Abstract
Cis-element identification is a prerequisite to understand transcriptional regulation of gene loci. From analysis of a limited number of conserved gene loci, sequence comparison has proved a robust and efficient way to locate cis-elements. Human and mouse GATA1 genes encode a critical hematopoietic transcription factor conserved in expression and function. Proper control of GATA1 transcription is critical in regulating myeloid lineage specification and maturation. Here, we compared sequence and systematically mapped position of DNase I hypersensitive sites, acetylation status of histone H3/H4, and in vivo binding of transcription factors over approximately 120 kilobases flanking the human GATA1 gene and the corresponding region in mice. Despite lying in approximately 10 megabase (Mb) conserved syntenic segment, the chromatin structures of the 2 homologous loci are strikingly different. The 2 previously unidentified hematopoietic cis-elements, one in each species, are not conserved in position and sequence and have enhancer activity in erythroid cells. In vivo, they both bind the transcription factors GATA1, SCL, LMO2, and Ldb1. More broadly, there are both species- and regulatory element–specific patterns of transcription factor binding. These findings suggest that some cis-elements regulating human and mouse GATA1 genes differ. More generally, mouse human sequence comparison may fail to identify all cis-elements.
Introduction
GATA1 is a key hematopoietic transcription factor and a member of a conserved family of GATA factors that execute a program of differentiation in diverse cell types.1 The precise pattern of GATA1 expression is critical for its function. Gain-of-function experiments demonstrate that GATA1 directs cell fate of myeloid progenitors in a manner dependent on the level of GATA1 expressed and that GATA1 expression has to be extinguished to allow neutrophil and macrophage differentiation.2-4 Loss-of-function experiments show that GATA1 is required for terminal maturation of erythroid cells, megakaryocytes, eosinophils, and mast cells.5-8 Moreover, reduction of GATA1 levels to 20% of normal is insufficient for erythroid maturation.9 Lastly, rescue experiments in GATA1-mutant hematopoietic cells10 and mice11 and analysis of knock-in mice12 show that GATA1 function is determined not just by specific GATA1 protein sequences (other GATA factors can, to a large extent, replace GATA1), but is also critically dependent on the precise pattern of GATA1 expression.
Consistent with this function, in both human and mouse definitive hematopoiesis, GATA1 expression is detected at a low level in the common myeloid progenitor (CMP)13,14 and is selectively maintained in erythroid cells, megakaryocytes, eosinophils, and mast cells (reviewed in Orkin15 ) and repressed in neutrophils and monocytes.4 Outside blood cells, GATA1 is expressed in testis where its role is unclear. Taken together, these observations argue that the time, place (cell-type), and level of GATA1 expression help regulate output of myeloid lineages from CMPs and allow their normal terminal maturation. Therefore, defining the molecular basis of GATA1 expression will be an important step to understanding the molecular control of lineage output from a CMP.
Identification of cis-acting sequences (cis-elements) regulating GATA1 is a necessary prerequisite to understanding how GATA1 expression is controlled. In murine blood cells, 3 GATA1 cis-elements regulate expression. They are: the hematopoietic exon 1 erythroid (IE) promoter16 ; an upstream enhancer, hypersensitive site (HS) I/G1HE, required to direct red cell and megakaryocyte reporter gene expression in transgenic mice;17,18 and an element in the first GATA1 intron, required with HS I to direct expression in definitive red cells.18
However, analysis of transgenic and germ-line mutant mice suggests that the full complement of GATA1 cis-elements has not been identified. Deletion of an approximately 8-kb region including HS I almost completely abolishes GATA1 expression only in megakaryocytes but does not affect expression in erythroid cells and eosinophils.6,19 Therefore, though HS I directs reporter-gene expression to both erythroid cells and megakaryocytes, it is nonredundant only for megakaryocyte-specific expression. Presumably, other unidentified sequences direct erythroid and eosinophil GATA1 expression in its absence.
Therefore, to define the molecular basis of GATA1 expression, we set out to identify all GATA1 cis-elements, and in particular those important for red cell GATA1 expression. We were also interested to determine whether the mouse was a faithful model to study transcriptional regulation of human GATA1 as this may have clinical implications. For example, if GATA1 expression was controlled by different cis-elements and trans-acting factors in the 2 species, changes in GATA1 expression in response to stress or growth factor therapy or in leukemia may differ between species. With the availability of the mouse and human genome sequence, and with the genome sequence of other species to follow, many have advocated that for genes with conserved function and expression, especially those encoding transcription factors, sequence comparison may be a rapid and comprehensive way of locating cis-elements.20-24 Therefore, we have compared sequence of the human and mouse GATA1 loci, together with analysis of chromatin structure, to pinpoint previously unidentified cis-elements in the GATA1 loci.
Materials and methods
Sequence analysis
Accession numbers of the human and mouse GATA1 loci sequence are NT 011568 and NT 039698, respectively (http://www.ncbi.nlm.nih.gov/). To search for conserved sequence, alignments were performed using 3 software programs: VISTA25,26 (both M-VISTA and R-VISTA; http://www-gsd.lbl.gov/vista), Pipmaker27 (http://bio.cse.psu.edu/pipmaker/), and MacVector (Accelrys, Cambridge, United Kingdom) with moving windows ranging from 30- to 100-bp in size. Whole genome alignment (including chaining and netting28 and phyloHMMcons) were performed using tools available on the University of California, Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/).
Primary cells and cell lines
Primary human erythroid cells (Io erythroid; > 95% purity) were obtained from culture of peripheral blood mononuclear cells.29 Primary human eosinophils (Io eos; > 90% purity) were obtained from peripheral blood of atopic individuals with eosinophil counts 2- to 3-fold above normal and peripheral blood and mouse eosinophils from spleen of interleukin-5 transgenic mice.30 References to cell lines can be found in Vyas et al31 and are available on request.
DNase I hypersensitive site analysis
Nuclei preparation, DNase I digestion, DNA extraction, and Southern blot analysis were performed as previously described.31 Details of single-copy probes used for Southern blot analysis are available on request.
Chromatin immunoprecipitation and analysis of material
Cells (1-3 × 107) were used to cross-link chromatin using the protocol from Upstate Biotechnology (Milton Keynes, United Kingdom) with minor modifications. Cells were incubated in 0.4% formaldehyde (1% formaldehyde was used for hGATA1 IP) for 10 minutes at room temperature. Glycine (0.125 M) was added to stop cross-linking. Cross-linked chromatin was sonicated 3 times for 200 seconds at 25% amplitude (Vibra-Cell sonicator; Sonics and Materials, Meryin/Satigny, Switzerland). Sonication produced DNA fragments of approximately 300 bp, with no fragments bigger than 800 bp. Polyclonal antibodies against all acetylated forms of H4 [αH4-Ac] (catalog number, 06-866) and against H3 acetylated at lysine 9 and 14 [αH3-Ac] (catalog number, 06-599) (Upstate Biotechnology) were used. Other antibodies used were as follows: GATA1 (N6 [no. sc-265] and C20 [no. sc-1233]), E2A, (no. sc-762) (Santa Cruz, Autogen Bioclear, Calne, United Kingdom), polyclonal anti–mouse SCL,32 polyclonal anti–mouse LMO2, and Ldb-1.33 Immunoprecipitation conditions were as suggested by the manufacturer's protocol (Upstate Biotechnology), except for N6 antibody,34 and C20 antibody where protein A/G agarose was used. For each cell type and antibody used, at least 3 independent chromatin preparations were used for immunoprecipitation. Real-time polymerase chain reaction (PCR) primers and 5′ 6-carboxy fluorescein–3′ 6-carboxytetramethyl rhodamine probes were designed with Primer Express 2.0 software (Applied Biosystems, Warrington, United Kingdom). Duplicate real-time PCR reactions on each immunoprecipitated template were performed on a Sequence Detection System 7000 thermocycler (Applied Biosystems) using Taqman universal PCR mastermix (Applied Biosystems), 400 nM of each primer, and 200 nM of probe in 25-μL reactions. Cycling conditions were 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. All primers and probes were validated over a range of genomic DNA dilutions (1-64 ng of genomic DNA) to ensure that the intensity of PCR product was proportional to the starting amount of DNA template. To calculate fold enrichment, a previously published method was used.35 In histone chromatin immunoprecipitation (ChiP) assays, but not the ChiP assays, this was normalized to the value obtained with glyceraldehyde-3-phosphate dehydrogenase (when mouse antibodies were used) or β-actin (when human antibodies were used) specific primers (Eurogentec, Romsey, United Kingdom). Sequences of primers and probes are available on request.
Stable transfection of MEL and L929 cells
L929 and MEL585 are adherent cell lines. MEL 585 cells (2 × 107) and L929 cells (1 × 107) were harvested in exponential growth phase, washed twice in 1x phosphate-buffered saline (PBS), resuspended in 0.8 mL of 1x PBS, and electroporated with 20 μg linearized DNA using a Bio-Rad Gene Pulse Electroporator (Bio-Rad, Hemel Hampstead, United Kingdom). Electroporation conditions for MEL and L929 cells were 960 μF and 220 V. Cells were plated in 5 × 10 cm plates in nonselective media for 2 days, and then 0.8 mg/mL G418 was added to MEL cells and 1.2 mg/mL G418, to L929 cells. Numbers of G418-resistant colonies were counted after a further 5 days (MEL cells) or 7 days (L929).
Results
Genomic structure of the human and mouse GATA1 loci
The mouse (m) (X 1.9 cM) and human (h) (Xp11.23) GATA1 genes lie in an approximately 10-Mb region of conserved synteny (National Center for Biotechnology Information [NCBI] Build 32 for mouse and NCBI Build 34.3 for human). Upstream of both GATA1 loci are the hematopoietic-expressed Wiskott-Aldrich syndrome (WAS) gene36 and the widely expressed histone lysine 9 methyl transferase SUV39H1 gene37 (Figure 1A-B). Downstream of GATA1, known genes include HDAC6, which is widely expressed,38 Eras (a RAS gene specifically expressed in mouse embryonic stem cells39 ), and PCSK1N (encoding an enzyme inhibitor predominantly expressed in brain and other neuroendocrine tissues40 ).
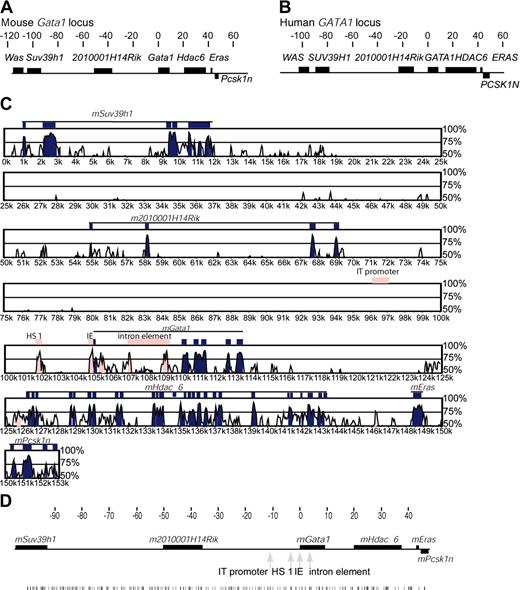
Organization of genes in the human and mouse GATA1 loci and sequence comparison between the mouse and human GATA1 loci. (A-B) Mouse (A) and human (B) GATA1 loci are illustrated. Above each locus, the scale is in kilobases, and coordinate 0 is at the mRNA cap site of GATA1 transcribed from the IE promoter. Genes are shown as dark boxes. Genes above the line are transcribed in the same direction as GATA1; PCSK1N is transcribed in the opposite direction and drawn below the locus. (C) Plot of sequence comparison of the mouse (reference sequence) and human GATA1 loci from SUV39H1 to PCSK1N. The y-axis is the percent sequence identity over a moving 100–base pair window. Different points along the Gata1 loci are represented on the x-axis. Different features in Gata1 loci are shown: genes (black lines), exons within genes (blue boxes), and regulatory elements (pink boxes). (D) Distribution of GATA sites (WGATAR) through the mGata1 locus. The coordinates are in kilobases with respect to the GATA1 IE promoter transcriptional start site. The genes are shown as black boxes, the positions of known mGATA1 cis-elements are marked by gray arrows, and the distribution of GATA sites is shown as black vertical lines below the rest of the panel.
Organization of genes in the human and mouse GATA1 loci and sequence comparison between the mouse and human GATA1 loci. (A-B) Mouse (A) and human (B) GATA1 loci are illustrated. Above each locus, the scale is in kilobases, and coordinate 0 is at the mRNA cap site of GATA1 transcribed from the IE promoter. Genes are shown as dark boxes. Genes above the line are transcribed in the same direction as GATA1; PCSK1N is transcribed in the opposite direction and drawn below the locus. (C) Plot of sequence comparison of the mouse (reference sequence) and human GATA1 loci from SUV39H1 to PCSK1N. The y-axis is the percent sequence identity over a moving 100–base pair window. Different points along the Gata1 loci are represented on the x-axis. Different features in Gata1 loci are shown: genes (black lines), exons within genes (blue boxes), and regulatory elements (pink boxes). (D) Distribution of GATA sites (WGATAR) through the mGata1 locus. The coordinates are in kilobases with respect to the GATA1 IE promoter transcriptional start site. The genes are shown as black boxes, the positions of known mGATA1 cis-elements are marked by gray arrows, and the distribution of GATA sites is shown as black vertical lines below the rest of the panel.
Immediately 5′ of GATA1 is a conserved gene of unknown function (LOC392465 in humans and 2010001H14Rik in mice, hereafter referred to as 2010001H14Rik). Its predicted mRNAs contain an open reading frame. There is 84% amino acid identity between the predicted proteins in the 2 species, which have motifs found in the glyoxalase/dioxygenase family. Given the proximity of 2010001H14 RIK to GATA1, we determined if it was coexpressed with GATA1, to understand if newly identified cis-elements could be important for regulating either GATA1 or 2010001H14 RIK or both genes. In both mice and humans, Northern blot analysis showed prominent 2010001H14 RIK expression only in megakaryocytic cell lines and not other GATA1-expressing cells or other nonhematopoietic primary cells or cell lines (data not shown). Furthermore, in mice, microarray analysis shows that this gene is expressed in the gastrointestinal tract (stomach and small and large intestine) (GNF1M mouse CHIP and U74b Mouse CHIP, http://genome.ucsc.edu).
Sequence comparison between the human and mouse GATA1 loci
As GATA1 function, pattern of expression, and sequence of known GATA1 cis-elements (HS 1, IE promoter, and the intron element) are conserved between humans and mice,41,42 we reasoned that additional functional cis-elements may also be conserved in position and sequence. Moreover, as currently complete sequence of only rodent and human GATA1 loci are available, we compared mouse and human sequences between SUV39H1 and PCSK1N genes, using software packages that aligned sequences between the GATA1 loci and also looked to see if sequences present in the GATA1 loci were present elsewhere in the genome (whole-genome alignments). For comparison of sequences between the GATA1 loci we used Vista,25,26 Pipmaker,27 and MacVector software packages with moving windows ranging between 30 to 100 bp. In Figure 1C, alignment analysis using the VISTA program is shown. The percentage sequence homology between species along the GATA1 loci is plotted with a window size of 100 bp. With decreasing window size, increasing numbers of regions of limited homology (< 50%) were seen that consisted of imperfectly matched simple repeats.
As expected, HS 1, IE promoter, the intron element, and exons of flanking genes were conserved in sequence. In addition, we noted 2 additional stretches of conserved sequence. The first, was within the first intron just after the IE exon and upstream of the previously defined intron element. This sequence is unlikely to contain a new GATA1 enhancer as constructs containing it do not direct reporter-gene expression to GATA1-expressing cells in transgenic mice.17,18 The second region lies just upstream of HDAC6 gene and will be discussed in “Identification of DNase I HSs in the hGATA1 locus”. Surprisingly, outside these regions, there was little sequence conservation. Strikingly, even sequence of the rodent IT promoter that transcribes GATA1 in testis43 is not conserved in humans. Similar results were obtained with Pipmaker and MacVector. Whole genome alignment tools (“Materials and methods”) showed that although there are additional regions with weak similarity between humans and mice, these are not subject to strong purifying selection and are certainly not as well-conserved as known GATA1 cis-elements (data not shown).
Cis-elements often contain multiple binding sites for transcription factors. In particular, GATA sites,42,44 especially closely spaced GATA sites, are important for GATA1 regulation.7,16 Therefore, 2 further approaches were taken to look for conserved cis-elements. First, we determined the position of all GATA sites (WGATAR), including clustered GATA sites (> 2 sites within 20 bp) in the mGata1 locus. There were 287 GATA sites present between Suv39h1 and Psck1N (Figure 1D), and 45 were clustered. However, sequences incorporating clustered GATA sites were not conserved in humans. Second, sequence of the mouse and human GATA1 loci was analyzed using the R-VISTA program26 to look for clustered binding sites for multiple transcription factors. This analysis revealed several regions with clustered binding sites, but again no previously unidentified conserved candidate cis-element sequences were uncovered (data not shown).
Identification of DNase I HSs in the mGata1 locus
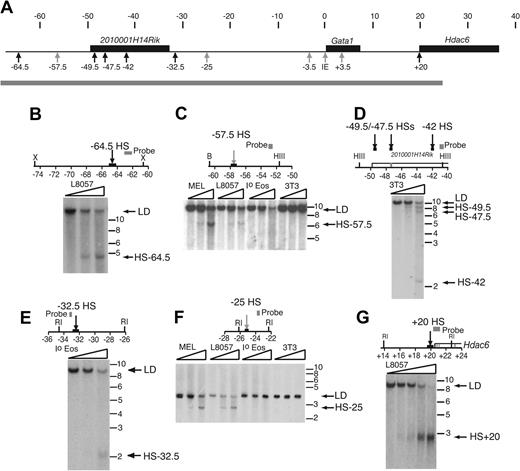
As sequence comparison did not locate previously unidentified cis-elements, we mapped DNase I hypersensitive sites (HSs) flanking the mouse and human GATA1 loci. DNase HSs almost always mark the position of important cis-elements.45 We searched for constitutive and hematopoietic DNase I HSs throughout chromatin from coordinates –68.2 to +24.4 in the mGata1 locus (Figure 2A). For the first time, hypersensitive sites mHS 1 (coordinate –3.5 and now referred to as mHS–3.5) and those associated with the IE promoter and the intron cis-element (coordinate +3.5 and now referred to as mHS+3.5) were detected in primary eosinophils. A summary of the cell type specificity of all sites is in Supplementary Table S1 (available on the Blood website; see the Supplemental Materials like at the top of the online article). Additionally, 2 hematopoietic sites (mHS–57.5 and mHS–25; Figure 2C and F, respectively) and 6 widely expressed sites (mHS–64.5, mHS–49.5, mHS–47.5, mHS–42, mHS–32.5, and mHS+20; Figure 2B, D, E, and G respectively), not previously reported, were detected. Hematopoietic sites mHS–57.5 and mHS–25 were present only in MEL and L8057 cells but not primary eosinophils. The constitutive site at mHS+20 colocalizes with a cytosine-phosphate-guanosine (CpG) island (data not shown) at the 5′ end of the widely expressed Hdac6 gene and may mark the promoter of this gene.
Examples of previously unidentified DNase HSs in the mGata1 locus. (A) The mGata1 locus is depicted as in Figure 1A. The positions of constitutive (black arrows) and hematopoietic (gray arrows) DNase I HSs are marked. Below, a gray bar indicates the region of chromatin, which was systematically surveyed throughout for DNase I HSs. (B-G) Maps of the relevant portions of the genomic mGata1 locus, together with examples of DNase I HSs, are shown. Gray boxes illustrate position of the probes used in the Southern blot analysis. A box and an arrow mark the position of the DNase I HS. Genes are depicted as open boxes; exons, as gray blocks. Restriction enzymes used include RI, EcoRI; X, XbaI; HIII, HindIII; and B, BamHI. Coordinates in kilobases are shown below each map with respect to GATA1 IE promoter transcriptional start site. Below, autoradiographs demonstrate DNase I HS. Nuclei from different primary cells and cell lines, as indicated in each panel, were digested with increasing concentrations of DNase I, as illustrated by triangles. Abbreviations for cells used are as follows: MEL, erythroleukemia cell line; L8057, megakaryocytic cell line; Io eos, primary eosinophils; and 3T3, fibroblast cell line. No exogenous DNase I was added to the first lane. Extracted DNA was then digested with different restriction enzymes as indicated in each panel. On the right of the autoradiograph are the positions of the limit digest (LD), DNase I hypersensitive sites, and DNA molecular weight markers in kilobases.
Examples of previously unidentified DNase HSs in the mGata1 locus. (A) The mGata1 locus is depicted as in Figure 1A. The positions of constitutive (black arrows) and hematopoietic (gray arrows) DNase I HSs are marked. Below, a gray bar indicates the region of chromatin, which was systematically surveyed throughout for DNase I HSs. (B-G) Maps of the relevant portions of the genomic mGata1 locus, together with examples of DNase I HSs, are shown. Gray boxes illustrate position of the probes used in the Southern blot analysis. A box and an arrow mark the position of the DNase I HS. Genes are depicted as open boxes; exons, as gray blocks. Restriction enzymes used include RI, EcoRI; X, XbaI; HIII, HindIII; and B, BamHI. Coordinates in kilobases are shown below each map with respect to GATA1 IE promoter transcriptional start site. Below, autoradiographs demonstrate DNase I HS. Nuclei from different primary cells and cell lines, as indicated in each panel, were digested with increasing concentrations of DNase I, as illustrated by triangles. Abbreviations for cells used are as follows: MEL, erythroleukemia cell line; L8057, megakaryocytic cell line; Io eos, primary eosinophils; and 3T3, fibroblast cell line. No exogenous DNase I was added to the first lane. Extracted DNA was then digested with different restriction enzymes as indicated in each panel. On the right of the autoradiograph are the positions of the limit digest (LD), DNase I hypersensitive sites, and DNA molecular weight markers in kilobases.
Sequence associated with 3 DNase I HSs (mHS–57.5, mHS–49.5, and mHS–32.5) is primarily or partly composed of repetitive DNA. A similar finding of DNase I HSs associated with repetitive sequence has been recently reported.46 Furthermore, global genome alignment shows that these repetitive sequences are not ancestral repeats, and thus orthologous copies are not present elsewhere in the genome.
Identification of DNase I HSs in the hGATA1 locus
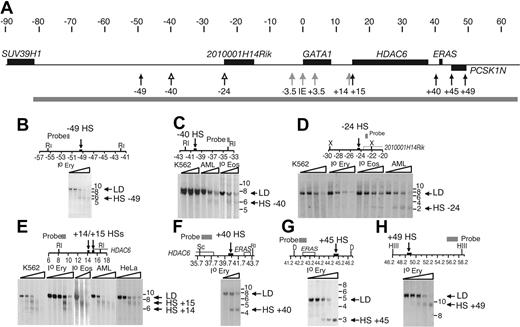
Chromatin throughout coordinates –81.5 and +65.3 in the hGATA1 locus (Figure 3A) was surveyed for DNase I HSs. Sites associated with hHS–3.5 (hHS 1), the IE promoter, and hHS+3.5 (intron element) were detected in human hematopoietic cells for the first time (Supplementary Table S1). Moreover, we detected 3 hematopoietic sites (hHS–40, hHS–24, and hHS+14; Figure 3C, D, and E, respectively) and 5 widely expressed sites (hHS–49, hHS+15, hHS+40, hHS+45, and hHS+49; Figure 3B, F-H) that had not been previously identified. There were 2 hematopoietic sites, hHS–40 and hHS–24, detected only in primary eosinophils and an eosinophil cell line (Figure 3C and D, respectively). hHS–24 is located close to the current predicted 5′end of 2010001H14 RIK. However, as expression of this gene was not detected in eosinophils (data not shown), it is unclear whether the site marks a regulatory element for this gene. The third hematopoietic site (hHS+14; Figure 3E) was detected only in primary erythroid cells and in hematopoietic K562 cells. Constitutive sites hHS–49 and hHS+15 were present in multiple cell lines and primary cells (Figure 3B and E, respectively). hHS+15 is located at the 5′end of the widely expressed hHDAC6 gene and may mark the gene's promoter. Although hHS+14 and hHS+15 are closely spaced (DNase I fine mapping indicates that they are only ∼ 400 bp apart, data not shown), they are separable (Figure 3E). DNase I HSs at coordinates +40, +45, and +49 were present in primary erythroid cells and neutrophils but not in the cell lines K562, HeLa, and AML14.3D10 (Figure 3F-H; Supplementary Table S1). All the constitutive sites colocalize with CpG islands (data not shown).
Examples of previously unidentified DNase HSs in the h GATA1 locus. (A) The hGATA1 locus, positions of constitutive (black arrows), hematopoietic (gray arrows), and eosinophil-specific (open arrows) DNase I HSs and the region of chromatin (gray bar) that was systematically surveyed throughout for DNase I HSs are depicted as in Figures 1B and 2A. (B-H) Maps of the relevant portions of the genomic hGATA1 locus, together with examples of DNase I HSs, are shown. The organization of these parts of the figure is the same as in Figure 2B-G. Abbreviations for cells used are as follows: Io Ery, primary erythroid cells; K562, myeloid/erythroleukemia cell line; Io eos, primary eosinophils; AML, eosinophil myelocyte cell line; and Hela, fibroblast cell line. Additional restriction enzymes used are Sc, ScaI; D, DraI; and HII, HindII.
Examples of previously unidentified DNase HSs in the h GATA1 locus. (A) The hGATA1 locus, positions of constitutive (black arrows), hematopoietic (gray arrows), and eosinophil-specific (open arrows) DNase I HSs and the region of chromatin (gray bar) that was systematically surveyed throughout for DNase I HSs are depicted as in Figures 1B and 2A. (B-H) Maps of the relevant portions of the genomic hGATA1 locus, together with examples of DNase I HSs, are shown. The organization of these parts of the figure is the same as in Figure 2B-G. Abbreviations for cells used are as follows: Io Ery, primary erythroid cells; K562, myeloid/erythroleukemia cell line; Io eos, primary eosinophils; AML, eosinophil myelocyte cell line; and Hela, fibroblast cell line. Additional restriction enzymes used are Sc, ScaI; D, DraI; and HII, HindII.
Re-analysis of sequence associated with all of the mouse and human hypersensitive sites revealed that there is prominent homology between sequence associated with mHS+20 and hHS+15 (regions of > 70% homology over a 500-bp window), which are both located at the 5′ end of HDAC6 and are likely to mark the promoter of this gene in mice and humans. There is weaker homology between mice and humans at sequence associated with mHS–47.5, mHS–42, hHS–24, and hHS+14. However, the level of homology is not suggestive of strong purifying selection and not comparable with that seen at previously identified GATA1 cis-elements.
In summary, it is striking that the location and cell-type specificity of some sites in the human and mouse GATA1 loci are different. In mouse, eosinophil-specific DNase I HSs were not present and a hematopoietic site was not found in an equivalent position to the erythroid hHS+14. Conversely, in humans, we did not detect erythroid/megakaryocytic sites 5′ of the GATA1 gene (compared with mHS–57.5 and mHS–25). Moreover, whereas there are 5 constitutive DNase I HSs 5′ of mGata1 there is only 1 5′ of hGATA1. More specifically, sequence of the previously unrecognized hematopoietic sites, mHS–25 (see “Sequence of mHS–25 and hHS+14”) and mHS–57.5 (data not shown), was not conserved and hHS+14 was poorly conserved (see “Sequence of mHS-25 and hHS + 14”).
Profile of histone H3 and H4 acetylation across the mouse and human GATA1 loci
Chromatin associated with cis-elements is acetylated at core histones H3/H4 in cell types where elements are active or poised to regulate transcription (reviewed in Grunstein47 and Turner48 ). To provide an independent means of validating the location and cell-type specificity of chromatin structure associated with cis-elements, we profiled acetylation status of H3/H4 between coordinate –67.8 to +22.2 in the mGata1 locus and coordinates –80.8 and +44.3 in the hGATA1 locus by chromatin immunoprecipitation, in GATA1-expressing and -nonexpressing cells. As H3 and H4 acetylation profiles are similar, H3 acetylation data in the mGata1 and hGATA1 loci are shown in Figures 4 and 5, respectively, and the H4 profiles are in Supplementary Figures S1-S2.
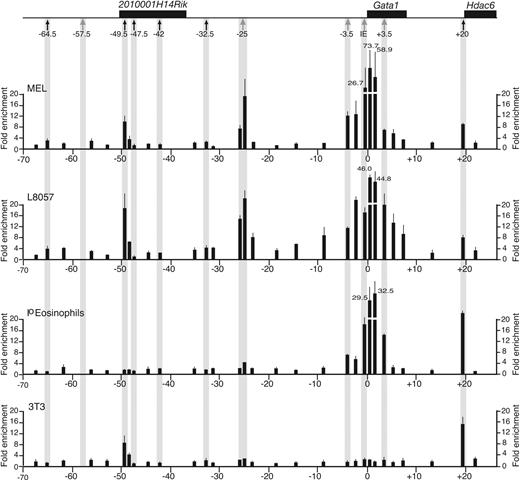
Acetylation profile of histone H3 in the mGata1 locus. At the top of the figure, the mGata1 locus is shown. Genes are depicted as black boxes and, below that, the positions of the hematopoietic DNase I HSs (gray arrows) and widely expressed DNase I HSs (black arrows) are marked. Panels below the locus show the profile of acetylated H3 at different points in the mGata1 locus as determined by ChiP using real-time Taqman PCR analysis in an erythroid cell line (MEL), megakaryocytic cell line (L8057), primary eosinophils (Io eosinophils), and a fibroblast cell line (NIH3T3 cells). The fold enrichment of acetylated H3 (y-axis) is shown at different points along the mGata1 locus (x-axis). The coordinates are in kilobases with respect to the transcriptional start site of the GATA1 IE promoter. The black bars show average fold enrichment results from duplicate real-time PCR reactions performed on 3 independent ChiP preparations. Error bars (1 SD) are shown. We were not able to assess histone acetylation status of chromatin overlying mHS–57.5 (note that the closest primer pair to mHS–57.5 is ∼ 1.0 kb away), as sequence encompassing mHS–57.5 is repetitive.
Acetylation profile of histone H3 in the mGata1 locus. At the top of the figure, the mGata1 locus is shown. Genes are depicted as black boxes and, below that, the positions of the hematopoietic DNase I HSs (gray arrows) and widely expressed DNase I HSs (black arrows) are marked. Panels below the locus show the profile of acetylated H3 at different points in the mGata1 locus as determined by ChiP using real-time Taqman PCR analysis in an erythroid cell line (MEL), megakaryocytic cell line (L8057), primary eosinophils (Io eosinophils), and a fibroblast cell line (NIH3T3 cells). The fold enrichment of acetylated H3 (y-axis) is shown at different points along the mGata1 locus (x-axis). The coordinates are in kilobases with respect to the transcriptional start site of the GATA1 IE promoter. The black bars show average fold enrichment results from duplicate real-time PCR reactions performed on 3 independent ChiP preparations. Error bars (1 SD) are shown. We were not able to assess histone acetylation status of chromatin overlying mHS–57.5 (note that the closest primer pair to mHS–57.5 is ∼ 1.0 kb away), as sequence encompassing mHS–57.5 is repetitive.
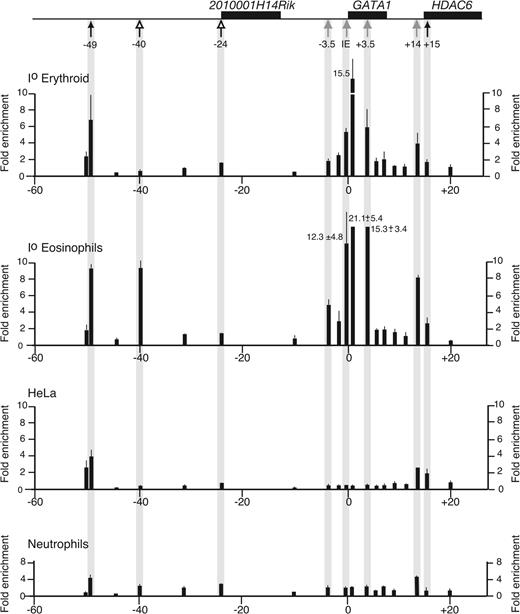
Acetylation profile of histone H3 in the hGATA1 locus. The organization of the figure is the same as in Figure 4. Top, the hGATA1 locus is drawn. Genes are depicted as black boxes. Below the locus, positions of hematopoietic (gray arrows), eosinophil-specific (open arrows), and widely expressed DNase I HSs (black arrows) are marked. Below that, the panels show the profile of acetylated H3 at different points along the hGATA1 locus in primary erythroid cells (Io erythroid), primary eosinophils (Io eosinophils), fibroblast cell line (HeLa), and Io neutrophils. Data are shown as average fold enrichments from duplicate real-time PCR reactions performed on 3 independent ChiP preparations. Error bars (1 SD) are shown.
Acetylation profile of histone H3 in the hGATA1 locus. The organization of the figure is the same as in Figure 4. Top, the hGATA1 locus is drawn. Genes are depicted as black boxes. Below the locus, positions of hematopoietic (gray arrows), eosinophil-specific (open arrows), and widely expressed DNase I HSs (black arrows) are marked. Below that, the panels show the profile of acetylated H3 at different points along the hGATA1 locus in primary erythroid cells (Io erythroid), primary eosinophils (Io eosinophils), fibroblast cell line (HeLa), and Io neutrophils. Data are shown as average fold enrichments from duplicate real-time PCR reactions performed on 3 independent ChiP preparations. Error bars (1 SD) are shown.
In mouse and human GATA1-expressing cells (erythroid cells [MEL], megakaryocytic cells [L8057], and primary eosinophils in mice, and primary erythroid cells and eosinophils in humans), chromatin throughout the region encompassing HS–3.5, IE promoter, and HS+3.5 was enriched for acetylated H3 and H4. By contrast, in GATA1-nonexpressing cells (fibroblasts [3T3] in mice and fibroblasts [HeLa] and primary neutrophils in humans), this region was not enriched for acetylated histone H3/H4.
Further away from the GATA1 gene, there were species- and cell type–specific differences in acetylation profiles. In mice, 3 areas were enriched for acetylated H3/H4 (–49.5, –25, and +20). Chromatin associated with the erythroid/megakaryocytic mHS–25 was specifically enriched for acetylated H3/H4 in cell types where the HS is present. Chromatin at +20 is associated with the widely expressed mHS+20 and is enriched for H3/H4 acetylated in all cell types. Chromatin at –49.5 is associated with 1 of 5 constitutive DNase I HSs upstream of Gata1, and is enriched for acetylated H3 in erythroid cells, megakaryocytic cells, and fibroblasts, but not eosinophils. Finally, we did not detect eosinophil-specific enrichment of acetylated H3/H4.
In humans, chromatin associated with eosinophil-specific hHS–40, but not hHS–24, was specifically enriched for acetylated H3/H4 only in eosinophils. In all cell types tested, the region covering hHS+14/+15 was enriched for acetylated H3/H4, though there was more marked enrichment for acetylated H3/H4 in primary erythroid cells and K562 with the region more closely associated with hHS+14. However, it is important to note that as the erythroid-specific hHS+14 and constitutive hHS+15 are only 300- to 400-bp apart and that chromatin used for immunoprecipitation was sheared to a modal value of 300 bp, it is not possible to clearly distinguish acetylation associated with hHS+14 compared with hHS+15. Finally, chromatin at –49 was enriched for acetylated H3/H4 in all cell types and is associated with the constitutive DNase I site hHS–49 and a CpG island (data not shown).
In summary, the histone acetylation profiles support DNase I HS mapping data in suggesting that, though chromatin structure immediately flanking GATA1 is similar in humans and mice, significant species- and cell type–specific differences exist further away from the GATA1 genes.
Enhancer activity associated with mHS–25 and hHS+14
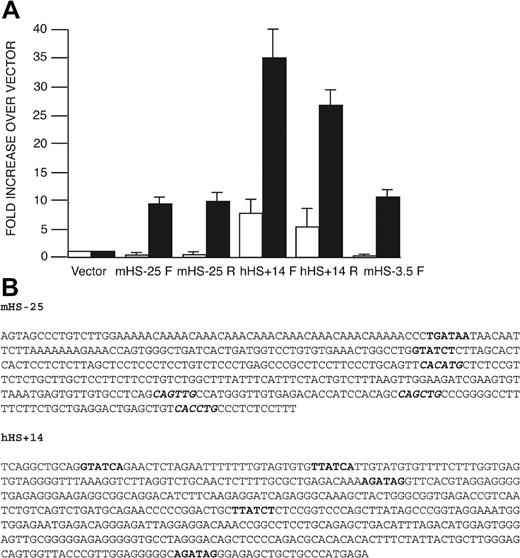
In “Introduction,” we reasoned that there were previously unidentified cis-elements important specifically for red cell GATA1 expression. Among all the DNase I HSs reported here, only mHS–25 and hHS+14 are hypersensitive to DNase I and enriched for acetylated H3/H4 specifically in red cells (and a megakaryocytic cell line in mice) but not other cell types. Thus, to prioritize functional analysis of potential cis-elements, we tested if sequence encompassing mHS–25 and hHS+14 had enhancer function in red cells. Sequences including both these sites were attached, in both orientations, to a neor (neomycin resistance) gene regulated by a thymidine kinase promoter. In addition, we compared activity of these sites with mHS–3.5, which has been shown to direct red cell GATA1-reporter gene expression in transgenic mice.17,18 Recombinant plasmids were stably transfected into semiadherent mouse erythroid cells (MEL) and mouse fibroblasts (L929). After antibiotic selection, numbers of resistant colonies were counted (Figure 6A). mHS–25 increased colony number over the baseline vector by approximately 10-fold in MEL cells, whereas no increase in colony numbers was seen in L929 cells. Similar results were obtained with mHS–3.5, which increased colony numbers approximately 10.5-fold in MEL cells and had no activity in L929 cells. In contrast, sequences containing hHS+14 increased colony numbers by 5- to 7-fold compared with vector in L929 cells. However, there was a further 5-fold increase in colony numbers in MEL cells compared with L929 cells (colony numbers were increased between 25- to 35-fold compared with vector alone). These findings suggest that both mHS–25 and hHS+14 contain sequences with enhancer function that is comparable with mHS–3.5 in this assay. For mHS–25, this activity appears red cell–specific. For hHS+14, the activity is more pronounced in red cells. Activity of hHS+14 in fibroblast cells may reflect inclusion of sequences located in the nearby constitutive hHS+15.
Transcriptional activity of mHS–25 and hHS+14 in the neor activation assay. (A) Fragments containing mHS–25, hHS+14, and mHS–3.5 were inserted either in the forward (F) or reverse (R) orientation into the neor-gene–containing vector, pMC1neo(polyA). The fold change in the number of G418-resistant colonies relative to the baseline vector, pMC1neo(polyA), upon transfection of L929 cells (□) and MEL cells (▪) is shown for each construct. Each bar represents average colony number from 5 experiments with each construct. Error bars (1 SD) are shown. Absolute average numbers of colonies were 14.3 colonies/1 × 107 L929 cells and 8.2 colonies/2 × 107 MEL cells when 20 μg linearized pMC1neo(polyA) was transfected. The amount of each recombinant plasmid DNA used for transfection was adjusted to ensure that equal molar ratios were transfected. (B) Sequence of mHS–25 and hHS+14. Sequence of GATA sites are marked in bold and E-boxes, in italics and bold.
Transcriptional activity of mHS–25 and hHS+14 in the neor activation assay. (A) Fragments containing mHS–25, hHS+14, and mHS–3.5 were inserted either in the forward (F) or reverse (R) orientation into the neor-gene–containing vector, pMC1neo(polyA). The fold change in the number of G418-resistant colonies relative to the baseline vector, pMC1neo(polyA), upon transfection of L929 cells (□) and MEL cells (▪) is shown for each construct. Each bar represents average colony number from 5 experiments with each construct. Error bars (1 SD) are shown. Absolute average numbers of colonies were 14.3 colonies/1 × 107 L929 cells and 8.2 colonies/2 × 107 MEL cells when 20 μg linearized pMC1neo(polyA) was transfected. The amount of each recombinant plasmid DNA used for transfection was adjusted to ensure that equal molar ratios were transfected. (B) Sequence of mHS–25 and hHS+14. Sequence of GATA sites are marked in bold and E-boxes, in italics and bold.
Sequence of mHS–25 and hHS+14
Inspection of sequence of mHS–25 and hHS+14 (Figure 6B) revealed DNA-binding motifs for transcription factors active in erythroid cells that could regulate GATA1 expression. These include GATA sites (both GATA1 and GATA2 have been implicated in promoting GATA1 expression16,49 ) and E-boxes that can bind the hematopoietic transcription factor SCL/Tal1 (hereafter referred to as SCL) and its heterodimeric partner E2A. GATA factors together with SCL/E2A and the non-DNA binding transcription factors LMO2 and Ldb1 form a complex on DNA in vitro that has been implicated in regulating GATA1 expression.42
mHS–25 has 2 GATA sites (only 1 with the extended consensus sequence of WGATAR) and 4 E-boxes. In contrast, hHS+14 has 5 GATA sites (of which 4 are WGATAR) and no E-boxes. Moreover, the location of binding sites with respect to each other and the immediate nucleotides flanking each site are not conserved. Thus, the 2 red cell/megakaryocyte DNase I sites exhibit enhancer activity in red cells and contain multiple binding sites for relevant regulators that may, in part, account for their activity, but do not share conserved motifs for these key red cell transcription factors.
In vivo transcription factor binding in the GATA1 locus
To study the potential for function of all the newly identified DNase HSs in erythroid cells and, specifically, to determine whether the GATA sites and E-box motifs identified in mHS–25 and hHS+14 (Figure 6B) were occupied by transcription factors in vivo, we studied transcription factor binding in MEL cells and primary human erythroblasts by chromatin immunoprecipitation (Figure 7).
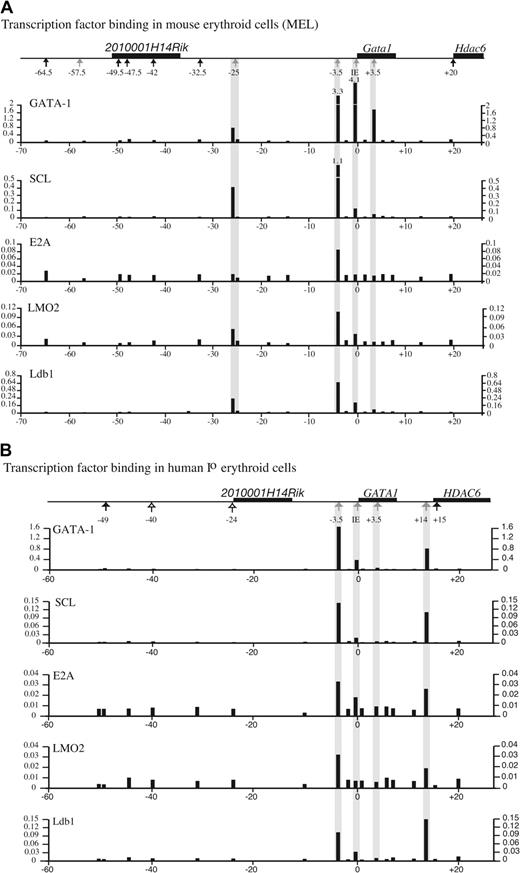
In vivo binding of known transcription factors upstream of GATA1 in mouse and human erythroid cells. Binding of the transcription factors GATA1, SCL, E2A, LMO2, and Ldb1 at different points along the mGata1 locus (A) and the hGATA1 locus (B). GATA1 loci are drawn above each panel, as in Figures 4-5. Transcription factor binding was determined by ChiP using Taqman real-time PCR analysis in a mouse erythroid cell line (MEL) and primary human erythroid cells. The black bars show the relative degree of binding (y-axis) at different points along the GATA1 loci (x-axis) from duplicate real-time PCR reactions. The analysis was performed on 3 independent ChiP preparations, and data from 1 representative experiment are shown.
In vivo binding of known transcription factors upstream of GATA1 in mouse and human erythroid cells. Binding of the transcription factors GATA1, SCL, E2A, LMO2, and Ldb1 at different points along the mGata1 locus (A) and the hGATA1 locus (B). GATA1 loci are drawn above each panel, as in Figures 4-5. Transcription factor binding was determined by ChiP using Taqman real-time PCR analysis in a mouse erythroid cell line (MEL) and primary human erythroid cells. The black bars show the relative degree of binding (y-axis) at different points along the GATA1 loci (x-axis) from duplicate real-time PCR reactions. The analysis was performed on 3 independent ChiP preparations, and data from 1 representative experiment are shown.
Specific GATA1 binding was seen at mHS–25, mHS–3.5, IE promoter, and mHS+3.5 in mice (Figure 7A), and hHS–3.5, the IE promoter, and hHS+14 but not hHS+3.5 in humans (Figure 7B). Prominent binding of SCL, LMO2, and Ldb-1 colocalize with GATA1 at mHS–25 and mHS–3.5 with weaker E2A binding at mHS–25. There is also weaker binding of SCL, LMO2, and Ldb1 at the mIE promoter, and binding is not detected at mHS+3.5. Similarly, strong SCL, E2A, LMO2, and Ldb1 binding was seen at hHS–3.5 and hHS+14, with weak binding of SCL, E2A, and Ldb1 at the hIE promoter and no binding at hHS+3.5. For the first time, we demonstrate in vivo colocalization of GATA1, SCL, E2A, LMO2, and Ldb1 at bona fide GATA1 regulatory sequences (HS–3.5) and potential GATA1 regulatory sequences (mHS–25 and hHS+14). These observations are consistent with data from in vitro electromobility shift analysis studies showing that these proteins form hematopoietic transcription factor complex(es).42,50 The regions of transcription factor binding colocalize with chromatin that is DNase I hypersensitive and enriched for acetylated H3/H4 only in hematopoietic cells. As both GATA1 and SCL associate with histone acetyltransferases (HATs),51,52 it is likely that they recruit HAT activity and that this, at least in part, results in the enrichment of histone acetylation that is detected.
Discussion
Lack of conservation of chromatin structure and cis-element organization between the human and mouse GATA1 loci
With the near-complete sequencing of the mouse and human genomes, and other genome sequences soon to follow, the identification of cis-elements, for genes whose function and expression is conserved through evolution, may become simpler by searching for conserved nonexonic sequence (reviewed in Hardison53 ; Frazer et al54 ; and Nobrega and Pennacchio55 ). In support of this, regulatory sequences at a few loci that were originally determined by conventional experimental approaches, such as some of the cis-elements in the globin loci in red cells,23,53 have been shown to be conserved between mice and humans. In some cases, regulatory elements were first noticed because their sequences were conserved. Examples here include loci encoding the interleukin genes,20 and the Hox22,56 and Pax6 transcriptional regulators.24 In blood cells, this approach has been successful in identifying cis-elements important in regulation of the transcription factor SCL21,57 and glycoprotein IIb in megakaryocytes.58
As functional studies in the mGata1 locus suggest that important red cell– and eosinophil-specific cis-elements remain uncharacterized (“Introduction”), we compared sequence of the human and mouse GATA1 loci as a first step to identify novel cis-elements. There were 2 regions of conserved sequence identified outside those corresponding to known GATA1 cis-elements and exons for flanking genes. The first lies between the IE exon and the intron element (HS+3.5). In transgenic mice, constructs containing this region together with 2.5 kilobases of sequences 5′ of the IE exon (but excluding HS–3.5) and HS+3.5 failed to direct reporter gene expression to red cells and megakaryocytes in transgenic mice.17,18 The second region lies just upstream of the HDAC6 gene and is most likely to correspond to the promoter of this gene. Outside these regions, the lack of highly conserved sequence between the mouse and human GATA1 is surprising. However, as sequence of the rodent testis-specific IT promoter59 is not conserved in humans (“Results”), it raised the possibility that other cis-elements in the GATA1 loci may also not be conserved in sequence. Therefore, we undertook a systematic examination of chromatin structure in the mouse and human GATA1 loci.
Our data demonstrate surprisingly marked differences in chromatin structure between the 2 GATA1 loci. In red cells, hHS+14 and mHS–25 are located in different positions with respect to the GATA1 gene and multiple sequence alignment programs reveal no homology between them (“Results” and data not shown). In marked contrast, 2 other cis-elements important for red cell GATA1 expression, HS–3.5 and HS+3.5, are highly conserved in position and sequence, between humans and mice.41,42 Although our data have not directly tested whether mHS–25 and hHS+14 regulate GATA1 itself, there are 3 reasons that argue that both DNase I HSs mark bona fide hematopoietic regulatory elements and are candidate GATA1 regulatory elements. First, they have the chromatin structure (DNase I HS and enriched for acetylated H3 and H4) associated with active cis-elements and this is detected only in red cells (and a megakaryocytic cell line in mice). Second, they function in an orientation-independent manner to augment transcription of a heterologous gene in red cells. Lastly, they bind a number of transcription factors in vivo important for red cell gene expression. Although these observations suggest that mHS–25 and hHS+14 are potential GATA1 regulatory elements, they do not constitute evidence that these elements regulate GATA1. Further experiments to test the physiologic role of mHS–25 and hHS+14 in regulating human and mouse GATA1 expression and genes neighboring GATA1 are now warranted (see “Possible functional roles in mHS–25 and +14”).
Similarly, chromatin structure of the GATA1 loci is quite different in mouse and human eosinophils. Eosinophil-specific DNase I HSs (hHS–40 and hHS–24) were detected only in human and not in mouse eosinophils. Additionally, compared with surrounding points in the hGATA1 locus, chromatin associated with hHS–40, but not hHS–24, was enriched for acetylated H3/H4 specifically in primary eosinophils. Although the function of hHS–40 in promoting eosinophil GATA1 expression remains to be established, our data would suggest that hHS–40 is a candidate eosinophil-specific GATA1 regulatory element and that a similar candidate eosinophil-specific regulatory element does not exist in an equivalent position in the mGATA1 locus.
Focusing on those DNase I HSs that may function to regulate GATA1 expression, whole genome alignments show that sequence corresponding to mHS–25 is not present in humans, and sequence corresponding to hHS–40 is not present in mice. Lastly, although the sequence corresponding to hHS+14 is present in an analogous position in the mouse genome 3′ of mGata1 adjacent to mHS+20, it is poorly conserved and in particular lacks the GATA binding sites seen in hHS+14. This would suggest that mice and humans could potentially use species-specific cis-elements to regulate GATA1.
Previous studies have hinted at differences in cis-elements regulating homologous genes in mice and humans. For example, in mice bearing large transgenes containing the human hematopoietic stem/progenitor cell marker CD34, the expression patterns of human and mouse CD34 are not equivalent.60 This suggests that cis-elements regulating CD34 expression, in the 2 species, may differ. However, this has yet to be formally demonstrated, as the precise position and sequence of CD34 cis-elements in both species,61,62 but especially mice, remain undefined. In comparison, our findings in the GATA1 locus are potentially even more striking than those in the CD34 locus, as, in contrast to the CD34 gene, GATA1 is expressed in an identical domain in the 2 species. Outside humans and mice, in different Drosphilia species, enhancers controlling the tightly regulated expression domains of even-skipped, Hoxa2, and Yolk protein genes have been shown to contain nucleotide substitutions in functionally important transcription factor binding sites. However, even in these cases the enhancers are conserved in equivalent genomic positions.56,63,64
Although our data suggest that success in identifying cis-elements by human and mouse sequence comparison alone may vary between loci, and that additional experimental approaches (eg, chromatin analysis and functional assays) may be required, given the paucity of gene loci where cis-elements have been adequately identified, it may be premature to estimate how generally successful mouse-human sequence comparisons will be in uncovering cis-elements. The systematic and detailed analysis of the GATA1 loci presented here will hopefully contribute to this assessment.
Possible functional roles of mHS–25 and hHS+14
In red cells, the chromatin structure and pattern of transcription factor binding of mHS–25 and hHS+14 are similar to HS–3.5. Although we have not established the physiologic role of mHS–25 and hHS+14 in regulation of GATA1, these observations together with the finding that mHS–25 and hHS+14 function as enhancers in red cells led us to hypothesize that mHS–25 and hHS+14 may work in concert with HS–3.5 to express GATA1 in red cells. If correct, it is possible that mHS–25 allows sustained red cell GATA1 expression in absence of mHS–3.5,9 suggesting that these 2 elements would be redundant, with respect to each other, for red cell GATA1 expression. To test this hypothesis, we are currently generating mice deleted for mHS–25, and mHS25 and mHS–3.5 together in cis.
Analysis of mice deleted for mHS–25 would also define the role of this element in megakaryocyte GATA1 expression. Given the features of active chromatin structure at mHS–25 in megakaryocytes and the observation that mHS–25 binds GATA1 and other regulators in megakaryocytic cells (B.G. and P.V., unpublished data, March 2004), we speculate that it functions to regulate megakaryocyte GATA1 expression. However, as megakaryocyte GATA1 expression is almost completely abolished in mice deleted for mHS–3.5,6 the functional relationship between mHS–3.5 and mHS–25 may differ in red cells and megakaryocytes. Lastly, mHS–25 resides between GATA1 and 2010001H14 RIK, and as the latter gene is expressed in megakaryocytes (data not shown), mHS–25 could regulate 2010001H14 RIK as well as GATA1. Mapping in vivo physical interactions65 between mHS–25 and the promoters of GATA1 and/or 2010001H14 RIK would directly resolve this question.
In vivo pattern of transcription factor binding at cis-elements in the GATA1 loci
A complex containing GATA1/SCL/E2A/LMO2/Ldb-1 was initially identified in erythroid cell (MEL) nuclear extracts in vitro using oligonucleotides with a composite GATA–E-box element.50 Subsequently, we showed that a composite GATA–E-box element within mHS–3.5 could bind in vitro such a complex that could include either GATA1 or GATA2.42 Others similarly demonstrated in vitro complex assembly on variations of the original GATA–E-box element or on GATA sites alone, at regulatory sequences of other genes.66-68 Finally, recent data suggest that all the components of this complex function with the ubiquitous transcription factor Sp1 to regulate expression of the red cell–specific glycophorin A gene.69
In the specific case of transcriptional control of GATA1, SCL, LMO2, and GATA2 all act upstream of GATA1 (reviewed in Orkin15 ) and are candidate GATA1 regulators. Our data now show, for the first time, that these classes of transcription factors colocalize in vivo at multiple GATA1 regulatory elements in red cells. Moreover, we have recently immunoprecipitated an endogenous complex containing, at a minimum, GATA1/SCL/E2A/LMO2/Ldb-1, from erythroid nuclear extracts (A. Schuh, P.V., and C.P., unpublished data, February 2004). The combination of these findings suggests that these proteins normally work together to regulate GATA1 expression. Interestingly, though all 5 transcription factors bind hHS+14, E-box sites are not present here, suggesting the complex may be anchored to this element by GATA site(s) alone. This observation is consistent with studies demonstrating that SCL does not have to bind DNA for many of its functions.32
Lastly, it is noteworthy that the pattern of transcription factor binding varies at different GATA1 cis-elements in red cells. At mHS+3.5 we detect only GATA1 but not SCL/E2A/LMO2/Ldb-1 binding. We have also recently seen differences in regulator binding at GATA1 cis-elements in eosinophils, where C/EBPϵ binding was detected only at mHS+3.5.19 These observations suggest that individual GATA1 cis-elements are likely to serve different roles in regulating GATA1 expression and provide a platform to more fully elucidate the full complement of regulator binding at GATA1 cis-elements as multipotential cells differentiate into GATA1-expressing lineages. These data are likely to be highly informative of how individual cis-elements function to express GATA1 in different myeloid lineages and of how GATA1 levels come to differ in GATA1-expressing cells.
Prepublished online as Blood First Edition Paper, July 20, 2004; DOI 10.1182/blood-2004-04-1333.
V.V.-G. and B.G. were funded by the Wellcome Trust and E.A. and I.H., by the MRC. C.P. is an MRC tenure-track scientist, and P.V. is a Wellcome Trust Senior Clinical Fellow.
V.V.-G. and B.G. contributed equally to this work.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs de Bruijn and Wood for comments, Dr de Gobbi for help with hGATA1 ChiP experiments, Dr Wood for providing primary erythroid cells, and Professor Orkin for LMO2 and Ldb1 antibodies.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal