Abstract
Hermansky-Pudlak syndrome (HPS), a disorder of organelle biogenesis, affects lysosomes, melanosomes, and platelet dense bodies. Seven genes cause HPS in humans (HPS1-HPS7) and at least 15 nonallelic mutations cause HPS in mice. Where their function is known, the HPS proteins participate in protein trafficking and vesicle docking/fusion events during organelle biogenesis. HPS-associated genes participate in at least 4 distinct protein complexes: the adaptor complex AP-3; biogenesis of lysosome-related organelles complex 1 (BLOC-1), consisting of 4 HPS proteins (pallidin, muted, cappuccino, HPS7/sandy); BLOC-2, consisting of HPS6/ruby-eye, HPS5/ruby-eye-2, and HPS3/cocoa; and BLOC-3, consisting of HPS1/pale ear and HPS4/light ear. Here, we report the cloning of the mouse HPS mutation reduced pigmentation (rp). We show that the wild-type rp gene encodes a novel, widely expressed 195-amino acid protein that shares 87% amino acid identity with its human orthologue and localizes to punctate cytoplasmic structures. Further, we show that phosphorylated RP is part of the BLOC-1 complex. In mutant rp/rp mice, a premature stop codon truncates the protein after 79 amino acids. Defects in all the 5 known components of BLOC-1, including RP, cause severe HPS in mice, suggesting that the subunits are nonredundant and that BLOC-1 plays a key role in organelle biogenesis.
Introduction
Hermansky-Pudlak syndrome (HPS) is an autosomal recessive disorder of organelle biogenesis that affects lysosomes and 2 lysosome-related organelles (LROs), melanosomes and platelet dense bodies.1,2 HPS occurs in all ethnic groups but is particularly prevalent in northwest Puerto Rico (1/1800) due to a founder effect involving HPS1.3 Melanosome defects led to oculocutaneous albinism causing nystagmus, decreased visual acuity, and skin damage on exposure to the sun.4 Platelet dense body defects result in prolonged bleeding, which can be severe. Some patients develop progressive pulmonary fibrotic disease, which typically leads to death in the fourth to fifth decades.4,5 The pathogenesis of lung fibrosis is poorly understood. It has been attributed to ceroid lipofuscin accumulation in defective lysosomes within alveolar macrophages.6 Recent studies in mice suggest that a defect in a fourth LRO, lamellar bodies of type II pneumocytes, may contribute to lung disease in HPS.7
HPS is clinically and genetically heterogeneous. Defects in 7 genes (HPS1-HPS7) cause HPS in humans.8,9 Patients with HPS-1 and HPS-4 are at high risk for pulmonary involvement.5,10 At least 15 nonallelic mutations cause HPS in mice, including the orthologues of HPS1-HPS7: pale ear (ep), pearl (pe), cocoa (coa), light ear (le), ruby-eye-2 (ru2), ruby-eye (ru), and sandy (sdy), respectively.8,11-16 Mutations at 6 loci in mice have been cloned for which no human HPS patients have yet been identified: mocha (mh), gunmetal (gm), buff (bf), pallid (pa), muted (mu), and cappuccino (cno).17-22
Where their function is known, the HPS proteins participate in sorting of cargo proteins and vesicle targeting/fusion events during the biogenesis of LROs. Genes encoding the β3A (Ap3b1) and δ (Ap3d) subunits of the adaptor complex AP-3, which functions in endosomal-lysosomal protein trafficking, 23 are defective in HPS-2/pearl and mocha, respectively. Buff is a defect in the gene encoding VPS33A, which is involved in protein transport to lysosomes in mammals as well as the yeast vacuole.24,25 Pallid is a defect in a novel protein, pallidin, which binds to syntaxin-13, a t-SNARE protein that mediates vesicle docking and fusion.22 Sandy encodes dysbindin (Dtnbp1), which binds to pallidin in addition to components of the dystrophin-associated protein complex.26 In gunmetal, the gene encoding the α subunit of Rab geranylgeranyl transferase (Rabggta) is mutated; rab proteins function in targeting and fusing transport vesicles to their acceptor membrane.18
The remaining HPS genes encode novel proteins containing no functional domains or structural motifs that give any clues as to their precise function. However, the phenotypic similarity among the mouse HPS models clearly points to a role in some aspect of organelle biogenesis. Moreover, the HPS proteins function within multiple subunit complexes. HPS2/pe and mocha are components of the AP-3 complex.27 Muted, pallidin, cappuccino, HPS7/sdy, and at least one additional unknown protein interact in a complex designated BLOC-1 (biogenesis of lysosome-related organelles complex 1).8,21,28 The presence of the syntaxin-binding protein pallidin suggests that BLOC-1 is involved in vesicle docking and fusion. HPS5/ru2, HPS6/ru, and HPS3/coa are components of BLOC-2, 16,29 whereas HPS1/ep interacts with HPS4/le in BLOC-3.30,31 Interestingly, unlike AP-3, the known components of BLOC-1, -2, and -3 are confined to metazoans, suggesting they serve specialized functions in the biogenesis of those LROs (eg, platelet dense bodies, melanosomes) that are specific to higher eukaryotes.
Of the known HPS mouse models, only subtle gray (sut)32 and reduced pigmentation (rp)33 have not been cloned. Here, we report the positional cloning of reduced pigmentation (GenBank accession no. AY515509) and show that wild-type rp encodes a novel, ubiquitously expressed 195-amino acid protein that in its phosphorylated form is part of the BLOC-1 complex. In mutant rp/rp mice a premature stop codon truncates the protein at 79 amino acids indicating that failure to produce an intact BLOC-1 complex underlies the HPS phenotype in rp/rp mice. Defects in the other known components of BLOC-1 (sdy, mu, pa, cno) all cause HPS as well, suggesting that these subunits are nonredundant and play key roles in the biogenesis of LROs.
Patients, materials, and methods
Animals
All animals were raised at The Jackson Laboratory, Bar Harbor, ME. The autosomal recessive rp mutation arose spontaneously on the C57BL/Tb (Tb) background in 1975.33 The mutation was back-crossed to C57BL/10ScSn (B10) for 2 generations and subsequently maintained on the Tb and B10 mixed genetic background by rp/+ × rp/rp matings. Heterozygotes, which show no phenotype, and closely related C57BL/6J (B6) mice served as normal controls in all studies. All animals were housed in humidity- and temperature-controlled rooms with a 12-hour light cycle with free access to acidified water and chow (NIH 5K52). The Jackson Laboratory Animal Care and Use Committee approved all protocols.
Blood analysis
Adult whole blood was collected in ethylenediaminetetraacetic acid (EDTA)–coated microtainer tubes (Becton Dickinson, Rutherford, NJ). Complete blood counts were obtained using an Advia 120 multispecies whole blood analyzer (Bayer, Tarrytown, NY) as described.34 Platelet dense bodies were enumerated in air-dried, unstained, unfixed whole platelets as described35 using a JEOL 100CXII transmission electron microscope (Jeol, Tokyo, Japan). Whole blood was collected in acid-citrate-dextrose (0.13 M citric acid, 0.15 M sodium citrate, 0.1 M dextrose) and centrifuged at 120g for 20 minutes at room temperature to obtain platelet-rich plasma. Bleeding times and platelet serotonin assays were performed as described.36,37
Fine-linkage mapping ofrp
An F2 intercross was established using rp/rp and Mus musculus castaneus (CAST/Ei) mice; 324 homozygous F2rp/rp mice, identified by their diluted coat color, were generated (648 meioses). DNA was prepared from spleens as described38 and from tail tissue using a commercial kit (Gentra Systems, Research Triangle Park, NC). Anonymous DNA microsatellite markers were mapped by polymerase chain reaction (PCR) using primers obtained from Research Genetics (Huntsville, AL). Products were resolved and scored on ethidium bromide-stained 1% to 5% NuSieve gels (Cambrex Bio Science Rockland, Rockland, ME). Polymorphic candidate genes were mapped by PCR or by Southern blot analysis using standard techniques.
Sequencing
gDNA was prepared from spleens of C57BL/6J (B6) and rp/rp mice. Approximately 1000 bp fragments within the rp critical interval on chromosome 7 and overlapping by 50 to 200 bp at either end were generated by PCR using high-fidelity Dynazyme EXT DNA polymerase according to the manufacturer's instructions (MJ Research, Watertown, MA). DNA fragments were sized by electrophoresis in 0.8% agarose gels, excised from the gel, and purified using a Spin-X column (Corning Costar, Corning, NY). Oligonucleotides were designed complementary to sequences publicly available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and were synthesized by Ransom Hill Bioscience (Ramona, CA). Sequencing was performed using the automated dye termination technique (ABI PRISM Model 3700 Genetic Analyzer, Applied Biosystems, Foster City, CA). Sequence was obtained with oligonucleotides used to generate PCR fragments and internal oligonucleotides where necessary. Sequencing reactions of templates with high GC content were supplemented with betaine to a final concentration of 100 mM.
gDNA and protein sequences were analyzed using the following programs and online sequence analysis resources: Sequencher version 4.1, Wisconsin GCG Package (version 10.3) and ExPASy Proteomics Tools (http://us.expasy.org/tools/). Protein identity/similarity analyses were carried out by GAP analyses in the Wisconsin GCG Package (version 10.3) using default parameters and no penalty for end gaps.
Northern and Southern blotting
Total RNA was prepared using TRIzol reagent (Invitrogen, Carlsbad, CA). Polyadenylated RNA was purified on oligo-dT columns from Stratagene (La Jolla, CA). Northern and Southern blotting were performed using standard techniques. Premade mouse multiple tissue Northern blots purchased from OriGene Technologies (Rockland, MD) were hybridized and washed as recommended by the manufacturer.
DNA constructs
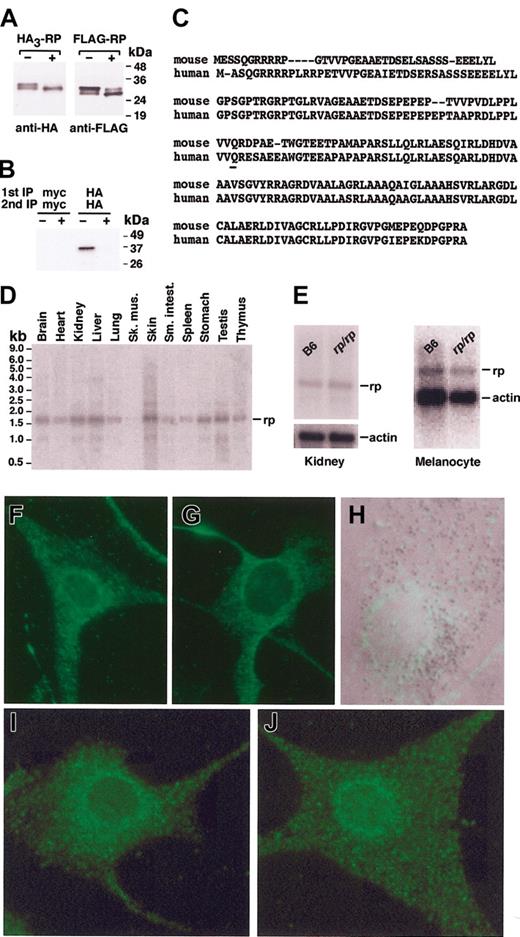
The construct pCI-HA3 was generated by cloning of annealed primers containing 3 copies of the HA epitope tag into XhoI-EcoRI sites of the mammalian expression vector pCI-neo (Promega, Madison, WI). The epitope-tagging at the amino-terminus of RP was performed through PCR amplification on a Marathon-Ready mouse 7-day embryo (Clontech, Palo Alto, CA) of the full-length rp cDNA, followed by in-frame cloning into EcoRI-XbaI sites of the pCI-HA3 (pCI-HA3-RP) vector. The insert of this plasmid was subcloned into XhoI-NotI sites of the vector pCDNA3.1-Hygromycin (Invitrogen) to generate pCDNA-Hygro-HA3-RP. The full-length cDNA encoding wild-type RP was also subcloned in-frame into EcoRI-EcoRV sites of the expression vector pFLAG-CMV-6c (Sigma Chemical, St Louis, MO) and the HindIII site of the expression vector pHM6 (pHM6-RP; Roche, Indianapolis, IN).
Cells cultures and transfections
The generation of cell lines derived from mouse skin fibroblasts and culture conditions for these and human melanoma MNT-1 cells have been described previously.30 MNT-1 cells were stably transfected with pCDNA-Hygro-HA3-RP using the FuGENE 6 reagent (Roche Diagnostics, Indianapolis, IN). Hygromycin-resistant clones expressing HA3-RP were selected using 200 μg/mL Hygromycin B. Transient expression of HA3-RP and FLAG-RP was also performed using FuGENE 6 reagent.
Melanocyte lines melan-a39 and lines derived from rp/rp animals were cultured according to the protocols of Bennett et al39 (available online at http://www.sghms.ac.uk/depts/anatomy/pages/dcbm&m.htm). Mutant rp/rp melanocyte cell lines were generated by mating rp/rp mice with C57BL6/J mice carrying an Ink4a-Arf exon 2 deletion40 at Texas A&M University. Litters homozygous for both mutations were obtained at the third generation, and trunk skins from neonatal (aged 0-2 days) mice were sent in chilled culture medium to St George's Hospital Medical School. There they were used as described previously for preparation of melanocyte cultures that were deficient in cell senescence to facilitate establishment as cell lines.41 Three such lines were derived: melan-rp1, melan-rp2, and melan-rp3. In the studies described here, unless otherwise specified, melan-rp1 cells were used. Digital images were taken at room temperature using a Zeiss Axioplan 2 microscope equipped with an Axiocam high resolution camera (12 megapixels) and captured with Axiovision Imaging software v. 3.1 (all from Carl Zeiss, Welwyn Garden City, Great Britain).
Transfection of melan-a and melan-rp1 cultures with pHM6-RP was performed using GeneJuice transfection reagent (Novagen, Madison, WI).
Melanin assays and DOPA histochemistry
Melanin assays were based on the optical density at 475 nm (OD475) of cell lysates, as described.42 Melanin content was normalized to protein content, which was determined using a bicinchoninic acid assay (Pierce, Chester, United Kingdom).42 Dihydroxyphenylalanine (DOPA) histochemistry was carried out by a method modified from that described by Boissy et al.43 In brief, cells were fixed in 2.5% glutaraldehyde, 2% paraformaldehyde in 0.2M sodium cacodylate buffer, pH 7.2, for 1 hour at room temperature. Cells were then washed 3 times for 5 minutes in 0.2 M sodium cacodylate buffer, pH 7.4, prior to incubation in l- or d-DOPA (0.1% in cacodylate buffer) for two 2.5-hour intervals at 37°C. (d-DOPA staining was used as a control and produced no stain.) Fresh DOPA solutions were used in the second incubation. The cells were washed as before and postfixed in 1% osmium tetroxide with 1.5% potassium ferrocyanide in cacodylate buffer for 1 hour at room temperature. Three final washes were carried out before dehydration and embedding for sectioning.
Antibodies
A peptide antibody to the N-terminus of RP (amino acids 1-12) was generated in chickens and affinity purified by the manufacturer (Aves Lab, Tigard, OR). The antibody performed well in immunofluorescence studies but proved unsuitable for Western blotting and immunoprecipitation. The generation and purification of rabbit antibodies to AP3 subunits σ3 and μ3, 44 HPS1, 45 HPS4, 30 and pallidin, cappuccino, and muted21 have been described previously. The following mouse monoclonal antibodies were also used: anti-HA (clone HA.11) and anti-Myc (clone 9E10) from Covance (Richmond, CA) and anti-FLAG (clone M2) and anti–α-tubulin (clone DM1A) from Sigma Chemical. Horseradish peroxidase-conjugated secondary antibodies for immunoblotting were from Amersham Biosciences (Piscataway, NJ). Alexa Fluor 488 goat anti–chicken and goat anti–rabbit secondary antibodies (Molecular Probes, Eugene, OR) and anti-HA (Sigma Chemical) were used in melanocyte immunofluorescence studies.
Sedimentation velocity analysis
Sedimentation velocity analysis was performed by ultracentrifugation on linear 2% to 15% (wt/vol) sucrose gradients (12 mL) prepared in buffer A (25 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.4, 150 mM NaCl, 1 mM EGTA [ethylene glycol tetraacetic acid], 1 mM dithiothreitol, 0.5 mM MgCl2, and protease inhibitor mixture). Cytosol (350 μL) from cells stably expressing HA3-RP, prepared in buffer A as described, 30 was layered on top of a linear sucrose gradient and centrifuged in a SW41 rotor (Beckman Coulter, Fullerton, CA) at 39 000 rpm for 17 hours at 4°C. Twenty 0.60-mL fractions were collected from the bottom of the tube and analyzed by immunoprecipitation and immunoblotting. The following protein standards from Amersham Biosciences were used (Svedberg coefficients in parentheses): chicken ovalbumin (3.6 S), bovine serum albumin (4.6 S), rabbit aldolase (7.3 S), and bovine catalase (11.3 S).
Immunoprecipitation-recapture assays
Metabolic labeling of cultured cells with [35S]methionine, preparation of Triton X-100 extracts, preparation and ultracentrifugation of detergent-free extracts, and immunoprecipitation-recapture were performed as described.30
Alkaline phosphatase treatment
Whole-cell extracts prepared from MNT-1 cells either mock transfected or transiently expressing HA3-RP or FLAG-RP were treated for 1 hour at 37°C in the presence or absence of alkaline phosphatase (Roche), electrophoresed, and immunoblotted using anti-HA and anti-FLAG antibodies. In other experiments, HA-tagged RP was isolated from 32P-orthophosphate–labeled MNT-1 cells expressing HA3-RP by immunoprecipitation-recapture. The immunoprecipitates were washed twice with phosphorylation buffer consisting of 50 mM Tris (tris(hydroxymethyl)aminomethane)–HCl (pH 8.5) and 0.1 mM EDTA and divided into 2 aliquots. One of the aliquots was treated with 0.2 U calf intestinal alkaline phosphatase (Roche) for 2 hours at 37°C; the other was mock-treated. Samples were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by fluorography.
Electrophoresis and immunoblotting
SDS-PAGE and immunoblotting analysis were performed as described.30 Horseradish peroxidase-labeled antibodies were detected by using the Western Lightning, Chemiluminescence Reagent Plus (Perkin Elmer Life Sciences, Boston, MA).
Immunofluorescence microscopy
Normal C57BL/6J and rp/rp melanocytes were plated on to coverslips in 6-well tissue culture plates at a density of 1 × 105 cells/well and allowed to grow for 48 hours. Then, 1 μg plasmid pHM6 or pHM6-RP per well was introduced into cells using GeneJuice transfection reagent (Novagen) according to the manufacturer's protocol. Twenty-four hours after transfection, the cells were washed in phosphate-buffered saline (PBS), fixed in 1 mL 4% paraformaldehyde for 5 minutes, and washed 2 times with methanol. The cells were then fixed in methanol for an additional 10 minutes and washed 5 × 5 minutes in PBS. All washes and fixation steps were carried out on ice with ice-cold reagents. Subsequent steps were carried out at room temperature. Prior to the application of the primary antibody, coverslips were incubated for 30 minutes in 1 × PBS containing 1% bovine serum albumin (BSA) and 0.1% Triton X-100. Coverslips were inverted onto a 50-μL drop of PBS/BSA/Triton buffer containing 1.6 μg/mL affinity-purified rabbit antihemagglutinin (HA; Sigma Chemical) and incubated in a humidified chamber for 1 hour. Coverslips were washed with PBS, followed by three 5-minute incubations in PBS/BSA/Triton buffer. Prior to application of the fluorescently labeled secondary antibody at 5 μg/mL (goat anti–rabbit Alexa Fluor 488, Molecular Probes), nonspecific binding was blocked by incubating the cells in 5% normal goat serum (NGS; Jackson ImmunoResearch, West Grove, PA) in PBS for 30 minutes. Following incubation for 1 hour with secondary antibody, coverslips were washed and mounted on glass slides using SlowFade Antifade (Molecular Probes) according to the manufacturer's instructions. Digital images were obtained at room temperature using a Leica DMRE microscope equipped with a Leica 40×/0.85 objective lens (Leica, Wetzlar, Germany). Images were captured with a Princeton Instruments (Princeton, NJ) camera model RTE/CCD 1300 Y/H using Basic Metamorph acquisition software (Universal Imaging, Downingtown, PA).
For direct staining of RP in melanocytes, a similar protocol was followed with these exceptions. Initial blocking of nonspecific binding was accomplished using a 1:3 dilution of BlokHen (Aves Labs), rinsed with 1 × PBS, and incubated with affinity-purified chicken anti-RP at 1.8 μg/mL. The secondary antibody used was Alexa Fluor 488-labeled goat anti–chicken (Molecular Probes) at 5 μg/mL.
Generation of double homozygotes
Mice doubly homozygous for rp and ruby-eye (ru) were generated by conventional interbreeding. The ruby genotype of F2 offspring was confirmed by PCR and restriction enzyme analysis. gDNA was amplified using ru-specific primers (forward 5′-GTT AAG GAC CAA CCA GAG CAG CTT TCA ACG-3′, reverse 5′-CAA GTG TCC CCT TGT TTG GGA TTC AGG-3′), and the reaction product digested with BsrDI. A single 296-bp band is seen in ru/ru mice due to deletion of a BsrD1 site, whereas wild-type DNA gives fragments of 123 bp and 182 bp. Because these experiments were performed prior to the identification of the rp gene, F2 mice were genotyped for rp by crossing to rp/rp homozygotes. The phenotypic distribution of the offspring was analyzed using a χ2 test to determine the probable genotype. To be classified as an rp/rp homozygote, we observed at least 6 rp/rp pups (and no other phenotypes) from each animal (.25 > P > .01).
Screening of human HPS patients
PCR amplification products from fibroblast DNA were sequenced using a Beckman CEQ 2000 and the CEQ Dye Terminator Cycle Sequencing kit according to the manufacturer's instructions.
Results
The rp mutation
The rp mutation (Figure 1A) was first reported in 1981 as a new coat color mutation with increased kidney lysosomal enzyme activity that mapped to mouse chromosome (Chr) 7.33 Since then rp has been classified as a mouse model of HPS.46 We have confirmed increased bleeding times in rp/rp mice (Table 1) and shown that the number of dense bodies is significantly decreased in rp/rp platelets compared to normal (Table 1; Figure 1B). Absent or decreased platelet dense bodies are considered the “gold standard” for a diagnosis of HPS.47 Consistent with decreased numbers of dense bodies and prolonged bleeding, homozygous rp platelet serotonin levels are significantly decreased (0.20 ± 0.05 μg/109 platelets versus 5.37 ± 0.46 in wild type [X ± SE], n = 4, P < .001]). All other hematologic parameters are normal (Table 1).
Characteristics of rp/rp mice. Wild-type (B6) data are shown in the left panels and rp/rp in the right. (A) Coat color dilution in rp/rp mice. (B) Lack of platelet dense bodies (arrows) in rp/rp platelets. Magnification × 14 000. (C) Morphology and pigmentation of wild-type (B6, melan-a) and rp/rp immortal mouse melanocytes (bright-field images). Note decreased melanin pigmentation in rp/rp cells. Bar represents 50 μM. (D) Electron microscopy. Melanosomes of rp/rp melanocytes are less numerous than in wild-type melanocytes. All stages of melanosome development are found, as in wild type, but immature stages are more frequent (arrows), and some clustering of melanosomes is seen (not shown). Bar represents 0.5 μM. (E) Tyrosinase localization shown by L-DOPA staining (bright-field microscopy). Tyrosinase accumulates in the perinuclear region in rp/rp melanocytes (arrow) but is well distributed across the cell in wild-type melanocytes. Bar represents 50 μM. (F) Electron microscopy of l-DOPA–stained melanocytes. Mutant melanocytes show DOPA-positive tubular structures (arrow) not seen in wild-type cells. Bar represents 1.0 μM.
Characteristics of rp/rp mice. Wild-type (B6) data are shown in the left panels and rp/rp in the right. (A) Coat color dilution in rp/rp mice. (B) Lack of platelet dense bodies (arrows) in rp/rp platelets. Magnification × 14 000. (C) Morphology and pigmentation of wild-type (B6, melan-a) and rp/rp immortal mouse melanocytes (bright-field images). Note decreased melanin pigmentation in rp/rp cells. Bar represents 50 μM. (D) Electron microscopy. Melanosomes of rp/rp melanocytes are less numerous than in wild-type melanocytes. All stages of melanosome development are found, as in wild type, but immature stages are more frequent (arrows), and some clustering of melanosomes is seen (not shown). Bar represents 0.5 μM. (E) Tyrosinase localization shown by L-DOPA staining (bright-field microscopy). Tyrosinase accumulates in the perinuclear region in rp/rp melanocytes (arrow) but is well distributed across the cell in wild-type melanocytes. Bar represents 50 μM. (F) Electron microscopy of l-DOPA–stained melanocytes. Mutant melanocytes show DOPA-positive tubular structures (arrow) not seen in wild-type cells. Bar represents 1.0 μM.
Hematologic parameters in adult rp/rp mice
. | WBCs . | RBCs . | HGB . | HCT . | PLT . | MPV . | BT . | . |
|---|---|---|---|---|---|---|---|---|
| GTP . | WBC count, × 109/L . | RBC count, × 1012/L . | HGB level, g/L . | HCT value . | PLT count, × 1012/L . | MPV, fL . | BT, min. . | DB*, no. per platelet . |
| +/rp | 10.7 ± 2.6 (5) | 9.9 ± 0.1 (5) | 149 ± 10 (5) | 423 ± 0.009 (5) | 0.99 ± 0.15 (5) | 4.3 ± 0.3 (5) | 1.3 ± 0.7 (6) | 10.8 ± 4.0 |
| rp/rp | 8.6 ± 1.3 (7) | 10.0 ± 0.3 (7) | 151 ± 4 (7) | 425 ± 0.010 (7) | 1.05 ± 0.13 (7) | 4.4 ± 0.3 (7) | 13.5 ± 3.1 (6†) | 0.8 ± 1.0† |
. | WBCs . | RBCs . | HGB . | HCT . | PLT . | MPV . | BT . | . |
|---|---|---|---|---|---|---|---|---|
| GTP . | WBC count, × 109/L . | RBC count, × 1012/L . | HGB level, g/L . | HCT value . | PLT count, × 1012/L . | MPV, fL . | BT, min. . | DB*, no. per platelet . |
| +/rp | 10.7 ± 2.6 (5) | 9.9 ± 0.1 (5) | 149 ± 10 (5) | 423 ± 0.009 (5) | 0.99 ± 0.15 (5) | 4.3 ± 0.3 (5) | 1.3 ± 0.7 (6) | 10.8 ± 4.0 |
| rp/rp | 8.6 ± 1.3 (7) | 10.0 ± 0.3 (7) | 151 ± 4 (7) | 425 ± 0.010 (7) | 1.05 ± 0.13 (7) | 4.4 ± 0.3 (7) | 13.5 ± 3.1 (6†) | 0.8 ± 1.0† |
All values are mean ± SD. Parenthetical values indicate the number of samples.
GTP indicates genotype; WBC, white blood cell; RBC, red blood cell; HGB, hemoglobin; HCT, hematocrit; PLT, platelet; MPV, mean platelet volume; BT, bleeding time; DB, dense bodies.
Average in 8 to 10 platelets from 3 different animals.
P < .001.
Melanosome defects are evident in cultured rp/rp melanocytes. By light microscopy rp/rp cells appear pale compared to melan-a cells, suggesting an overall decrease in pigmentation (Figure 1C). Transmission electron microscopy confirms that rp/rp melanosomes are abnormal. All stages of melanosome development are found, as in wild type, but immature stages are more frequent, and some clustering of melanosomes is seen (Figure 1D). Melanin assays support the visual evidence. Melanin content was 143.82 ± 12.43 μg melanin/mg protein in rp/rp melanocytes versus 225.72 ± 12.96 μg melanin/mg protein in wild-type melanocytes (X ± SE, P < .02 in 3 assays from each of 3 cell lines of each genotype). Tyrosinase distribution within rp/rp melanocytes shown by l-DOPA staining is abnormal as well. Tyrosinase accumulates in the perinuclear region in rp/rp melanocytes but is well distributed across the cell in wild-type melanocytes (Figure 1E). Perinuclear tyrosinase accumulation has been noted in all HPS mouse mutants studied (E.V.S. and D.C.B., unpublished observations, 2004). Mislocalization of tyrosinase has also been noted in human HPS.48,49 In addition, mutant melanocytes show DOPA-positive tubular structures not seen in wild-type cells (Figure 1F). We conclude from these analyses that rp is a classic model of HPS displaying significantly decreased numbers of platelet dense bodies and defective tyrosinase transport in melanocytes.
Positional cloning ofrp
We fine mapped rp to a 0.5-cM interval between the Rtn2 (reticulon 2) gene and the anonymous sslp marker D7Mit57 on Chr 7 (Figure 2A).50 This interval encompasses 300 kb and contains more than 30 known or predicted transcripts (Celera Discovery System and Celera Genomics associated databases). No strong candidate genes were contained within this interval. For that reason and because most HPS genes are novel, we chose to directly sequence the interval. We identified a C>T substitution in rp/rp DNA compared to B6 DNA that creates a premature stop codon in one predicted coding sequence, BC043666 (Figure 2B). This substitution was subsequently confirmed in +/+ and +/rp animals derived from the rp line by intercrossing obligate heterozygotes. The mutation removes an Fnu4HI site, changing 63- and 71-bp fragments to a 134-bp fragment. The polymorphism was used to demonstrate that the mutation segregates with the rp phenotype (Figure 2C). From this and other evidence described herein, we conclude that BC043666 is the rp gene (GenBank accession nos. AY515509 and BK005392).
Identification of therpgene defect. (A) Fine-linkage map of the rp critical interval on Chr 7. (B) A C>T transition creates a premature stop codon in the rp gene. (C) A 672-bp fragment within exon 2 was amplified from gDNA using forward primer 5′-AATGCTACCACGCATTGGAGGCCT-3′ and reverse primer GCCATCTCGTGT-GACCTAGTTCTA-3′ and digested with Fnu4HI. The rp mutation removes an Fnu4HI site, changing 63- and 71-bp fragments to a 134-bp fragment that was used to genotype mice.
Identification of therpgene defect. (A) Fine-linkage map of the rp critical interval on Chr 7. (B) A C>T transition creates a premature stop codon in the rp gene. (C) A 672-bp fragment within exon 2 was amplified from gDNA using forward primer 5′-AATGCTACCACGCATTGGAGGCCT-3′ and reverse primer GCCATCTCGTGT-GACCTAGTTCTA-3′ and digested with Fnu4HI. The rp mutation removes an Fnu4HI site, changing 63- and 71-bp fragments to a 134-bp fragment that was used to genotype mice.
The rp gene structure, expression, and protein distribution
The wild-type rp mRNA contains 1867 bp consisting of 2 exons, separated in the gDNA by a 694-bp intron. Exon 1 encodes 61 bp of 5′ noncoding sequence. Exon 2 encodes 9 bp of 5′ noncoding sequence, the entire coding sequence of 585 bp, and 1212 bp of 3′ untranslated sequence. The gene encodes a 195-amino acid protein with a predicted molecular weight of 20.4 kDa and a pI of 4.84. There are no predicted transmembrane domains or coiled-coil domains. PROSCAN predicts a single protein kinase C phosphorylation site and several casein kinase II phosphorylation sites. Immunoblotting analysis using extracts of MNT-1 cells transiently expressing RP tagged with either a triple HA epitope (HA3-RP) or a FLAG epitope (FLAG-RP) shows the presence of 2 polypeptide bands with molecular masses of 30 kDa and 33 kDa for HA3-RP and 28 kDa and 30 kDa for FLAG-RP, respectively (Figure 3A). Alkaline phosphatase treatment of the extracts increases the electrophoretic mobility of the 33 kDa (HA3-RP) and 30 kDa (FLAG-RP) bands, suggesting that these correspond to phosphorylated forms of RP (Figure 3A). The phosphorylation of RP was confirmed by metabolic labeling of HA3-RP–expressing MNT-1 cells with 32P-orthophosphate, followed by immunoprecipitation with antibodies to the myc (nonspecific control) or HA epitopes (Figure 3B). Together, these experiments reveal that HA3-RP is specifically labeled with 32P and migrates as a 33-kDa species. The apparent molecular mass of the dephosphorylated band is larger than that predicted from its amino acid sequence, even accounting for the contribution of the triple-HA (24 kDa) or FLAG (21.5 kDa) epitopes, indicating that RP migrates anomalously on SDS-PAGE. No other motifs are present that might give clues to the function of RP. The wild-type mouse RP protein shows 87% identity with the human orthologue (GenBank accession no. BK005393; Figure 3C) and 94% with the rat (GenBank accession no. XM_218422; not shown). In rp/rp the premature stop codon occurs at nucleotide 308, truncating the protein at 79 amino acids (Figure 3C). Searches of BLASTn, BLASTp, and Swiss-Prot indicate the gene is confined to higher eukaryotes; no orthologues are seen in yeast, flies, or worms.
rp encodes a phosphorylated, conserved, widely expressed cytoplasmic protein. (A) Phosphorylation of wild-type RP. Whole cell extracts prepared from MNT-1 cells transiently expressing HA3-RP or FLAG-RP were treated for 1 hour at 37°C in the presence (+)orabsence(–) of alkaline phosphatase. Samples were then subjected to immunoblotting analysis as indicated. The positions of molecular mass markers are indicated on the right. (B) The phosphorylation of RP was confirmed by metabolic labeling of HA3-RP–expressing MNT-1 cells with 32P-orthophosphate in the presence (+) or absence (–) of alkaline phosphatase, followed by immunoprecipitation-recapture with antibodies to the myc (nonspecific control) or HA epitopes. The positions of molecular mass markers are indicated on the right. (C) Comparison of human and mouse RP amino acid sequence reveals an overall identity of 87% (shaded residues). The C>T transition converts amino acid 80 (underlined, glutamine) to a stop codon. (D) OriGene Technologies Northern blot showing expression of rp in mouse tissues. The blot was hybridized with the same 672-bp fragment described for Figure 2C. Molecular weight marker positions are indicated on the left. (E) rp message levels appear normal in kidney (left) and cultured melanocyte (right) mRNA. Note that a lower amount of rp/rp melanocyte mRNA was loaded, as judged by the intensity of the actin signal. Each lane contains 2 μg mRNA. (F-G) Immunofluorescence image of B6 (F) and rp/rp (G) melanocytes transfected with pHM6-RP and stained with anti-HA. No staining was evident in melanocytes transfected with vector alone (not shown). (H) Merged Nomarski and stained (affinity-purified chicken anti-RP peptide antibody recognizing amino acids 1-12 of the RP protein) image showing that RP-containing vesicles (green structures) and melanosomes (dark structures) do not colocalize extensively. (I-J) B6 (I) and rp/rp (J) melanocytes stained with affinity-purified chicken anti-RP peptide antibody. No staining was seen when the primary antibody (affinity-purified chicken anti-RP) was eliminated (not shown). Original magnification × 400 for panels F-J.
rp encodes a phosphorylated, conserved, widely expressed cytoplasmic protein. (A) Phosphorylation of wild-type RP. Whole cell extracts prepared from MNT-1 cells transiently expressing HA3-RP or FLAG-RP were treated for 1 hour at 37°C in the presence (+)orabsence(–) of alkaline phosphatase. Samples were then subjected to immunoblotting analysis as indicated. The positions of molecular mass markers are indicated on the right. (B) The phosphorylation of RP was confirmed by metabolic labeling of HA3-RP–expressing MNT-1 cells with 32P-orthophosphate in the presence (+) or absence (–) of alkaline phosphatase, followed by immunoprecipitation-recapture with antibodies to the myc (nonspecific control) or HA epitopes. The positions of molecular mass markers are indicated on the right. (C) Comparison of human and mouse RP amino acid sequence reveals an overall identity of 87% (shaded residues). The C>T transition converts amino acid 80 (underlined, glutamine) to a stop codon. (D) OriGene Technologies Northern blot showing expression of rp in mouse tissues. The blot was hybridized with the same 672-bp fragment described for Figure 2C. Molecular weight marker positions are indicated on the left. (E) rp message levels appear normal in kidney (left) and cultured melanocyte (right) mRNA. Note that a lower amount of rp/rp melanocyte mRNA was loaded, as judged by the intensity of the actin signal. Each lane contains 2 μg mRNA. (F-G) Immunofluorescence image of B6 (F) and rp/rp (G) melanocytes transfected with pHM6-RP and stained with anti-HA. No staining was evident in melanocytes transfected with vector alone (not shown). (H) Merged Nomarski and stained (affinity-purified chicken anti-RP peptide antibody recognizing amino acids 1-12 of the RP protein) image showing that RP-containing vesicles (green structures) and melanosomes (dark structures) do not colocalize extensively. (I-J) B6 (I) and rp/rp (J) melanocytes stained with affinity-purified chicken anti-RP peptide antibody. No staining was seen when the primary antibody (affinity-purified chicken anti-RP) was eliminated (not shown). Original magnification × 400 for panels F-J.
The rp gene is expressed ubiquitously (Figure 3D). In homozygous rp/rp tissues, there is no apparent loss of message, suggesting the mRNA is not subject to nonsense-mediated decay (Figure 3E). By immunofluorescence, the RP protein shows a punctate, cytoplasmic distribution in B6 melanocytes transfected with the normal RP cDNA in expression vector pHM6 (Figure 3F). The positive-staining structures extend well into the dendrites of the cells. A similar distribution is seen in transfected rp/rp melanocytes (Figure 3G). No staining was evident when cells were transfected with pHM6 vector alone (not shown). The RP-positive vesicles do not appear to colocalize with mature melanosomes (Figure 3H). Staining nontransfected cells using an affinity-purified peptide antibody raised in chickens that recognizes amino acids 1 to 12 of RP also reveals a similar distribution pattern in B6 and rp/rp melanocytes (Figure 3I-J). No such signal is obtained in the absence of primary antibody. We presume that a truncated protein is being detected in rp/rp cells. However, definitive proof of this is not currently available because attempts to generate peptide antibodies to the C-terminus of RP have been unsuccessful. Further studies will be required to identify the precise structures containing RP. From the current analyses, we can only conclude that these structures appear normally distributed in rp/rp cells.
RP assembles with components of BLOC-1 but not AP-3 or BLOC-3
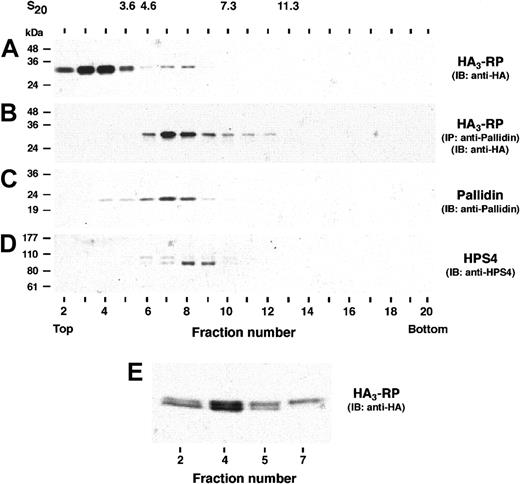
Loss of one BLOC-1 component has been shown to destabilize other subunits of the complex.21,28,51 Pallidin, a component of BLOC-1, is decreased in rp/rp fibroblasts compared to normal C57BL/6 (B6) fibroblasts (Figure 4A), suggesting that RP may be a BLOC-1 component. To examine this possibility directly, we stably transfected MNT-1 cells with HA3-RP and performed immunoprecipitation-recapture assays. Proteins were first immunoprecipitated with anti-HA, denatured, and then subjected to a second immunoprecipitation (“recapture”)27 using antisera to the BLOC-1 components, pallidin, cappuccino, muted, and to the HA epitope. In each case, coimmunoprecipitation of HA3-RP with the BLOC-1 components was evident (Figure 4B). We also demonstrated coprecipitation by using antipallidin and anti-HA in the first and second rounds of immunoprecipitation, respectively (Figure 5). Furthermore, HA3-RP failed to coimmunoprecipitate with components of the AP-3 complex (σ3, μ3) and BLOC-3 (HPS1, HPS4; Figure 5). The interaction of HA3-RP and pallidin was further examined by sedimentation velocity analysis on sucrose gradients using cytosol from MNT-1 cells stably expressing HA3-RP. Gradient fractions were assayed by immunoblotting with anti-HA (Figure 6). The distribution of HA3-RP on the gradient exhibited 2 peaks in fractions number 4 and 7-8, respectively (Figure 6A). Immunoprecipitation with antibodies against pallidin followed by immunoblotting with antibodies to HA (to reveal assembled HA3-RP) revealed a single peak (fraction 7) corresponding to a sedimentation coefficient of 5.2 S (Figure 6B). This peak coincided with the pallidin peak (Figure 6C). Therefore, the HA3-RP-pallidin complex sediments as a 5.2 S species, whereas the peak of HA3-RP in fraction 4 likely represents excess, unassembled HA3-RP. The sedimentation coefficient of the HA3-RP-pallidin complex was distinct from that of BLOC-3 as shown by immunoblotting with anti-HPS4 antibody (6.3 S, Figure 6D and Gautam et al29 ). Significantly, the electrophoretic mobility of the HA3-RP protein that cosediments with pallidin in fraction 7 (Figure 6E) is shifted upward. Similar results were obtained using Myc3-tagged RP (not shown). Together with the altered mobility on alkaline phosphatase treatment (Figure 3A) and 32P-incorporation studies (Figure 3B), these data indicate that the phosphorylated form of RP is a component of BLOC-1.
RP interacts with pallidin, cappuccino, and muted. (A) The expression of pallidin was assessed in whole-cell extracts prepared from wild-type (C57BL/6J), reduced pigmentation (RP), and pallid (PA) mouse fibroblasts by immunoblotting using an antipallidin antibody. Immunoblotting with anti–α-tubulin provided a control for sample loading. The positions of molecular mass markers are indicated on the right. (B) RP coimmunoprecipitates with the BLOC-1 components pallidin, cappuccino, and muted. MNT-1 cells stably expressing HA3-RP were metabolically labeled with [35S]methionine for 22 hours and extracted with lysis buffer containing 0.5% (wt/vol) Triton X-100. The extracts were then subjected to a first immunoprecipitation (1st IP) with mouse monoclonal anti-HA (lanes 5-8) or anti-Myc (lanes 1-4, controls). Washed immunoprecipitates were subsequently denatured by heating at 95°C for 5 minutes in the presence of SDS and dithiothreitol, diluted with lysis buffer, and subjected to a recapture immunoprecipitation step (2nd IP) using either rabbit antisera to pallidin (PA, lanes 1 and 5), cappuccino (CNO, lanes 2 and 6), or muted (MU, lanes 3 and 7) or mouse anti-HA antibody (lanes 4 and 8). The resulting immunoprecipitates were analyzed by SDS-PAGE on 4% to 20% gradient gels followed by fluorography. In all cases, the expected product was obtained indicating an association (*). The positions of molecular mass markers are indicated on the right. The slower migrating bands (arrows) likely represent nonspecific products because they are present in all immunoprecipitates including the controls.
RP interacts with pallidin, cappuccino, and muted. (A) The expression of pallidin was assessed in whole-cell extracts prepared from wild-type (C57BL/6J), reduced pigmentation (RP), and pallid (PA) mouse fibroblasts by immunoblotting using an antipallidin antibody. Immunoblotting with anti–α-tubulin provided a control for sample loading. The positions of molecular mass markers are indicated on the right. (B) RP coimmunoprecipitates with the BLOC-1 components pallidin, cappuccino, and muted. MNT-1 cells stably expressing HA3-RP were metabolically labeled with [35S]methionine for 22 hours and extracted with lysis buffer containing 0.5% (wt/vol) Triton X-100. The extracts were then subjected to a first immunoprecipitation (1st IP) with mouse monoclonal anti-HA (lanes 5-8) or anti-Myc (lanes 1-4, controls). Washed immunoprecipitates were subsequently denatured by heating at 95°C for 5 minutes in the presence of SDS and dithiothreitol, diluted with lysis buffer, and subjected to a recapture immunoprecipitation step (2nd IP) using either rabbit antisera to pallidin (PA, lanes 1 and 5), cappuccino (CNO, lanes 2 and 6), or muted (MU, lanes 3 and 7) or mouse anti-HA antibody (lanes 4 and 8). The resulting immunoprecipitates were analyzed by SDS-PAGE on 4% to 20% gradient gels followed by fluorography. In all cases, the expected product was obtained indicating an association (*). The positions of molecular mass markers are indicated on the right. The slower migrating bands (arrows) likely represent nonspecific products because they are present in all immunoprecipitates including the controls.
Coimmunoprecipitation of HA3-RP and BLOC-1. Whole cell extracts of metabolically labeled MNT-1 cells stably expressing HA3-RP were subjected to immunoprecipitation-recapture analysis, as described in the legend of Figure 4. Rabbit antisera to either irrelevant proteins (C, controls), the σ subunit of AP-3 (σ3), pallidin (PA), the Hermansky-Pudlak syndrome 1 (HPS1) protein, the Hermansky-Pudlak syndrome 4 (HPS4) protein, and mouse monoclonal anti-myc (control) or anti-HA were used in the 1st IP. Mouse monoclonal anti-HA was used in the 2nd IP step. To confirm the presence of AP-3, BLOC-1, and BLOC-3 complexes after the first immunoprecipitation (1st IP), the supernatant obtained after the recapture step (2nd IP) were subjected to 2 additional IP steps (3rd IP and 4th IP) using antibodies to σ3 and μ3, pallidin and cappuccino, or HPS1 and HPS4 or HPS4 and HPS1 in the consecutive IP steps (3rd IP and 4th IP), respectively. The positions of molecular mass markers are indicated on the right.
Coimmunoprecipitation of HA3-RP and BLOC-1. Whole cell extracts of metabolically labeled MNT-1 cells stably expressing HA3-RP were subjected to immunoprecipitation-recapture analysis, as described in the legend of Figure 4. Rabbit antisera to either irrelevant proteins (C, controls), the σ subunit of AP-3 (σ3), pallidin (PA), the Hermansky-Pudlak syndrome 1 (HPS1) protein, the Hermansky-Pudlak syndrome 4 (HPS4) protein, and mouse monoclonal anti-myc (control) or anti-HA were used in the 1st IP. Mouse monoclonal anti-HA was used in the 2nd IP step. To confirm the presence of AP-3, BLOC-1, and BLOC-3 complexes after the first immunoprecipitation (1st IP), the supernatant obtained after the recapture step (2nd IP) were subjected to 2 additional IP steps (3rd IP and 4th IP) using antibodies to σ3 and μ3, pallidin and cappuccino, or HPS1 and HPS4 or HPS4 and HPS1 in the consecutive IP steps (3rd IP and 4th IP), respectively. The positions of molecular mass markers are indicated on the right.
Sedimentation velocity analysis of HA3-RP. Cytosol prepared from MNT-1 cells stably expressing HA3-RP was fractionated by ultracentrifugation on a 2% to 15% (wt/vol) linear sucrose gradient. (A) Samples representing 3.3% of the fraction volume were analyzed for the presence of HA3-RP by immunoblotting (IB) using monoclonal anti-HA antibody. (B) The presence of the HA3-RP - pallidin complex was assessed on samples (∼ 40% of the volume of each fraction) by immunoprecipitation using antibody to pallidin and immunoblotting using anti-HA antibody. Note that RP and pallidin cosediment in fractions 6 to 8 and that the mobility of RP is shifted in these fractions. (C) The sedimentation of endogenously expressed pallidin was analyzed by immunoblotting using antibodies to pallidin. Note that HA3-RP and pallidin cosediment in fractions 6 to 8 with a peak in fraction 7. (D) The sedimentation of endogenously expressed HPS4 was assessed by immunoblotting; HPS4 peaked in fraction 8. (E) Samples from fractions 2, 4, 5, and 7 representing 2.5%, 0.8%, 2.5%, and 5.8% of the fraction volume, respectively, were analyzed by IB using anti-HA antibody. Notice that the upper band of HA3-RP (phosphorylated form) is predominant in fraction 7. The positions of standard proteins (sedimentation coefficients given in Svedberg units) in the gradient are indicated on the top. The positions of molecular mass markers are indicated on the left.
Sedimentation velocity analysis of HA3-RP. Cytosol prepared from MNT-1 cells stably expressing HA3-RP was fractionated by ultracentrifugation on a 2% to 15% (wt/vol) linear sucrose gradient. (A) Samples representing 3.3% of the fraction volume were analyzed for the presence of HA3-RP by immunoblotting (IB) using monoclonal anti-HA antibody. (B) The presence of the HA3-RP - pallidin complex was assessed on samples (∼ 40% of the volume of each fraction) by immunoprecipitation using antibody to pallidin and immunoblotting using anti-HA antibody. Note that RP and pallidin cosediment in fractions 6 to 8 and that the mobility of RP is shifted in these fractions. (C) The sedimentation of endogenously expressed pallidin was analyzed by immunoblotting using antibodies to pallidin. Note that HA3-RP and pallidin cosediment in fractions 6 to 8 with a peak in fraction 7. (D) The sedimentation of endogenously expressed HPS4 was assessed by immunoblotting; HPS4 peaked in fraction 8. (E) Samples from fractions 2, 4, 5, and 7 representing 2.5%, 0.8%, 2.5%, and 5.8% of the fraction volume, respectively, were analyzed by IB using anti-HA antibody. Notice that the upper band of HA3-RP (phosphorylated form) is predominant in fraction 7. The positions of standard proteins (sedimentation coefficients given in Svedberg units) in the gradient are indicated on the top. The positions of molecular mass markers are indicated on the left.
Ruby-eye interacts semidominantly with rp
Genetic complementation studies in mice frequently provide valuable insights into the functional interdependence of proteins or protein complexes. We produced mice doubly homozygous for rp and cno and rp and pa by conventional interbreeding for 2 generations. No exacerbation of the HPS phenotype was seen in either case (not shown). These results are consistent with the biochemical data and provide further proof that RP is a component of BLOC-1. To test for an interaction of BLOC-1 and BLOC 2, we produced mice carrying various combinations of HPS5/ru and rp. Doubly homozygous offspring (rp/rp, ru/ru) showed an exacerbated coat-color phenotype compared to either single homozygote alone (Figure 7). Notably, one copy of ru is sufficient to enhance the pigmentation defect in rp/rp mice; rp/rp, +/ru showed a phenotype intermediate to that of rp/rp, +/+ and rp/rp, ru/ru mice. These data indicate that BLOC-1 and BLOC-2 define functionally independent pathways in the biogenesis of LROs.
ru acts semidominantly to exacerbate the rp phenotype. Genotypes are indicated. Note the more severe coat color dilution in the double homozygotes (ru/ru, rp/rp) compared to single homozygotes and that one copy of ru (+/ru, rp/rp) suffices to exacerbate the rp coat color defect.
ru acts semidominantly to exacerbate the rp phenotype. Genotypes are indicated. Note the more severe coat color dilution in the double homozygotes (ru/ru, rp/rp) compared to single homozygotes and that one copy of ru (+/ru, rp/rp) suffices to exacerbate the rp coat color defect.
Human mutation screening
We screened for RP mutations 15 patients with HPS for which the genetic basis of their disease is unknown. No defects were detected.
Discussion
Increased bleeding times, coat color dilution, and elevated kidney lysosomal enzyme concentrations in rp/rp mice have been previously described.33,46 Here, we extend the observations to demonstrate a marked decrease in the number of platelet dense bodies, the hallmark of HPS, with concomitant decrease in platelet serotonin levels. In addition, a detailed analysis of cultured rp/rp melanocytes revealed abnormalities characteristic of HPS, specifically decreased numbers of melanosomes with decreased pigment content and increased numbers of small and immature melanosomes. In addition, tyrosinase trafficking is abnormal as evidenced by its accumulation in the perinuclear region and the presence of abnormal DOPA-positive tubular structures. Of note, rp/rp mice were originally reported to have normal melanosomes in the eye. However, a detailed electron microscopic analysis indicates this is not the case (B.G. and L.L.P., manuscript in preparation).
By fine-linkage mapping, we localized rp to a 300-kb interval on mouse Chr 7. Direct sequencing identified rp as a novel gene encoding a highly acidic (pI 4.84) protein that is strongly conserved with the human protein (87% identity). There are no predicted transmembrane domains, suggesting that the protein is cytosolic. Wild-type RP protein localizes to punctate structures within the cytoplasm, reminiscent of other HPS proteins. In homozygous mutants, a premature stop codon is present at amino acid 80. However, mRNA levels are not affected, and the protein distribution appears normal at the light microscopy level, suggesting that the truncated protein is stable.
Loss of one subunit of a multiprotein complex frequently results in deficiency of other subunits. For example, loss of β3A destabilizes the AP-3 complex in HPS227 /pearl, 52 and loss of the CNO protein in cappuccino results in deficiencies of the BLOC-1 components pallidin and muted.21 In rp/rp fibroblasts, we observed that pallidin was decreased compared to wild-type fibroblasts, suggesting that RP might be a component of BLOC-1. Coprecipitation and cosedimentation results indicate that RP associates with the known BLOC-1 components, pallidin, cappuccino, and muted. Notably, the migration of the RP protein suggests that it is the phosphorylated form that specifically assembles within BLOC-1 (Figures 3A and 6E).
Wild-type RP has a predicted molecular weight of 20.4 kDa, but migrates at a higher molecular weight (Figures 4, 5, 6), as do cappuccino, muted, and pallidin.21,51 This is likely related to the highly acidic nature of the proteins. In addition, the predicted secondary structure of RP is similar to that of pallidin, muted, and cappuccino.21,51 An unstructured amino terminal domain is followed by a domain of high α-helical content (44% in RP). Dysbindin (sandy) also shows a high α-helical content. RP differs from these other known BLOC-1 components, however, in that RP has no predicted coiled-coil domains, whereas cappuccino, pallidin, muted, and sandy all have 2. Unlike AP-3 subunits, the 5 known BLOC-1 proteins are found only in metazoans, indicating that BLOC-1 evolved to serve functions specific to melanosomes and platelet granules, which are confined to higher eukaryotes.
Long before the HPS1/ep and HPS4/le genes were identified at the molecular level, the genes were surmised to function within the same protein complex or pathway based on the observation that double homozygotes showed a phenotype identical to that of the single homozygotes, that is, no phenotypic exacerbation occurred.53 Their mutual participation in BLOC-3 was recently demonstrated biochemically.30,31 On the other hand, mice doubly homozygous for HPS1/ep and Ap3b1/pe show an exacerbated phenotype, as 2 functionally distinct pathways are disrupted (BLOC-3, adaptor protein 3 complex).54 Our immunoprecipitation assays revealed that rp is a BLOC-1 component and does not interact with components of the AP-3 complex or BLOC-3. To test for a possible interaction between BLOC-1 and BLOC-2, we generated rp and ru double homozygotes. An exacerbated phenotype is observed in ru/ru, rp/rp mice as well as in +/ru, rp/rp mice, indicating that ru and rp (BLOC-1 and BLOC-2) participate in functionally independent processes during the biogenesis of LROs. Interestingly, mice doubly homozygous for pallid and pale ear, components of BLOC-1 and BLOC-3, respectively, do not show an exacerbated phenotype, suggesting that BLOC-1 and BLOC-3 are functionally interdependent.31
The mouse models of HPS have been invaluable in identifying gene defects in human HPS and in dissecting the protein building blocks participating in the fundamental processes of organelle biogenesis. Their participation in functionally distinct subcellular protein complexes is now well established. However, the precise interactions of most of the HPS proteins within these complexes and the interactions and functions of the complexes themselves remain to be established.
Prepublished online as Blood First Edition Paper, July 20, 2004; DOI 10.1182/blood-2004-04-1538.
Supported by National Institutes of Health grant HL55321 (L.L.P.) and RR01183 (The Jackson Laboratory Mouse Mutant Resource), National Cancer Institute CA34196 (The Jackson Laboratory), and Program Grant 064583 from the Wellcome Trust (E.V.S.).
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Note added in proof. While this manuscript was being submitted, Starcevic and Dell'Angelica identified the same mutation in rp/rp mice using a biochemical approach.55
We thank Jane Barker and Linda Washburn for critical review of the manuscript.



![Figure 4. RP interacts with pallidin, cappuccino, and muted. (A) The expression of pallidin was assessed in whole-cell extracts prepared from wild-type (C57BL/6J), reduced pigmentation (RP), and pallid (PA) mouse fibroblasts by immunoblotting using an antipallidin antibody. Immunoblotting with anti–α-tubulin provided a control for sample loading. The positions of molecular mass markers are indicated on the right. (B) RP coimmunoprecipitates with the BLOC-1 components pallidin, cappuccino, and muted. MNT-1 cells stably expressing HA3-RP were metabolically labeled with [35S]methionine for 22 hours and extracted with lysis buffer containing 0.5% (wt/vol) Triton X-100. The extracts were then subjected to a first immunoprecipitation (1st IP) with mouse monoclonal anti-HA (lanes 5-8) or anti-Myc (lanes 1-4, controls). Washed immunoprecipitates were subsequently denatured by heating at 95°C for 5 minutes in the presence of SDS and dithiothreitol, diluted with lysis buffer, and subjected to a recapture immunoprecipitation step (2nd IP) using either rabbit antisera to pallidin (PA, lanes 1 and 5), cappuccino (CNO, lanes 2 and 6), or muted (MU, lanes 3 and 7) or mouse anti-HA antibody (lanes 4 and 8). The resulting immunoprecipitates were analyzed by SDS-PAGE on 4% to 20% gradient gels followed by fluorography. In all cases, the expected product was obtained indicating an association (*). The positions of molecular mass markers are indicated on the right. The slower migrating bands (arrows) likely represent nonspecific products because they are present in all immunoprecipitates including the controls.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/10/10.1182_blood-2004-04-1538/6/m_zh80220469540004.jpeg?Expires=1766051675&Signature=RqpAf0AGdpRNr7yHRoSTSYN3Woq~23NPnaII8LRdDiVz5pJt8o50U6M6kOAECAFpKAGd0L~IkAX4YGu1Xv4D61iVybaHylUstggkuxLV5qOEiCJdSUW-VhAjlufwkQRIjHzXQilD~LCNLlQNq9a1hnHJzzqDUhTOEV2RRlaf6Oe42idFTh4LI2naMGgPtwfcYjwCKUKGxJMmwweigkr-yY7l6KZSoLvOzgxwdWA4lksfnXRcHgstvjlq9Rsi~c7r-WNCUqnlIEWAc7qSko8eQ0bGcgRkMYM57KG1hzXWVb8c3fX0be8c40GLZTWPvRfucUNS0OYwAuSmQzt6puD-zg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal