suppressing Ig heavy (H)-chain expression. Chromatin immunoprecipitation (ChIP) was used to analyze transcription factor binding to octamer motifs present in the IgH regulatory regions. Transcription factors were bound to these motifs in control cell lines, however, they were absent in the cHL-derived cell lines KMH2, L1236, and L428. Ectopic expression of octamer-binding transcription factor (Oct2) and/or B-cell Oct binding protein/Oct-binding factor (BOB.1/OBF.1) did not result in any measurable binding to these sites. Increased histone 3 Lysine 9 (H3-K9) methylation was observed in the promoter region of the IgH locus in L428 and L1236 cells. This is a typical feature of heterochromatic, transcriptionally silent regions. Treatment of cHL-derived cell lines with the DNA demethylating agent 5-aza-2′-deoxycytidine (5-aza-dC) partially reactivated IgH transcription and affected chromatin modifications. Our results suggest an important role of epigenetic silencing in the inhibition of IgH transcription in HRS cells. (Blood. 2004;104:3326-3334)
Introduction
B-cell-type Hodgkin and Reed-Sternberg (HRS) cells of classical Hodgkin lymphoma (cHL) are unable to produce both immunoglobulin (Ig) light (L) chain1-3 and heavy (H) chain,4 despite the presence of clonally rearranged IgL and IgH genes.4,5 Two explanations have been considered so far for this Ig down-regulation: “crippling” mutations within the transcription control regions and/or in the coding sequence6 ; and the absence or diminished production of critical transcription factors.7-9
Crippling mutations in the Ig coding regions can hardly be considered to be a main cause of Ig down-regulation in HRS cells because the vast majority of cHL cases (75%) harbor functionally rearranged Ig genes without mutations.4 Likewise, mutations in the Ig regulatory elements are also not responsible for the absence of Ig transcription because they were found in only one cHL-derived cell line, L1236 (mutation in the first position of the octamer motif of the variable region of the Ig heavy chain gene [VH] promoter), but not in isolated primary HRS cells, except in one cHL case.7,10,11
In contrast, the absence or substantial down-regulation of B-cell-specific transcription factors, such as octamer-binding transcription factor (Oct2) and B-cell Oct binding protein/Oct-binding factor (BOB.1/OBF.1)7-9 as well as the Ets family member PU.1,12-14 was observed in all cHL cell lines as well as in primary HRS cells of all cHL cases. Forced expression of these factors in cHL-derived cell lines resulted in the activation of cotransfected Ig promoter constructs, underscoring their decisive role in the regulation of Ig transcription.14 Their important role for B lymphopoiesis and Ig gene transcription was also shown in knock-out experiments where PU.1 is crucial for the development of several hematopoietic lineages15 and BOB.1/OBF.1 is critical for high titers of secondary Ig isotypes.16,17 Taking into account that in cHL-derived cell lines class-switch recombination is often targeted to IgG4 or IgA,18 down-regulation of BOB.1/OBF.1 could be the explanation for decreased Ig transcription in many cHL cases. Therefore, it is an unexpected finding that simultaneous cotransfection of Oct2 and BOB.1/OBF.1 expression plasmids in cHL-derived cell lines is unable to reactivate endogenous Ig production.14 This fact implies the existence of alternative mechanisms of Ig suppression in HRS cells, which are different from genetic mutations and down-regulation of transcription factors.
It has become clear that loss of gene function in cancer cells is mediated at least as often by epigenetic factors as by gene mutations.19 Epigenetic silencing in cancer cells generally proceeds through aberrant promoter cytidine guanidine dinucleotide (CpG) methylation, followed by the binding of methyl-binding proteins and the recruitment of transcriptional corepressors, chromatin remodeling proteins, and histone deacetylases.20,21 Finally, modification of histones, induced by such complexes, results in the formation of a condensed chromatin structure known as a “heterochromatin,” which is associated with transcriptionally inactive regions.22 Epigenetic silencing, in particular, was found to be responsible for down-regulation of numerous tumor suppressor genes such as BRCA1,23 p16/CDKN2,24 VHL,25 and p15/INK4B1.26 In B-cell-derived tumors, gene silencing by epigenetic mechanism was shown for p7327 and B29 genes.28 In the present study we show that epigenetic silencing also contributes to the down-regulation of Ig expression in cultured and primary HRS cells.
Materials and methods
Cell lines
The human Burkitt lymphoma cell lines Namalwa and B-JAB, as well as the cHL cell lines L1236, KM-H2, and L428, were cultured in RPMI 1640 medium (Life Technologies, Karlsruhe, Germany) supplemented with 10% fetal bovine serum (PAN Biotech GmbH, Aidenbach, Germany), antibiotics, l-glutamine, and 50 μM of 2-mercaptoethanol at 37°C and 5% CO2. The L428-Oct2 cell line was obtained by stable cotransfection of the L428 cells with a mammalian expression vector containing the full-length human Oct2A29 together with the pcDNA-neo vector. Positive colonies were selected with 1.3 mg/mL of G-418 (Calbiochem, Bad Soden, Germany). Control cell line L428-neo was transfected only with the pcDNA-neo vector and selected as described above. The BOB.1/OBF.1 expression vectors pcDNA-BOB.130 or pCFG5-BOB.1, created by the cloning of full-length BOB.1/OBF.1 coding sequence into pCFG5 IEGZ vector,31 which was more efficient in transfection of 5-aza-2′-deoxycytidine (5-aza-dC)-treated cells, were transfected using the Nucleofector device (Nucleofector Kit V, program Q-07 or T-13; Amaxa GmbH, Cologne, Germany). For L1236 cells and KM-H2 cells, the Oct2 expression vector was also transfected using the nucleofection strategy. Typically we took 1 × 107 cells and 5 μg of expression vector(s) per transfection. On the next day, cells were harvested and used for Western blot and chromatin immunoprecipitation (ChIP) assays. Life cell purification by ficoll gradient centrifugation was performed when necessary. Nucleofection efficiencies were tested in cHL cell lines using cotransfection of BOB.1/OBF.1 and/or Oct2-expressing vectors with pcDNA-enhanced green fluorescent protein (EGFP) vector (better than 80% for L428, lower efficiencies for KM-H2 and L1236). The cells were harvested and used for ChIP assay the day after nucleofection.
Western blot
For Western blot analysis, 5 × 106 cells were washed twice with phosphate-buffered saline (PBS), lysed on ice in 500 μL of 2 × concentrated sample buffer (70 mM Tris [tris(hydroxymethyl)aminomethane]; 11.15% vol/vol glycerol; 0.0015% bromphenol blue; 3% sodium dodecyl sulfate [SDS]; and 5% vol/vol 2-mercaptoethanol, pH 6.8), sonicated, and boiled for 5 minutes. Twenty-five microliters of lysates were separated on a 10% polyacrylamide gel and electroblotted onto an Immobilon-P polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA). Membranes were blocked overnight at 4°C with 5% skimmed milk powder in Tris-buffered saline (TBS; pH 8.0) and 0.1% Tween-20 (TBST). The primary antibodies specific for Oct2 (affinity-purified rabbit polyclonal antibodies against Oct2-sc-233), BOB.1/OBF.1 (sc-955), Oct1 (sc-232), or nuclear factor κB (NFκB) p65 (sc-372) were purchased from Santa Cruz Biotechnology (Heidelberg, Germany). Rabbit anti-human IgG antibody, specific for gamma chains (no. A0423), was obtained from DAKO A/S (Glostrup, Denmark). After 2 washing steps with TBST, the membranes were incubated for 1 hour at room temperature with goat anti-rabbit IgG-horseradish peroxidase (HRP; sc-2004; Santa Cruz Biotechnology). Signals were visualized using the SuperSignal West Dura extended-duration substrate (Perbio Science Deutscland GmbH, Bonn, Germany).
Chromatin immunoprecipitation (ChIP)
We used the ChIP assay kit (no. 17-295; Biomol GmbH, Hamburg, Germany) according to the manufacturer's instructions, with some minor modifications. Briefly, 3 × 107 cells were collected in culture medium and then 37% formaldehyde stabilized with 10% of methanol was added to a final concentration of 1% formaldehyde. After 12 minutes of incubation at 37°C, cells were washed twice with cold PBS and resuspended in 900 μL lysis buffer containing 1 mM PMSF, 10 μg/mL aprotinin, 10 μg/mL leupeptin, and 1 μg/mL pepstatin. After 10 minutes of incubation on ice, the lysate was divided into 300-μL aliquots and sonified with a Branson Sonifier 250 equipped with a 2-mm tip (Branson Sonic Power, Danbury, CT). Four series of pulses (10 seconds each, 80% duty, 20% of maximum power) sheared DNA into fragments that were 200 to 1000 bp in length. Samples were centrifuged for 10 minutes at 4°C at 13 000g, and the supernatant was diluted 1:10 with ChIP dilution buffer and precleared with salmon sperm DNA/protein A agarose-50% slurry (1 h at 8°C). Precleared chromatin solutions were divided into aliquots (1.5 mL each) and incubated overnight on an overhead shaker with one of the following antibodies: antidimethyl-histone 3 Lys 9 (H3-K9), rabbit affinity-purified Ig, 5 μL ab7312 (Abcam, Cambridge, United Kingdom), or 10 μL no. 07-212 (Upstate, Lake Placid, NY). The Oct1, Oct2, and BOB.1/OBF.1 immunoprecipitations were performed with 1 μg the respective affinity-purified polyclonal rabbit antibodies described above (Santa Cruz Biotechnology). As a control, 1 μg affinity-purified polyclonal antibody against phospholipase Cγ1 (PLCγ1; sc-81; Santa Cruz Biotechnology) was used. Immunocomplexes were precipitated with 55 μL salmon sperm DNA/protein A agarose-50% slurry for 2 hours. The beads were collected by mild centrifugation. Five hundred-microliter aliquots were used from the “no antibody” control sample to serve as an “input” control and to determine the DNA content. Beads were washed sequentially with low-salt buffer, high-salt buffer, LiCl buffer, and tris(hydroxymethyl)aminomethane buffer containing ethylenediaminetetraacetic acid (TE). Immunocomplexes were eluted from the beads using 500 μL elution buffer (1% SDS, 0.1 M NaHCO3) for 30 minutes. After reversion of protein-DNA cross-linking by mild heating (6 h or overnight at 65°C), DNA was recovered by proteinase K digestion and phenol/chloroform extraction. Samples were redissolved in 25 μL TE (100 μL for total inputs). The concentration of DNA in the input samples ranged from 80 to 120 μg/mL. Samples were analyzed by polymerase chain reaction (PCR). The amount of input per reaction was adjusted to 20 ng. The samples were amplified by Taq DNA polymerase no. 27-0799-01 (Amersham Pharmacia Biotech, Piscataway, NJ) using the primers and conditions listed in Table 1.
Primers and amplification conditions for ChIP and RT-PCR
Primer set . | Primer sequence, 5′-3′ . | Annealing temperature, °C . | No. cycles . | Product, no. bp's . |
|---|---|---|---|---|
| VH-3 (B-JAB), ChIP | F: GGCCCTCCCTCTACTGATGAAAACCA | 60 | 35 | 498 |
| R: ATTGTAGCCATCTCCCGTGCCCTCTT | ||||
| VH-1 (L1236), ChIP | F: TGGGACATCTATCTTCTTTCTCAACCTC | 63 | 35 | 636 |
| R: ACTGCGCCACCTGCCACCTCTC | ||||
| VH-3 (KM-H2), ChIP | F: GTTAATCAAGGCACAATCATA | 54.8 | 35 | 731 |
| R: GCACCCCCTCGCACAGAAATAAA | ||||
| VH-4 (Namalwa), ChIP | F: CCTCCCTCATCCCTTTTCACCTCT | 62 | 35 | 622 |
| R: GTCGCCACCACTCCAAAAATCGTA | ||||
| VH-5 (L428), ChIP | F: GCAACTATGCAAATGCAAGTGGGG | 63 | 35 | 500 |
| R: AGATAGGCAGTGGAGACGGATTTGTC | ||||
| Internal enhancer, ChIP | F: AGGCCAGATCTGAAAGTGCT | 57 | 35 | 320 |
| R: TAATGCTCTGAGGTATCGAAAAAGTA | ||||
| Cα1-enhancer, ChIP | F: GGGGGAGGGGGCGGGAGAAT | 60 | 35 | 278 |
| R: GGGGCAAGCTGGTGAGGAAAGTGG | ||||
| Cα2-enhancer, ChIP | F: CCAAGTGGGCCTGTCCTG | 60 | 35 | 408 |
| R: GGGCTCTGATCTGTTTCTCCTT | ||||
| Cα2-enhancer KM-H2, ChIP | F: GCCTGGCCACGCTGTTG | 58.7 | 35 | 296 |
| R: AGGGCTCTGATCTGTTTCTCC | ||||
| H2B histone promoter, ChIP | F: GGGCGCGAAAAAGAAGACGTGTTGTTG | 65 | 35 | 284 |
| R: GAGGTGATGGTCGAGCGCTTGTTGTAA | ||||
| Involucrin promoter, ChIP | F: TGAGCATGGCATTCCTGAGA | 54.8 | 35 | 423 |
| R: GGCATCCTACTGTTTTGTGACCT | ||||
| KMH2 leader, RT-PCR | F: TGGGCTGACCTGGGTTTTCCTCGTT | 59.3 | 35 | 494/392 |
| R: CCTTGGCCCCAGTGGTCAGATA | ||||
| L428 leader, RT-PCR | F: AACCGCCATCCTCGCCCTCCTC | 61.0 | 35 | 444/360 |
| R: CCCCATCATCTGACTATGTCTCCCACAG | ||||
| L1236 leader, RT-PCR | F: CCTGGAGGGTCTTCTGCT | 55.6 | 35 | 410/324 |
| R: ACGGCCGTGTCCTCAAA | ||||
| PBDG30 | F: AGCTGCAGAGAAAGTTCCC | 60 | 30 | 437 |
| R: GTTACGAGCAGTGATGCC |
Primer set . | Primer sequence, 5′-3′ . | Annealing temperature, °C . | No. cycles . | Product, no. bp's . |
|---|---|---|---|---|
| VH-3 (B-JAB), ChIP | F: GGCCCTCCCTCTACTGATGAAAACCA | 60 | 35 | 498 |
| R: ATTGTAGCCATCTCCCGTGCCCTCTT | ||||
| VH-1 (L1236), ChIP | F: TGGGACATCTATCTTCTTTCTCAACCTC | 63 | 35 | 636 |
| R: ACTGCGCCACCTGCCACCTCTC | ||||
| VH-3 (KM-H2), ChIP | F: GTTAATCAAGGCACAATCATA | 54.8 | 35 | 731 |
| R: GCACCCCCTCGCACAGAAATAAA | ||||
| VH-4 (Namalwa), ChIP | F: CCTCCCTCATCCCTTTTCACCTCT | 62 | 35 | 622 |
| R: GTCGCCACCACTCCAAAAATCGTA | ||||
| VH-5 (L428), ChIP | F: GCAACTATGCAAATGCAAGTGGGG | 63 | 35 | 500 |
| R: AGATAGGCAGTGGAGACGGATTTGTC | ||||
| Internal enhancer, ChIP | F: AGGCCAGATCTGAAAGTGCT | 57 | 35 | 320 |
| R: TAATGCTCTGAGGTATCGAAAAAGTA | ||||
| Cα1-enhancer, ChIP | F: GGGGGAGGGGGCGGGAGAAT | 60 | 35 | 278 |
| R: GGGGCAAGCTGGTGAGGAAAGTGG | ||||
| Cα2-enhancer, ChIP | F: CCAAGTGGGCCTGTCCTG | 60 | 35 | 408 |
| R: GGGCTCTGATCTGTTTCTCCTT | ||||
| Cα2-enhancer KM-H2, ChIP | F: GCCTGGCCACGCTGTTG | 58.7 | 35 | 296 |
| R: AGGGCTCTGATCTGTTTCTCC | ||||
| H2B histone promoter, ChIP | F: GGGCGCGAAAAAGAAGACGTGTTGTTG | 65 | 35 | 284 |
| R: GAGGTGATGGTCGAGCGCTTGTTGTAA | ||||
| Involucrin promoter, ChIP | F: TGAGCATGGCATTCCTGAGA | 54.8 | 35 | 423 |
| R: GGCATCCTACTGTTTTGTGACCT | ||||
| KMH2 leader, RT-PCR | F: TGGGCTGACCTGGGTTTTCCTCGTT | 59.3 | 35 | 494/392 |
| R: CCTTGGCCCCAGTGGTCAGATA | ||||
| L428 leader, RT-PCR | F: AACCGCCATCCTCGCCCTCCTC | 61.0 | 35 | 444/360 |
| R: CCCCATCATCTGACTATGTCTCCCACAG | ||||
| L1236 leader, RT-PCR | F: CCTGGAGGGTCTTCTGCT | 55.6 | 35 | 410/324 |
| R: ACGGCCGTGTCCTCAAA | ||||
| PBDG30 | F: AGCTGCAGAGAAAGTTCCC | 60 | 30 | 437 |
| R: GTTACGAGCAGTGATGCC |
F indicates forward; R, reverse; and PBDG, porphobilinogen deaminase.
RT-PCR
Total RNA was isolated from 1 × 106 cells using High Pure RNA Isolation Kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. Two micrograms RNA and 0.5 μg dT15 primer (MWG-Biotech, Ebersberg, Germany) were heated for 5 minutes at 70°C. After rapid cooling on ice, the first-strand cDNA was synthesized by Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT); (Promega, Madison, WI). Primers for the detection of IgVH transcripts were designed to bind to the leader sequence (forward primer) and to the complementarity determining region 3 (CDR3; reverse primer) in order to distinguish correctly spliced mRNA from a possible contamination with genomic DNA. Sizes of DNA- and mRNA-derived amplification products are given in Table 1 (when 2 fragments are indicated, the larger one corresponds to genomic DNA, the smaller one to mRNA-derived cDNA).
IgG ELISA and immunocytochemistry
To quantify IgG protein expression, an IgG enzyme-linked immunosorbent assay (ELISA) kit (catalog no. K 6510; Cell Concepts, Umkirch, Germany) was used according to the manufacturer's instructions. Cells (2 × 107) were incubated in the presence of 500 nM 5-aza-dC for 72 hours. Cell lysates containing 100 μg protein or 200 μL cell culture supernatants, respectively, were added to the wells of a microtiter plate precoated with a polyclonal anti-human IgG rabbit antibody. After washing, a peroxidase-conjugated polyclonal anti-human IgG rabbit antibody was added. Bound antibody was detected by the addition of trimethylbenzidine (TMB) substrate and measuring the extinction in an MRX ELISA reader (Dynatech Laboratories, Denkendorf, Germany) at 450 nm.
For immunostaining, the acetone-fixed cytospin preparations of cHL cells were incubated with the same polyclonal rabbit antibody against human IgG as described in “Western blot.” Slides were washed in PBS and incubated with an antirabbit horseradish peroxidase polymer (EnVision System; DAKO A/S). Peroxidase activity was visualized with the substrate 3-amino-9-ethylcarbazole (0.1 mg/mL in 0.17 M sodium acetate [pH 5.2] plus 0.01% H2O2). Nuclear counterstaining was done with hematoxylin.
Results
Transcription factors interact with Ig octamer motifs in Burkitt lymphoma control cell lines
To find out whether regulatory regions for IgH transcription are accessible for the octamer-binding transcription factors Oct1 and Oct2, as well as for the BOB.1/OBF.1 coactivator, we used ChIP assays to investigate protein-DNA interactions in living cells.32 The choice of transcription factors was based on their important role in the regulation of Ig transcription and because octamer motif(s) are present in all 3 regulatory regions: the VH promoter, the internal enhancer, and the 3′ enhancers.
Because of the high homology among the many IgVH promoters, the primers for amplification of immunoprecipitated DNA were located within 150 bp upstream of the octamer motif and within the CDR3 region (Figure 1A).
Oct1, Oct2, and BOB.1 transcription factors interact with the octamer motifs in Ig regulatory elements of immunoglobulin-producing Burkitt lymphoma cell lines. (A) Maps of the IgH and histone H2B showing the location of primers used for ChIP analysis. Oct indicates octamer motif; CDR3, complementarity determining region 3; intE, internal enhancer; and LCR, locus control region. Primer locations are depicted by arrows. (B) Binding of Oct1, Oct2, and BOB.1/OBF.1 to IgH regulatory regions of B-JAB and Namalwa cell lines was detected by chromatin immunoprecipitation (ChIP) assays. Primers specific for the histone H2B promoter were used as positive control. Negative controls were chromatin immunoprecipitated with anti-PLC-γ1 antibody as well as samples to which no antibody was added (No Ab). DNA purified from the lysates incubated without antibody was used as input control (Input). Amplification products were detected by ethidium bromide staining on agarose gel. All experiments were performed at least in triplicate. prom indicates promoter; and Int Enh, internal enhancer.
Oct1, Oct2, and BOB.1 transcription factors interact with the octamer motifs in Ig regulatory elements of immunoglobulin-producing Burkitt lymphoma cell lines. (A) Maps of the IgH and histone H2B showing the location of primers used for ChIP analysis. Oct indicates octamer motif; CDR3, complementarity determining region 3; intE, internal enhancer; and LCR, locus control region. Primer locations are depicted by arrows. (B) Binding of Oct1, Oct2, and BOB.1/OBF.1 to IgH regulatory regions of B-JAB and Namalwa cell lines was detected by chromatin immunoprecipitation (ChIP) assays. Primers specific for the histone H2B promoter were used as positive control. Negative controls were chromatin immunoprecipitated with anti-PLC-γ1 antibody as well as samples to which no antibody was added (No Ab). DNA purified from the lysates incubated without antibody was used as input control (Input). Amplification products were detected by ethidium bromide staining on agarose gel. All experiments were performed at least in triplicate. prom indicates promoter; and Int Enh, internal enhancer.
In all cases, these primer combinations selectively amplified only the rearranged allele in the respective cell line. The internal enhancer region containing a cluster of 2 imperfect octamer motifs was amplified by flanking primers. The 3′ enhancer hs1,2 located in the Cα1 and Cα2 locus control regions (LCRs) was reported to be more important for the regulation of IgG transcription than other 3′ IgH enhancers (hs3A and hs4).33,34 Therefore, we amplified the octamer-containing fragment of hs1,2 from the Cα1 and Cα2 LCRs using the respective specific set of primers (Table 1). Due to the allelic polymorphism of hs1,2,34,35 we used different primers for the amplification of the Cα2 hs1,2 from KM-H2 cells (Table 1). As a control, we amplified the octamer-containing promoter region of the ubiquitously expressed histone H2B gene.
There are, as yet, no data available on the binding of octamer-specific transcription factors to regulatory regions of human IgH in vivo. We therefore determined the binding patterns of Oct1, Oct2, and BOB.1/OBF.1 in the Burkitt lymphoma cell lines Namalwa and B-JAB (Figure 1B), which are known to produce Ig and express Oct1, Oct2, and BOB.1/OBF.1.4,11
The results obtained in both cell lines were virtually identical. All 3 proteins were bound to the octamer motif of the IgVH promoters roughly equally. The binding pattern to the internal enhancer region containing 2 imperfect octamer motifs was slightly different. Whereas Oct2 and BOB.1/OBF.1 were present at the internal enhancer, the Oct1 binding signal was nearly undetectable. All 3 transcription factors were again bound to the Cα1 and Cα2 enhancers in both cell lines. However, the amplification signals were stronger in case of the Cα1 enhancer. In addition, Oct1, Oct2, and BOB.1/OBF.1 were bound to the H2B control promoter in both Burkitt lymphoma cell lines. The fact that Oct2 signals are stronger than Oct1 signals in B-JAB cells compared with Namalwa cells is most likely due to the fact that B-JAB cells express large amounts of Oct2.
Ig regulatory elements are not occupied by transcription factors in cHL cell lines
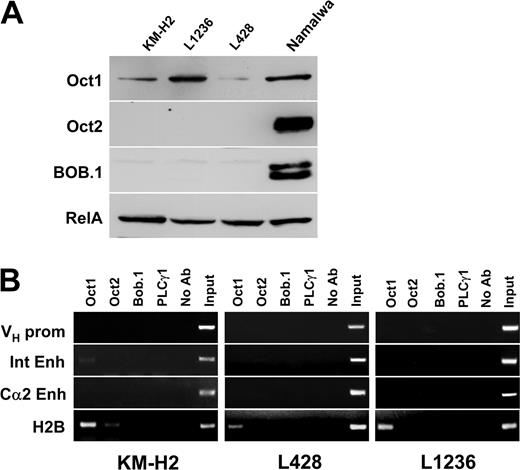
We then investigated whether these regulatory elements are also bound by transcription factors in the cHL-derived cell lines KM-H2, L428, and L1236. We first reanalyzed the status of Oct1, Oct2, and BOB.1/OBF.1 expression in these 3 cell lines. Oct1 was expressed in all 3 cHL cell lines. Oct2 and BOB.1/OBF.1 were not detected on normal exposures of the Western immunoblot. (Figure 2A). On long exposures, some Oct2 was detected in KM-H2 cells and low levels of BOB.1/OBF.1 were seen in L1236 cells (data not shown). When chromatin immunoprecipitation was performed in these cHL lines it became clear that even Oct1, which is present in all cHL cell lines, did not bind to any of the IgH regulatory regions (Figure 2B). Similarly, the low levels of Oct2 in KM-H2 cells and of BOB.1/OBF.1 in L1236 cells did not result in measurable binding to the Ig regulatory elements.
Absence of transcription factors at IgH regulatory regions in cHL cell lines. (A) Expression of Oct1, Oct2, and BOB.1/OBF.1 transcription factors in cHL cell lines. The proteins were detected by Western blot as described in “Materials and methods.” Equal loading was controlled for using antibodies specific for RelA (Rel family member p65). (B) ChIP experiments were performed as described in Figure 1B. Due to the switch recombination to IgG4 in Hodgkin cell lines, only the Cα2 3′ enhancer was assayed.
Absence of transcription factors at IgH regulatory regions in cHL cell lines. (A) Expression of Oct1, Oct2, and BOB.1/OBF.1 transcription factors in cHL cell lines. The proteins were detected by Western blot as described in “Materials and methods.” Equal loading was controlled for using antibodies specific for RelA (Rel family member p65). (B) ChIP experiments were performed as described in Figure 1B. Due to the switch recombination to IgG4 in Hodgkin cell lines, only the Cα2 3′ enhancer was assayed.
Because class-switch recombination in KM-H2, L1236, and L428 cell lines was shown to be specifically targeted to IgG4,18 which proceeds through the removal of Cα1 as well as the Cα1 LCR by deletional recombination, the germ line Cα1 LCR-hs1,2 enhancer product was not amplified in cultured HRS cells (data not shown). To test the binding of these proteins to a control promoter region of a gene expressed in these cells, we again analyzed the H2B promoter. In L428 and L1236 cells, only Oct1 binding could be scored. In contrast, in the KM-H2 cell line, which expresses low amounts of Oct2 protein, we found both Oct1 and Oct2 associated with the H2B promoter. This result confirms that the endogenous proteins are capable of binding to promoter regions. However, the octamer motif(s) of IgH gene regulatory regions are not occupied by these proteins in cHL cell lines.
Ectopically expressed Oct2 and BOB.1/OBF.1 do not bind Ig regulatory regions in cHL cells
Earlier work has shown that Oct1 is as active as Oct2 in the activation of IgVH promoters yet it was shown to be unable to activate the internal enhancer29 and the 3′ enhancer.36 Indeed, we found almost no interaction between Oct1 and the internal enhancer, even in our control cell lines (Figure 1B). However, we had shown earlier that immunoglobulin gene promoter activity can be mediated by Oct1 plus BOB.1/OBF.1, even in the absence of Oct2. We therefore asked whether our failure to detect binding to the enhancer regions resulted from a lack of Oct2, and whether the failure to detect binding to the promoter might be due to a lack of BOB.1/OBF.1.
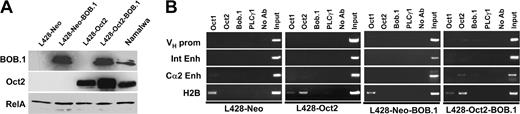
We therefore ectopically expressed these proteins in cHL cell lines and asked whether this would affect transcription factor occupancy at the respective Ig regulatory region octamer motifs. L428 cells were first stably transfected with an Oct2 expression vector and then supertransfected with the BOB.1/OBF.1 expression vector using the nucleofection (Amaxa) strategy. This resulted in the generation of L428 derivatives expressing Oct2, BOB.1/OBF.1, or both proteins at levels comparable to the Namalwa control cell line (Figure 3A).
Ectopically expressed Oct2 and BOB.1/OBF.1 do not bind to cognate octamer motifs in L428 cells. (A) Western immunoblot with extracts from L428 cells expressing transfected BOB.1/OBF.1 and/or Oct2. L428-Oct2 cells stably transfected with an Oct2 expression vector or control cells and transfected with the neomycin resistance gene only (L428-Neo) were supertransfected with a BOB.1/OBF.1-expression vector as indicated in “Cell lines.” One day after nucleofection, cells were harvested and used for Western blot or ChIP assays. Extracts from Namalwa cells were used as positive control. RelA-specific antibodies were used as loading control. (B) ChIP analysis of BOB.1/OBF.1, Oct1, and Oct2 binding to Ig regulatory regions in L428 cells overexpressing Oct2 and/or BOB.1/OBF.1. See Figure 1 legend.
Ectopically expressed Oct2 and BOB.1/OBF.1 do not bind to cognate octamer motifs in L428 cells. (A) Western immunoblot with extracts from L428 cells expressing transfected BOB.1/OBF.1 and/or Oct2. L428-Oct2 cells stably transfected with an Oct2 expression vector or control cells and transfected with the neomycin resistance gene only (L428-Neo) were supertransfected with a BOB.1/OBF.1-expression vector as indicated in “Cell lines.” One day after nucleofection, cells were harvested and used for Western blot or ChIP assays. Extracts from Namalwa cells were used as positive control. RelA-specific antibodies were used as loading control. (B) ChIP analysis of BOB.1/OBF.1, Oct1, and Oct2 binding to Ig regulatory regions in L428 cells overexpressing Oct2 and/or BOB.1/OBF.1. See Figure 1 legend.
We then asked whether these ectopically expressed proteins were capable of interaction with the endogenous Ig regulatory elements. Interestingly, we could not detect any significant interaction of these proteins with the Ig regulatory elements either in the cells transfected individually with Oct2 or BOB.1/OBF.1 or in double-positive cells. However, the transfected Oct2 was readily able to interact with the octamer motif in the histone H2B control promoter (Figure 3B). In addition to these experiments measuring protein-DNA binding in vivo, we also analyzed whether the expression of the endogenous Ig gene was increased in these transfectants. However, we were unable to detect any significant levels of Ig mRNA by RT-PCR (Figure 4B). In case of the KM-H2 cell line, which contains low levels of functional Oct2, we overexpressed only the BOB.1/OBF.1 protein. Again, these transfectants failed to show any measurable binding of transcription factors to the Ig octamer motifs (data not shown). These results suggest that it is not only the absence of these proteins that explains the failure to detect transcription factor binding to these control regions but also that these DNA elements are inaccessible to endogenous and ectopic transcription factors in cHL cell lines.
Oct2 and BOB.1/OBF.1 overexpression potentiates 5-aza-dC-induced Ig transcription in cHL cells. L428-neo or L428-Oct2 cells were pretreated for 24 hours with 1 μM 5-aza-dC. After an additional 24 hours in normal growth medium, cells were nucleofected with the BOB.1/OBF.1-expressing vector pCFG5-BOB.1 or by the empty vector pCFG5-IEGZ. On the next day cells were harvested and used for RNA or protein analysis as described in “Materials and methods.” (A) Expression of Oct2, BOB.1/OBF.1, and IgG gamma chains in cell lysates were assessed by Western blots. (B) RT-PCR analysis of Ig transcription was done as described in the legend for Figure 5 and in “Materials and methods.”
Oct2 and BOB.1/OBF.1 overexpression potentiates 5-aza-dC-induced Ig transcription in cHL cells. L428-neo or L428-Oct2 cells were pretreated for 24 hours with 1 μM 5-aza-dC. After an additional 24 hours in normal growth medium, cells were nucleofected with the BOB.1/OBF.1-expressing vector pCFG5-BOB.1 or by the empty vector pCFG5-IEGZ. On the next day cells were harvested and used for RNA or protein analysis as described in “Materials and methods.” (A) Expression of Oct2, BOB.1/OBF.1, and IgG gamma chains in cell lysates were assessed by Western blots. (B) RT-PCR analysis of Ig transcription was done as described in the legend for Figure 5 and in “Materials and methods.”
Inhibition of DNA methylation partially reactivates IgH transcription
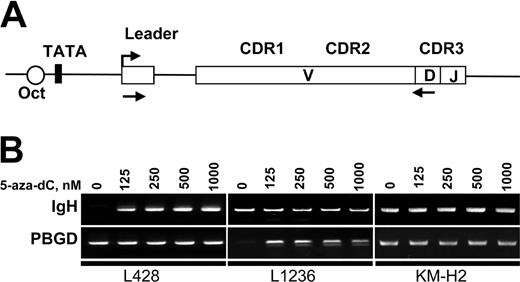
To assess IgH transcription in KM-H2, L1236, and L428 cells we used RT-PCR. The cDNA was amplified with sets of primers specific for each cell line (Figure 5A). The RT-PCR included the leader region to discern contaminating DNA from spliced mRNA amplification signals. All sets of primers showed similar amplification efficiencies (not shown). In untreated cells, we only found IgH expression (albeit at a very low level) in KM-H2 cells (Figure 5B). After 72 hours of treatment with 5-aza-dC, we observed a reactivation of Ig transcription in L428 and L1236 cell lines at all 5-aza-dC concentrations (Figure 5B). Semiquantitative RT-PCR experiments revealed an up-regulation of at least 27-fold compared with the untreated controls. Nevertheless, the levels of Ig transcription obtained in these 2 cell lines after 5-aza-dC treatment were still very low and comparable to the low basal levels detected in KM-H2 cells. Treatment with the demethylating reagent did not increase this low-level IgH transcription in KM-H2 cells. We also investigated the effect of histone deacetylase inhibitor trichostatin A (TSA) on gene transcription. Control experiments revealed that TSA concentrations above 250 nM were toxic to all 3 cHL cell lines. Cells were therefore incubated for 24 hours with different concentrations of TSA up to 250 nM in the presence or absence of 5-aza-dC. However, TSA did not induce any significant increase of spliced mRNA transcription, either alone or in combination with 5-aza-dC (data not shown).
Inhibition of DNA methylation results in reactivation of IgH transcription. (A) Map of the VH region showing the location of primers used for RT-PCR analysis of IgH expression. Location of the forward primer in the leader regions enables the discrimination between spliced mRNA and genomic DNA contamination. V indicates variable; D, diversity; and J, joining. (B) Detection of Ig transcription in cHL cells by RT-PCR. L428, L1236, and KM-H2 cells were incubated for 72 hours in the presence of the indicated concentrations of 5-aza-dC. Total RNA was extracted from 1 × 106 cells and converted to cDNA. The specifically amplified products were detected by ethidium bromide staining on agarose gels. Sizes of the PCR products are indicated in Table 1.
Inhibition of DNA methylation results in reactivation of IgH transcription. (A) Map of the VH region showing the location of primers used for RT-PCR analysis of IgH expression. Location of the forward primer in the leader regions enables the discrimination between spliced mRNA and genomic DNA contamination. V indicates variable; D, diversity; and J, joining. (B) Detection of Ig transcription in cHL cells by RT-PCR. L428, L1236, and KM-H2 cells were incubated for 72 hours in the presence of the indicated concentrations of 5-aza-dC. Total RNA was extracted from 1 × 106 cells and converted to cDNA. The specifically amplified products were detected by ethidium bromide staining on agarose gels. Sizes of the PCR products are indicated in Table 1.
The IgH gene promoters show features of inactive chromatin that can be reactivated by inhibition of DNA methylation
Unlike genetic mutations, epigenetic silencing is potentially reversible.19 DNA methylation is one of the most common mechanisms, which, directly or through formation of a repressive chromatin structure, is involved in the suppression of gene function.19 The observed partial reactivation of Ig gene transcription by 5-aza-dC in L428 and L1236 cells suggested that indeed the gene silencing in cHL cell lines might be a consequence of such chromatin modifications. To monitor the chromatin structure, we used ChIP with antibodies against methylated lysine 9 in histone H3 (H3-K9Me). Previous work had demonstrated that the local density of H3-K9Me is a reliable marker of “closed” inactive chromatin structure.37,38 Methylation of H3-K9 correlates better with heterochromatin formation than histone H3 lysine 4 methylation37 or the general histone acetylation.39 As a positive control we amplified immunoprecipitated DNA with primers specific for the promoter region of involucrin. Involucrin is a tissue-specific gene involved in terminal differentiation of keratinocytes and it is not expressed in lymphoid cells. When we analyzed the histone H3 methylation status at the involucrin promoter or the transcriptionally active Ig heavy-chain promoter regions in B-JAB and Namalwa cells, we got the expected results (Figure 6B). Whereas the involucrin promoter showed methylated histone H3, the Ig promoters were not precipitated by this antibody. Also as expected, the methylated histones were found at the involucrin promoter in all cHL cell lines. In addition, in L428 and L1236 cells we also detected methylated histone H3 in the Ig promoters (Figure 6A). This chromatin modification could not be detected in KM-H2 cells. Importantly, treatment of L428 and L1236 cells with 500 nM of 5-aza-dC for 72 hours resulted in the disappearance of H3-K9Me from both promoters. All these results are in agreement with the observed reactivation of Ig transcription by 5-aza-dC (Figure 5). Binding of the H3-K9Me-specific antibody was not detected at the internal enhancer or at the 3′-enhancer region in any of the cell lines (data not shown). This suggests that inhibition of Ig transcription in HRS cells may be due to the chromatin-dependent silencing of the promoter region.
Chromatin in IgH gene promoters of cHL cells has a “closed” structure that can be opened by inhibition of DNA methylation. (A) L428, L1236, or KM-H2 cells were incubated for 72 hours in the presence of 500 ng of DNA methylation inhibitor 5-aza-dC (right). Local methylation at the VH promoter chromatin was analyzed by antidimethylated histone 3 lysine 9 antibodies by ChIP analysis. Signals obtained for the IgH promoter, the involucrin promoter, and the histone H2B promoter are shown. (B) Histone H3-K9 methylation was assessed by ChIP at the VH promoter regions of Ig-producing B-JAB and Namalwa cells. As controls, the histone H2B promoter and the involucrin promoter were used.
Chromatin in IgH gene promoters of cHL cells has a “closed” structure that can be opened by inhibition of DNA methylation. (A) L428, L1236, or KM-H2 cells were incubated for 72 hours in the presence of 500 ng of DNA methylation inhibitor 5-aza-dC (right). Local methylation at the VH promoter chromatin was analyzed by antidimethylated histone 3 lysine 9 antibodies by ChIP analysis. Signals obtained for the IgH promoter, the involucrin promoter, and the histone H2B promoter are shown. (B) Histone H3-K9 methylation was assessed by ChIP at the VH promoter regions of Ig-producing B-JAB and Namalwa cells. As controls, the histone H2B promoter and the involucrin promoter were used.
Overexpression of Oct2 and BOB.1/OBF.1 in 5-aza-dC-treated cHL cells increases Ig transcription but does not result in Ig protein synthesis
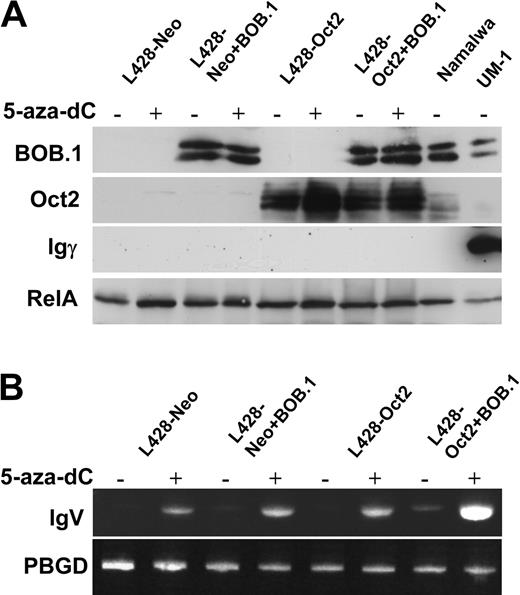
Reactivation of Ig transcription and decrease of histone H3-K9 methylation in the IgH promoter region in cHL cells treated with 5-aza-dC indicated the induction of an open chromatin structure in the IgH locus, which is accessible for transcription factors. This suggested that reconstitution of Oct2 and BOB.1/OBF.1 in cHL cells might further increase Ig transcription and might even result in detectable Ig protein expression.
To test this hypothesis, we assessed Ig production in L428-neo and L428-Oct2 cell lines treated with 5-aza-dC and nucleofected with BOB.1/OBF.1 expression vector. We measured Ig expression by RT-PCR, Western blot, ELISA, and immunocytochemistry. BOB.1/OBF.1 levels in transfected cells were comparable to the positive control cell lines (Namalwa, UM-1), whereas Oct2 levels in L428-Oct2 cells was higher than in Namalwa cells. Under these experimental conditions we were able to detect weak Ig RNA expression in L428-Oct2 cells overexpressing BOB.1/OBF.1 by RT-PCR even without 5-aza-dC treatment (Figure 4B). Importantly, in all cases RNA transcription levels were increased by 5-aza-dC and, in addition, highest levels were observed in cells expressing both Oct2 and BOB.1/OBF.1. We then checked whether these levels of RNA were sufficient to yield detectable Ig protein levels. We assessed Ig protein production using IgG-specific Western blots (Figure 4A), immunocytochemistry, and ELISA (data not shown). Namalwa cells producing IgM were used as a negative control, whereas the IgG-producing lymphoblastoid cell line UM-1 served as a positive control. None of these assays detect Ig protein expression. Similarly, the combination of 5-aza-dC treatment with BOB.1/OBF.1 and Oct2 overexpression did not induce Ig protein production as it was shown in KM-H2 (ELISA and Western blot) or L1236 (Western blot) cells (data not shown).
These data on stimulation of Ig transcription in cHL cell lines by a combination of DNA demethylation and Oct2/BOB.1 overexpression support our hypothesis considering the lack of octamer-binding transcription factors and epigenetic silencing of the IgH promoter as possible mechanisms involved in Ig down-regulation in cHL. At the same time, the inability of this complex treatment to induce Ig protein production indicates that additional mechanisms are involved. This could be either the lack of further positively acting factors or the presence of additional suppressing factors.
Discussion
The purpose of the present study was to clarify whether epigenetic mechanisms are involved in the silencing of Ig transcription in HRS cells. To answer this question, we compared the binding pattern of the transcription factors Oct1, Oct2 and BOB.1/OBF.1 to regulatory sequences in the IgH locus of Ig-producing B-cell lines and Ig-negative cHL-derived cell lines and analyzed the transcriptional effect of demethylation agents in the latter cell lines.
Transcription factor occupancy of IgH regulatory regions
Binding of Oct1 and Oct2 to the 3 transcription regulatory regions of IgH locus (promoter, internal enhancer, 3′ enhancers) had been shown in the past by electrophoretic mobility shift assay (EMSA) with extracts from many B-cell lines. In addition, functional data had left little doubt on the physical interaction of these transcription factors with the octamer motifs of the Ig promoters.40 However, it was not clear whether Oct1 and Oct2 would interchangeably bind to the same regions or whether there would be transcription factor selectivity in vivo. The observed binding of all 3 transcription factors to the promoter and the 3′ enhancer in Burkitt lymphoma cells suggests that for these regulatory regions, there is no such selectivity. This result is in perfect agreement with published data on the ability of the ubiquitous Oct1 to substitute for the B-cell-specific Oct2 in activating Ig promoters.41
In contrast, the situation at the internal enhancer does suggest some level of selectivity, which is, however, not absolute. The role of octamer-binding factors in regulating the human internal enhancer was unclear in the past. Whereas the mouse counterpart contains a perfect octamer motif,42,43 the human enhancer has 3 octamer-like sequences with lower affinity to Oct transcription factors.44 The 2 sites analyzed in the present study show the best conservation to the functional sites.45 Our ChIP assays reveal that Oct2 and BOB.1/OBF.1 but only very little Oct1 were bound to the internal enhancer of Ig-producing B-cell lines Namalwa and B-JAB. This implies that Oct2 predominantly regulates the function of this motif in the human Ig internal enhancer. This observation is consistent with earlier data where reporter constructs containing the mouse internal enhancer sequence were activated by Oct2 but not by Oct1.29
The 3′ IgH enhancer elements lie downstream of the Cα1 and Cα2 regions (Figure 1A). Because transgenic constructs containing the 3′ IgH enhancer elements were found to express the reporter gene in position-independent manner, they were referred to as locus control regions (LCRs).46 In humans, both Cα1 and Cα2 LCRs contain 3 DNAse I hypersensitivity sites (HS3A; HS1,2; and HS4), which are responsible for enhancer activity, and they all contain octamer motifs.47,48 Although these elements show strong functional synergy only when combined, HS1,2 is assumed to play the most important role in the control of transcription.33,34 Our finding that all 3 transcription factors do interact with the HS1,2 enhancer of Cα1 and Cα2 LCRs in Burkitt lymphoma cell lines is consistent with the observed ability of both Oct1 and Oct2 to bind to murine hs1,2.49 It is also in line with the reported ability of BOB.1/OBF.1 to strongly enhance the activity of the 3′ enhancer induced by interleukin 4 (IL-4) and anti-CD40 antibody.50
It has recently been shown that Oct1 or Oct2 preferentially recruit BOB.1/OBF.1 when they bind to a subset of octamer motifs, which have been called palindromic Oct factor recognition elements (POREs).51,52 The octamer motifs like those in the IgH promoter have been considered to be the prototypes of octamer motifs that do not recruit BOB.1/OBF.1 (more POREs [MOREs]). This concept contradicted numerous functional assays of Ig gene regulation.53,54 Furthermore, our ChIP data presented here disagree with a simple version of the PORE/MORE concept. Neither the human IgH regulatory elements nor the H2B promoter region contain PORE-type octamer motifs, yet they are apparently bound by BOB.1/OBF.1 in vivo. We have recently identified novel genes regulated by BOB.1/OBF.1 using expression profile analysis. The best regulated gene was found to contain a perfect octamer motif in its promoter and this motif again did not resemble the PORE consensus.55
Octamer motifs in Ig regulatory elements are inaccessible in cHL-derived cell lines
The absence of transcription factor binding to Ig regulatory regions in cHL-derived cell lines is in striking contrast to Burkitt lymphoma cell lines. Although it was demonstrated that the lack of transcription factors mainly contributes to the down-regulation of Ig expression in cHL-derived cell lines,7 the inability of Oct1 (in all cHL-derived cell lines) and Oct2 (in KM-H2 and in L428-Oct2 cells) to bind to any of the IgH regulatory regions in cultured HRS cells suggests, in addition, an epigenetic mechanism. This epigenetic mechanism could explain not only the down-regulation of Ig transcription but also the absence of many other B-cell characteristic genes in cultured and primary HRS cells.
Inhibition of DNA methylation results in reactivation of IgH transcription
Somewhat conflicting data have been published with respect to the expression of IgH RNA in different cHL cell lines.4,8,18,56 In line with recent results from Affymetrix GeneChip assays, we only found spliced Ig transcripts by RT-PCR in KM-H2 cells.57
We hypothesized that epigenetic mechanisms could be responsible for the down-regulation of IgH expression. As outlined in the “Introduction,” epigenetic silencing of so-called tumor suppressor genes has been identified as a common oncogenic mechanism contributing to tumorigenesis.19 Many of these genes could be reactivated by an inhibitor of the DNA methyltransferase 5-aza-dC. Interestingly, the DNA demethylating agent 5-aza-dC reactivated Ig transcription not only in L428, with the intact octamer motif in the VH5′ regulatory region, but also in the L1236 cells, which carry a mutated promoter octamer motif.11 This observation is in accordance with the reported octamer-binding transcription factor-independent transcriptional activity VH6 gene-containing mutated octamer motif in the promoter region.58 One should stress that the level of Ig expression detected by RT-PCR in L428 and L1236 after reactivation by 5-aza-dC cells was very low and comparable to that in KM-H2 cells but much lower than in B-cell lines such as Namalwa. The slightly increased levels of Ig transcription in 5-aza-dC-treated cells coexpressing Oct2 and BOB.1/OBF.1 stress the importance of these factors for IgH promoter activity41 and Ig production.16 Nevertheless, the observed inability of the combination of DNA demethylation and the octamer-binding transcription factors/cofactors to reconstitute measurable Ig protein expression in cHL implies the existence of additional regulatory events. Indeed, these data suggest that even when the chromatin structure is open and octamer-binding factors are present, the full set of B-cell-specific factors is required to completely reactivate Ig production.
In HRS cells, IgH chain gene chromatin has a “closed” structure only in the promoter region and can be opened by inhibition of DNA methylation
In most cases, DNA methylation does not inhibit transcription per se but rather plays a pivotal role in a complex mechanism of chromatin-dependent gene silencing.59 Indeed, in mammalian cells, the principal processes involved in epigenetic silencing are chromatin modifications. They include DNA methylation and histone modification (acetylation, phosphorylation, methylation, and ubiquitination).60 Interplay between DNA methylation and core histone modifications resulting in epigenetic silencing can be described by a bidirectional positive feedback loop model. DNA methylation directs histone deacetylation, and subsequently gene silencing or, vice versa, histone methylation directs DNA methylation. This includes de novo methylation of DNA followed by binding of methyl-CpG-binding proteins (eg, methyl-CpG binding protein [MeCP2] and the recruitment of a histone deacetylase complex such as MeCP2-transcriptional repressor histone deacetylase complex [Sin3a]). Deacetylation of core histones (histone H3 and histone H4) by this complex results in gene silencing. The deacetylated chromatin is specifically methylated at H3-K9 by the histone methyltransferase suppressor of position effect variegation (Suv 39H1) or G9a to stabilize the inactive state of chromatin.61 Alternatively, H3-K9 methylation can serve as a signal to recruit the methyl-binding protein heterochromatin protein 1 (HP1), which might in turn attract DNA methyltransferases, resulting in DNA methylation.62 Our findings of H3-K9 methylation in the VH promoter region of L428 cells and its reversion by 5-aza-dC treatment, paralleled by the reactivation of Ig transcription, are in keeping with this model.
The absence of H3-K9 methylation at the enhancer regions was unexpected because it obviously contradicted the observed absence of transcription factor binding in ChIP experiments. One explanation for this could be the involvement of alternative chromatin modification and gene-silencing pathways. It has recently been reported that some genes can be silenced by chromatin-dependent mechanisms relying primarily on methylation of lysine 27 in histone H3 by polycomb group proteins human enhancer of zeste (E(Z)/Ezh2).63
In summary, our data indicate that (1) transcription factors fail to bind to their target sites in Ig regulatory elements of cHL cell lines; (2) some level of Ig transcription can be reactivated by the 5-aza-dC; (3) histone H3 is methylated at lysine 9 in the IgVH promoter region; and finally, (4) even if chromatin in the Ig locus has an open structure, octamer-binding transcription factors can only activate a low level of Ig transcription, which is not sufficient to reveal Ig protein production. These results suggest an important role of epigenetic silencing in suppression of Ig transcription in cHL-derived cell lines.
Prepublished online as Blood First Edition Paper, July 29, 2004; DOI 10.1182/blood-2003-04-1197.
Supported by grants from the Deutsche Forschungsgemeinschaft (DFG SFB497, C5) and the Fonds der Chemischen Industrie (T.W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank A. Kick and O. Sakk for excellent technical assistance.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal