Abstract
Notch1 is known to play a critical role in regulating fates in numerous cell types, including those of the hematopoietic lineage. Multiple defects exhibited by Notch1-deficient embryos confound the determination of Notch1 function in early hematopoietic development in vivo. To overcome this limitation, we examined the developmental potential of Notch1–/– embryonic stem (ES) cells by in vitro differentiation and by in vivo chimera analysis. Notch1 was found to affect primitive erythropoiesis differentially during ES cell differentiation and in vivo, and this result reflected an important difference in the regulation of Notch1 expression during ES cell differentiation relative to the developing mouse embryo. Notch1 was dispensable for the onset of definitive hematopoiesis both in vitro and in vivo in that Notch1–/– definitive progenitors could be detected in differentiating ES cells as well as in the yolk sac and early fetal liver of chimeric mice. Despite the fact that Notch1–/– cells can give rise to multiple types of definitive progenitors in early development, Notch1–/– cells failed to contribute to long-term definitive hematopoiesis past the early fetal liver stage in the context of a wild-type environment in chimeric mice. Thus, Notch1 is required, in a cell-autonomous manner, for the establishment of long-term, definitive hematopoietic stem cells (HSCs).
Introduction
In the adult mammal, definitive, or adult-type, hematopoietic cells arise in a sequential process of differentiation from a common precursor, the hematopoietic stem cell (HSC). This definitive HSC has the unique capability of long-term reconstitution of all lymphoid and myeloid lineages in adult recipients. In contrast, during embryogenesis, development of the mammalian hematopoietic system is characterized by sequential waves of hematopoietic progenitor formation with limited reconstitution capacity that proceeds to definitive (adultlike) HSC development only later. The first transient wave of hematopoiesis during development arises in the blood islands of the extraembryonic yolk sac and consists of primitive erythroid colony-forming unit (CFU) progenitors. These CFU progenitors are detected during a short window after gastrulation from about 7 days after coitum (dpc) to 8.5 dpc, and generate primitive nucleated erythrocytes that express a distinct, embryonic-type globin and constitute the first circulating cells of the developing embryo.1 The second wave, which begins slightly later, involves formation of definitive hematopoietic progenitors that give rise to enucleated erythrocytes, macrophages, and granulocytes, cell types similar to those observed in the adult.1,2 Although these definitive progenitors are first detected in the yolk sac, they also appear later in the embryo proper, reflecting migration of yolk sac cells through the newly established circulation and/or autonomous intraembryonic initiation of definitive hematopoiesis in a region first referred to as the para-aortic splanchnopleura (P-Sp).3-6 At this early stage, these definitive hematopoietic progenitors have limited selfrenewal capacity in that they are unable to reconstitute long-term hematopoiesis in adult recipients. Thus, it has been proposed that these progenitors are the first to seed the fetal liver, where they differentiate in mass to form the first functioning definitive hematopoietic cells in the circulation, supplanting the primitive erythrocytes.1 Only later is a definitive HSC with full hematopoietic repopulating capability detected.7-9 After development and/or maturation in the yolk sac and aorta-gonadmesonephros region (AGM), the definitive HSC, which homes first to the fetal liver and eventually the bone marrow, provides multilineage hematopoiesis throughout the remainder of development and in the adult. The exact relationship between the early definitive CFU progenitors and the later multilineage repopulating hematopoietic stem cell (HSC) remains unclear (for review see Palis and Yoder10 ). Specifically, are these early definitive progenitors the progeny of an immature, embryonic HSC that attains adult-repopulating capacity only later in development, or do they arise from a population of cells distinct from the definitive HSC?
The diverse stages of developmental hematopoiesis are dependent on a variety of critical signal pathways that are both overlapping and distinct from those in the adult. The Notch receptor pathway has well-established roles in various aspects of embryonic and adult development, including cell fate decisions through control of differentiation, survival, and/or proliferation in numerous tissues.11-16 Notch1, as well as the other 3 mammalian Notch homologues and their respective ligands, is expressed in both overlapping and distinct patterns in bone marrow, thymus, fetal liver hematopoietic and/or stromal cells, the developing yolk sac vasculature, and peripheral circulation, suggesting roles for Notch proteins in various aspects of hematopoiesis17-23 (for reviews see Allman et al24 and Milner and Bigas25 ). Activation of the Notch pathway in established cell lines and bone marrow–derived precursors suggest that, at the level of the hematopoietic progenitor, Notch signaling can regulate lineage determination or promote maintenance of a precursor fate at the expense of lineage-specific differentiation21,26-37 (for review see Ohishi et al38 ). Studies to address the in vivo role of Notch1 signaling in adult hematopoiesis have used an inducible deletion in newborn mice of Notch1 or of RBP-Jκ, which acts in a nuclear complex with the cleaved intracellular domain of all 4 Notch receptors.15,39-41 These experiments showed that Notch1 signaling, though critical for T versus B lymphoid differentiation, does not have an essential, physiologic role in other aspects of adult hematopoiesis, such as maintenance of HSCs or differentiation of erythro-myeloid lineages. However, embryonic sites of hematopoiesis represent environments distinct from adult hematopoiesis, with unique regulatory requirements.42-46 Thus, studies of this nature, conducted in the mouse postnatally, do not preclude the possibility that Notch1 signaling has additional functions during embryonic development of the hematopoietic system.
A recent study suggested that Notch1 plays a critical role during definitive hematopoietic development.47 Notch1-deficient or γ-secretase inhibitor–treated 9.5-dpc P-Sp tissues were cultured in vitro, generating adherent beds of endothelial cells but severely reduced numbers of definitive CFUs. Furthermore, long-term definitive HSC development was found lacking in the absence of Notch1, as determined by reconstituting hematopoiesis in conditioned newborn mice with cells isolated from 9.5-dpc Notch1-deficient embryos. Given a critical role for Notch1 in various aspects of endothelial cell fate (for review see Lawson and Weinstein48 and Rossant and Hirashima49 ) and function50-52 (for review see Iso et al13 ), the use of the already defective Notch1-deficient tissues from 9.5-dpc embryos in a reconstitution assay to examine their developmental potential in hematopoiesis poses problems in the interpretation of Notch1 functions. For example, Notch1-deficient embryos at 9.5 dpc already exhibit major defects in the vasculature, including collapse of the dorsal aorta and defects in vascular morphogenesis in the yolk sac53 ; this may disturb HSC development as it is thought to occur in close association with, or potentially directly from, the arterial endothelium at these sites.54,55 Thus, it remains to be determined if inability to detect long-term HSCs from Notch1-deficient embryos is a cell-intrinsic defect or a consequence of the defective environment.
A second important unanswered question is how Notch1-deficient cells contribute to the early waves of extraembryonic primitive and definitive CFU progenitor formation prior to HSC development. Notch1-deficient embryos contain primitive erythrocytes in the yolk sac prior to lethality.53 A temporal analysis of primitive erythropoiesis in Notch1-deficient embryos, to rule out quantitative differences, has not been undertaken. Furthermore, it is not clear whether Notch1-deficient cells lack the ability to give rise to all definitive progenitors, including those detected prior to HSC development.56
To address these critical questions regarding the function of Notch1 in developmental hematopoiesis while circumventing the complications of early lethality in Notch1-deficient mice, we generated Notch1-deficient embryonic stem (ES) cells for in vitro and in vivo analysis. For in vivo analysis, we used a somatic chimera approach in which LacZ-tagged (ROSA26) Notch1-deficient ES cells were injected into wild-type host blastocysts and allowed to progress through the full course of hematopoietic development, thereby determining the potential of Notch1 null hematopoietic progenitors to contribute to yolk sacs, fetal liver, and bone marrow hematopoiesis. In this system, extrinsic requirements for Notch1 would be provided by wild-type cells, thus, defects exhibited by the Notch1-deficient cells would be indicative of the intrinsic function of Notch1.
Materials and methods
Derivation of ES cells
Murine ES cells homozygous for a Notch1 null mutation (the Notch1in32 allele57 ) were constructed by sequential gene targeting of the remaining wild-type allele in ES cells heterozygous for this Notch1 mutant allele.57 There were 4 ES cell lines derived from the CJ7 parental cell line (1 wild-type, 1 heterozygous, and 2 independently derived homozygous null Notch1 cell lines) used for in vitro differentiation assays. All ES cells were maintained in an undifferentiated state on mitomycin-C–treated STO fibroblast feeder cells or murine embryonic fibroblasts (MEFs; Stem Cell Technologies, Vancouver, BC) or freshly prepared in media, consisting of Dulbecco modified Eagle medium (DMEM), 15% ES-screened fetal bovine serum (FBS; Hyclone, Logan, UT), 2 mM glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 0.14 mM monothioglycerol, and 1000 U/mL leukemia inhibitory factor (LIF; Chemicon, Temecula, CA), at 37°C, 5% CO2. All chemicals were from Sigma (St Louis, MO), unless stated otherwise.
Differentiation of ES cells into embryoid bodies (EBs)
ES cells were predifferentiated for 2 days without feeder cells in ES media (Iscoves modified Dulbecco medium [IMDM] was substituted for DMEM). Subsequently, cells were trypsinized to obtain a single-cell suspension, washed twice, seeded in Petri dishes in IMDM with 15% FBS (Hyclone, defined), 2 mM glutamine, 0.14 mM monothioglycerol, and 50 μg/mL ascorbic acid, and harvested at the indicated day of differentiation for hematopoietic colony assays.
Embryoid body (EB) formation was characterized by comparing plating efficiency of EBs and EB size at days 6 and 12 of differentiation in methylcellulose medium (M3234; Stem Cell Technologies) supplemented with 50 μg/mL ascorbic acid.
Colony-forming unit (CFU) assays in methylcellulose
EBs collected at various days of differentiation were pooled, trypsinized, passaged through a 21-gauge (21G) needle to obtain a single-cell suspension, and resuspended in IMDM. Embryos were dissected at various stages of development and trypsinized or treated with collagenase (Stem Cell Technologies) and then passaged through a 21G needle to obtain a single-cell suspension. A small percentage of cells was used for genotyping, with primers previously described.57,58 For older embryos, the yolk sac was dissected from the embryo prior to treatment with trypsin or collagenase. The embryo was used for genotyping. For chimeric embryos, the date of blastocyst implantation was considered day 3.5 (3.5 days after coitum [dpc]). For CFU progenitor analysis from chimeras, the yolk sac (9.5 dpc to 12.5 dpc) or fetal liver (13.5 to 17.5 dpc) was dissected, treated with collagenase, and dissociated to a single-cell suspension. Bone marrow samples from postnatal mice were prepared similarly, but without collagenase treatment.
EBs or embryo cells were assayed for hematopoietic progenitors as described.1,59 The following reagents and cytokines were used: M3234 methylcellulose medium (Stem Cell Technologies); murine interleukin-3 (IL-3, 10 ng/mL), IL-6 (10 ng/mL), IL-11 (5 ng/mL), stem cell factor (SCF, 50 ng/mL), granulocyte colony-stimulating factor (G-CSF, 30 ng/mL), macrophage colony-stimulating factor (M-CSF, 5 ng/mL), granulocyte-macrophage colony-stimulating factor (GM-CSF, 3 ng/mL), and vascular endothelial growth factor (VEGF, 10 ng/mL) (all from Peprotech, Rocky Hill, NJ); and human erythropoietin (3 to 5 U/mL; Amgen). In preliminary experiments (not shown), colony identities were confirmed by Wright-Giemsa staining and/or reverse-transcriptase–polymerase chain reaction (RT-PCR) to detect expression of β-H1 and β-major globins with primers/conditions as described.60
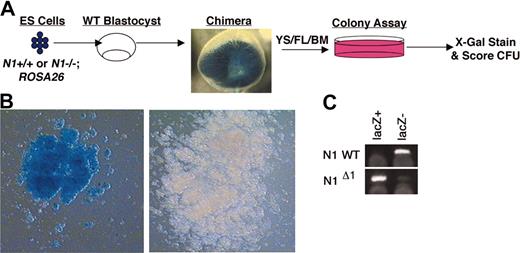
Colonies generated from chimeric embryos were stained with 5-bromo-4-chloro-3findolyl β-d-galactopyranoside (X-gal) stain and scored: blue (LacZ+ ES derived) and nonblue (LacZ– embryo derived) (Figure 5). In control experiments, no LacZ+ colonies were detected from wild-type embryos, and 100% of myeloid and erythroid colonies derived from ROSA26 embryos stained blue.
Generation and hematopoietic analysis of embryos chimeric for Notch1-deficient (Notch1Δ1 allele) or wild-type (control) cells marked by ROSA26. (A) Schematic representation of generation of chimeric embryos. Primary Notch1–/– or wild-type ES cell lines containing the ROSA26 gene were independently derived from blastocysts. These ES lines were injected into wild-type blastocysts to obtain chimeric embryos. At various stages of development (Table 1), the yolk sac (YS), a portion of fetal liver (FL), or bone marrow (BM) was dissected out and dispersed cells were plated for hematopoietic CFU activity and the remaining embryo (or dissected organs) was stained with X-gal. Only those embryos that contained widespread, substantial contribution of LacZ+ cells by gross visualization of stained tissues were included in the hematopoietic analysis. Hematopoietic colonies were stained with X-gal and CFUs scored in order to determine relative percent contribution of ES-derived (LacZ+) and blastocyst-derived (LacZ–) cells. (B) A representative LacZ+ colony from Notch1-deficient (ES derived) cells from a chimeric embryo and LacZ– colony from Notch1+/+ (embryo derived) cells from a chimeric embryo. Images were visualized using Olympus 60 ×/1.4 oil objective lenses. (C) DNA samples from single LacZ– (embryo-derived Notch1+/+) or LacZ+ (ES-derived Notch1–/–) colonies were isolated and used for PCR to genotype colonies.
Generation and hematopoietic analysis of embryos chimeric for Notch1-deficient (Notch1Δ1 allele) or wild-type (control) cells marked by ROSA26. (A) Schematic representation of generation of chimeric embryos. Primary Notch1–/– or wild-type ES cell lines containing the ROSA26 gene were independently derived from blastocysts. These ES lines were injected into wild-type blastocysts to obtain chimeric embryos. At various stages of development (Table 1), the yolk sac (YS), a portion of fetal liver (FL), or bone marrow (BM) was dissected out and dispersed cells were plated for hematopoietic CFU activity and the remaining embryo (or dissected organs) was stained with X-gal. Only those embryos that contained widespread, substantial contribution of LacZ+ cells by gross visualization of stained tissues were included in the hematopoietic analysis. Hematopoietic colonies were stained with X-gal and CFUs scored in order to determine relative percent contribution of ES-derived (LacZ+) and blastocyst-derived (LacZ–) cells. (B) A representative LacZ+ colony from Notch1-deficient (ES derived) cells from a chimeric embryo and LacZ– colony from Notch1+/+ (embryo derived) cells from a chimeric embryo. Images were visualized using Olympus 60 ×/1.4 oil objective lenses. (C) DNA samples from single LacZ– (embryo-derived Notch1+/+) or LacZ+ (ES-derived Notch1–/–) colonies were isolated and used for PCR to genotype colonies.
γ-Secretase inhibitor treatment
A single dose of γ-secretase inhibitor was added to the media of differentiating EBs at the indicated time of differentiation. For Compound no. 11 (Cpd no. 11, a generous gift from M. Wolfe61 ), a final concentration of 50 μM was used.62 For DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-[s]-phenylglycine butylester)63-65 (a generous gift from T. Golde, currently available from Calbiochem, San Diego, CA), a concentration of 1 μM was used. Both inhibitors were delivered in dimethyl sulfoxide (DMSO). For control samples the carrier DMSO was added alone.
Generation of LacZ-tagged ES cells and chimeras
CD1 mice harboring one copy of a null mutation of the Notch1 receptor (Notch1Δ1)58 were crossed to CD1 mice harboring the Rosaβgeo26 gene (ROSA26)66 to obtain mice heterozygous at each allele. These mice were then crossed one generation into the SV129 background to increase efficiency of ES derivation. Notch1Δ1/+; ROSA26/+(SV129 generation 1) mice were then intercrossed. Blastocysts were flushed from the uterus of pregnant mice at 3.5 dpc using prewarmed M2 medium (Specialty Media, Philipsburg, NJ) and transferred to in vitro fertilization culture dishes (Costar 3260; Costar, Cambridge, MA). Blastocysts were washed in blastocyst medium (ES medium, but containing 25% FBS and 2000 U/mL LIF) and transferred to individual drops of blastocyst medium under mineral oil. Alternatively, blastocysts were transferred to 60-mm tissue culture plates treated with 0.1% gelatin in blastocyst medium. After 3 to 4 days of incubation, the inner cell mass of individual, hatched blastocyst was removed, transferred to a 96-well plate containing 35 μL trypsin, and incubated for 5 minutes at 37°C. After repeated pipetting, cells were transferred to fresh 96-well plates containing inactivated MEFs and 200 μL blastocyst medium. Fresh medium was added daily. After 4 to 6 days of incubation, ES cell colonies were transferred to a new well containing MEFs. When necessary, individual ES colonies were picked and subcloned to remove contaminating cell types. Cells were passaged every 2 to 3 days until in T25 flasks, upon which time LIF and FBS concentrations were reduced to that of regular ES medium. All ES lines used were mouse antibody production (MAP) tested and karyotyped.
To generate chimeric embryos and adult mice, ROSA26+; Notch1Δ1–/– and ROSA26+; Notch1+/+ ES cells were injected into wild-type C57Bl6 or CD1 blastocysts and reimplanted into pseudo-pregnant mice. To determine overall contribution from LacZ+ ES-derived cells, embryos or individual organs were fixed and stained with X-gal solution (2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6 × 3 H2O, 0.1% Triton-X, and 1 mg/mL X-gal, in phosphate-buffered saline [PBS]).
Notch1 protein expression analysis
Samples of pooled EBs at various stages of differentiation were lysed in hot Laemmli buffer and boiled for 5 minutes. Protein levels were semiquantitatively analyzed by serial sample dilution and Coomassie staining. Equalized protein samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to membrane, and immunoblotted with anti-Notch1 antibody.67
Staining for activated Notch1 (NICD) in embryo sections
Embryos at approximately 7.5 dpc were fixed in Bouin fixative, embedded in paraffin, and sectioned serially at 6 μm. Sections were stained for the Notch1 intracellular domain (NICD) using antibody Val1744 (Cell Signaling Technology, Beverly, MA) as described previously.68 For nuclear counterstaining, sections were incubated in Bis-benzimide (0.2 μg/mL in PBS). Fluorescent microscopy was performed on an Olympus IX70 microscope (Olympus, Melville, NY). Images were captured using a CoolSnap HQ cooled CCD camera with Metamorph 6.0 software (Universal Imaging, Downington, PA).
Flow cytometric analysis of EB and embryo cells
EBs or embryos were incubated in collagenase or trypsinized, passaged several times through a 21G needle, and passed through a 70-μm nylon filter (Fisher, Hampton, NH) to attain a single-cell suspension. Per sample, 5 × 105 cells were washed twice with Stain Buffer (Pharmingen, San Diego, CA) and blocked with antimouse CD16/32 (Pharmingen). For detection of flk-1, cells were incubated with the directly conjugated primary antibody flk-1 phycoerythrin (PE; Pharmingen). For detection of Notch1, cells were incubated with a primary rabbit antibody to the extracellular domain of Notch1 (catalog no. 06-809; Upstate Biotechnology, Lake Placid, NY), followed by antirabbit biotin (Vector, Burlingame, CA), and finally with Streptavidin PE (Pharmingen). Samples were analyzed with a Becton Dickinson FACScan flow cytometer (San Jose, CA) using CellQuest software.
Statistical analyses
Where indicated, the Student t test was used to determine statistical significance.
Results
Lack of Notch1 did not affect ES cell proliferation, EB formation, or differentiation into flk-1+ mesodermal cells
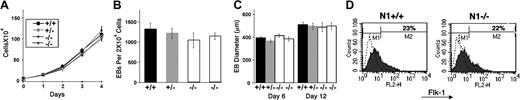
To address a potential role for Notch1 in various aspects of early developmental hematopoiesis, and circumvent the early lethality of Notch1-deficient embryos, we first adopted a routinely deployed model: in vitro ES cell differentiation into hematopoietic precursors. There were 2 independently derived Notch1–/– ES cell lines, a Notch1+/– ES cell line and a Notch1+/+ wild-type parental CJ7 ES cell line, plated at equal densities onto STO fibroblast feeder layers, subsequently trypsinized and counted. All ES cell lines exhibited a similar rate of expansion (Figure 1A). ES cells, predifferentiated for 2 days, were plated at equal densities in methylcellulose medium, and total numbers of EBs formed were scored at day 6 of differentiation. Notch1–/– ES cell lines exhibited a similar efficiency of embryoid body formation as Notch1+/– and Notch1+/+ ES cells (Figure 1B). In addition, embryoid bodies derived from Notch1–/– ES cells allowed to differentiate in liquid culture were of similar size at days 6 and 12 of differentiation as those derived from Notch1+/– and Notch1+/+ ES cells (Figure 1C). The earliest marker of differentiation of the mesodermal population that includes the hematopoietic and endothelial lineages is flk-1, a receptor for VEGF.69 To determine whether the differentiation of this early population of cells was affected by the absence of Notch1, we examined the expression of flk-1 by fluorescence-activated cell-sorter (FACS) analysis. Day-3.75 Notch1-deficient EBs contained a similar percentage of flk-1–expressing cells as a Notch1 heterozygous control (Figure 1D). Together, these results indicated that ES proliferation, EB plating efficiency and growth, and differentiation of early flk-1+ progenitors were not significantly altered in the absence of Notch1.
Notch1-deficient ES cell lines (Notch1in32 allele) exhibit no defect in proliferation, embryoid body (EB) plating efficiency or growth, or differentiation of the early flk1+ population. (A) ES cell proliferation curve. ES cells were plated on day 0 at a density of 5 × 104 cells/mL in 24-well plates. On subsequent days, samples were trypsinized and total cell numbers counted. Values represent the average of 3 samples and error bars indicate standard deviation (SD). Wild-type (+/+), heterozygous (+/–), and 2 independently derived null ES lines (–/–) are shown. (B-C) ES cells, predifferentiated for 2 days without the STO-neo feeder layer but in the presence of LIF, were trypsinized and plated in methylcellulose without exogenous cytokines (B) or in liquid suspension culture (C), at a density of 2 × 104 cells/mL. Total number of EBs were counted at day 4 (B). At days 6 and 12, EBs were pooled and the maximum diameters of at least 30 EBs were measured for each sample (C). Values represent the average of 3 replicate platings from one experiment and error bars indicate SD. Similar results were obtained for several independent experiments. Wild type (+/+), Notch1 heterozygous (+/–), and 2 independently derived Notch1 null (–/–) ES cell lines. (D) FACS analysis of flk-1 expression in Notch1–/– and Notch1+/– EBs differentiated for 3.75 days. EBs were dissociated to single-cell suspension and stained with an antibody to flk-1, conjugated to phycoerythrin. The profile indicated by a dotted line represents unstained EB cells. Similar results were obtained in 2 independent experiments, and with additional Notch1–/– and control cell lines. Percentage of cells in the M2 window is shown.
Notch1-deficient ES cell lines (Notch1in32 allele) exhibit no defect in proliferation, embryoid body (EB) plating efficiency or growth, or differentiation of the early flk1+ population. (A) ES cell proliferation curve. ES cells were plated on day 0 at a density of 5 × 104 cells/mL in 24-well plates. On subsequent days, samples were trypsinized and total cell numbers counted. Values represent the average of 3 samples and error bars indicate standard deviation (SD). Wild-type (+/+), heterozygous (+/–), and 2 independently derived null ES lines (–/–) are shown. (B-C) ES cells, predifferentiated for 2 days without the STO-neo feeder layer but in the presence of LIF, were trypsinized and plated in methylcellulose without exogenous cytokines (B) or in liquid suspension culture (C), at a density of 2 × 104 cells/mL. Total number of EBs were counted at day 4 (B). At days 6 and 12, EBs were pooled and the maximum diameters of at least 30 EBs were measured for each sample (C). Values represent the average of 3 replicate platings from one experiment and error bars indicate SD. Similar results were obtained for several independent experiments. Wild type (+/+), Notch1 heterozygous (+/–), and 2 independently derived Notch1 null (–/–) ES cell lines. (D) FACS analysis of flk-1 expression in Notch1–/– and Notch1+/– EBs differentiated for 3.75 days. EBs were dissociated to single-cell suspension and stained with an antibody to flk-1, conjugated to phycoerythrin. The profile indicated by a dotted line represents unstained EB cells. Similar results were obtained in 2 independent experiments, and with additional Notch1–/– and control cell lines. Percentage of cells in the M2 window is shown.
Notch1 regulated the numbers of primitive erythroid colony-forming progenitors during differentiation of embryoid bodies in vitro, but not in vivo
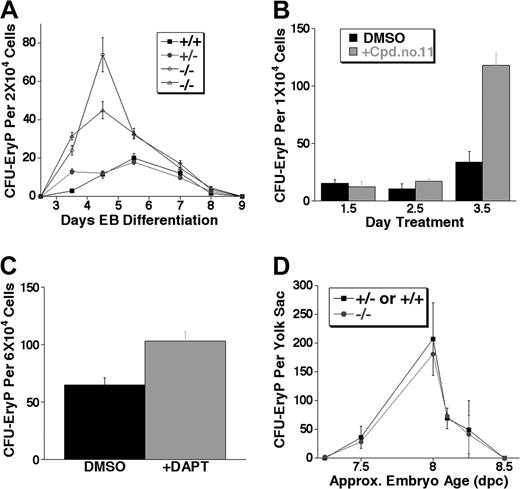
The first detectable hematopoietic progenitors in vivo and in the ES differentiation system are those of the primitive erythroid lineage. To quantitatively test if Notch1 regulates these progenitors, equal numbers of cells from EBs were replated in methylcellulose medium and development of primitive erythroid colony-forming progenitors (CFU-EryPs) scored. For 2 different Notch1–/– ES cell lines, Notch1–/– EBs consistently gave rise to several-fold more primitive erythroid CFU-EryPs than either Notch1+/+ or Notch1+/– EBs, throughout the wave of CFU-EryP formation (Figure 2A).
Notch1-deficient (Notch1in32 allele) and γ-secretase inhibitor–treated embryoid bodies (EBs), but not Notch1-deficient embryos (Notch1Δ1 allele), produce expanded primitive erythroid colony-forming unit (CFU-EryP) progenitors. (A) Kinetic analysis of primitive erythroid progenitor formation from EBs at various stages of differentiation. EBs differentiated in liquid culture for 2.5 to 9 days were assayed for CFU-EryP. Values represent the average of 3 replicate platings. Similar results were obtained in at least 4 independent experiments performed in EBs differentiated for 4 to 6 days. Wild type (+/+), Notch1 heterozygous (+/–), and 2 independently derived Notch1 null (–/–) ES cell lines were examined. (B) Wild-type EBs were treated at 1.5, 2.5, or 3.5 days of differentiation with a single dose of the γ-secretase inhibitor Cpd no. 11 (50 μM) or with the carrier alone, DMSO, as a control. Primitive erythroid progenitors (CFU-EryP) were then assayed at day 5 of differentiation. Values represent the average of 3 replicate platings. Similar results were obtained in an independent experiment. (C) Wild-type EBs were treated at day 3.5 of differentiation with a single dose of the γ-secretase inhibitor, DAPT (1 μM), or with the carrier, DMSO, as a control. Primitive erythroid progenitors (CFU-EryP) were then assayed at day 4.75 of differentiation (P < .01, Student t test). Values represent the average of 3 replicate platings. (D) CFU-EryPs were assayed from Notch1-deficient CD1 embryos (–/–) or heterozygous and wild-type (+/– or +/+) littermate controls from approximately day 7.25 to 8.5 dpc. Each time point represents the average number of CFUs from Notch1-deficient (–/–) or control (+/– or +/+) littermate embryos whose approximate chronologic age was based on morphology and/or somite numbers. Similar results not shown in this graph were obtained in additional experiments performed at various embryonic stages. Approx indicates approximately.
Notch1-deficient (Notch1in32 allele) and γ-secretase inhibitor–treated embryoid bodies (EBs), but not Notch1-deficient embryos (Notch1Δ1 allele), produce expanded primitive erythroid colony-forming unit (CFU-EryP) progenitors. (A) Kinetic analysis of primitive erythroid progenitor formation from EBs at various stages of differentiation. EBs differentiated in liquid culture for 2.5 to 9 days were assayed for CFU-EryP. Values represent the average of 3 replicate platings. Similar results were obtained in at least 4 independent experiments performed in EBs differentiated for 4 to 6 days. Wild type (+/+), Notch1 heterozygous (+/–), and 2 independently derived Notch1 null (–/–) ES cell lines were examined. (B) Wild-type EBs were treated at 1.5, 2.5, or 3.5 days of differentiation with a single dose of the γ-secretase inhibitor Cpd no. 11 (50 μM) or with the carrier alone, DMSO, as a control. Primitive erythroid progenitors (CFU-EryP) were then assayed at day 5 of differentiation. Values represent the average of 3 replicate platings. Similar results were obtained in an independent experiment. (C) Wild-type EBs were treated at day 3.5 of differentiation with a single dose of the γ-secretase inhibitor, DAPT (1 μM), or with the carrier, DMSO, as a control. Primitive erythroid progenitors (CFU-EryP) were then assayed at day 4.75 of differentiation (P < .01, Student t test). Values represent the average of 3 replicate platings. (D) CFU-EryPs were assayed from Notch1-deficient CD1 embryos (–/–) or heterozygous and wild-type (+/– or +/+) littermate controls from approximately day 7.25 to 8.5 dpc. Each time point represents the average number of CFUs from Notch1-deficient (–/–) or control (+/– or +/+) littermate embryos whose approximate chronologic age was based on morphology and/or somite numbers. Similar results not shown in this graph were obtained in additional experiments performed at various embryonic stages. Approx indicates approximately.
We have previously demonstrated that a γ-secretase inhibitor, which reduces Notch signaling by inhibiting the γ-secretase–dependent intramembranous cleavage of the Notch protein, can recapitulate a biologic effect of decreased Notch1 signaling in a relevant assay, the fetal thymus organ culture.62 To test if γ-secretase inhibitors affected primitive erythroid progenitor expansion, we examined the effect of these inhibitors on the hematopoietic potential of wild-type EBs. EBs were treated at a single time point with the inhibitor Compound no. 1161,62 or carrier (DMSO) control, either at day 1.5, 2.5, or 3.5 of differentiation, and were then assayed at day 5 for CFU-EryP activity. Treatment with Cpd no. 11 at day 3.5 of differentiation, a time point near the beginning of primitive erythroid progenitor activity, resulted in the significant expansion of CFU-EryPs assayed at day 5 (Figure 2B). A significant expansion of CFU-EryPs was also observed when day-3.5 EBs were treated with a different γ-secretase inhibitor, DAPT,63,64,68 and assayed at day 4.75 (Figure 2C). The expansion of EB-derived CFU-EryPs in the presence of γ-secretase inhibitors was consistent with the phenotype observed during Notch1-deficient ES cell differentiation, demonstrating that CFU-EryPs were expanded when Notch1 signals are reduced or absent during ES cell differentiation into EBs.
Primitive erythroid progenitor (CFU-EryP) activity occurs during a short wave of early development from approximately 7.0 dpc to 8.5 dpc.1 Since vascular defects and eventual lethality in Notch1-deficient embryos occur after this phase of hematopoiesis, we wished to determine if expansion of primitive erythroid progenitors, observed during the differentiation of Notch1-deficient ES cells, was recapitulated in embryos lacking Notch1. Original experiments in the C57Bl6 background revealed that the CFU-EryP wave of activity in this strain was more transient than that reported previously (in Swiss Webster mice),1 complicating a statistical comparison of CFU-EryP activity between Notch1in32/in32 embryos and littermate controls in this strain. To circumvent this problem, we examined CFU-EryPs from mice of the CD1 strain harboring a deletion allele of Notch1Δ,1 which exhibits an identical phenotype to that of the Notch1in32/in32 mice.58 Surprisingly, we found that at various stages of the primitive erythroid wave examined, Notch1-deficient embryos produced similar numbers of primitive erythroid CFU progenitors as wild-type and heterozygous littermate controls (Figure 2D).
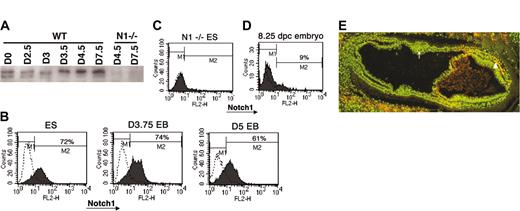
One possibility explaining the discordant effects of Notch1 deletion on primitive erythropoiesis observed during in vitro differentiation of ES cells and in vivo development could be differences in how Notch1 expression or activation is regulated in these 2 systems. It is possible that ectopic expression and activation of Notch1 outside of its normal developmental context can lead to phenotypes that are not physiologically relevant. In the developing embryo, it has been shown that Notch1 mRNA expression was not detectable at 6.5 dpc but became evident by 7.0 to 7.5 dpc, at which time it was primarily restricted to specific subsets of mesodermal tissues.17,23 To characterize Notch1 expression during the differentiation of ES cells, we first performed Western blot analysis using a specific anti-Notch1 antibody.67 ES cells and EBs differentiated for various days were analyzed for Notch1 expression. Notch1 protein was readily detected in undifferentiated ES cells and throughout EB differentiation (Figure 3A). To determine if Notch1 is widely expressed in ES and EB cells in culture or if it is expressed only in a selected subset of cells, we analyzed the Notch1-expressing population using FACS. In contrast to early embryo cells, the majority of ES cells continuously expressed Notch1, both prior to differentiation and during early EB differentiation when the primitive erythroid wave of CFU progenitor formation begins (Figure 3B, D). These results suggested that the temporal and tissue-specific modulation of Notch1 expression during normal development was not accurately reproduced during EB differentiation in vitro.
Expression of Notch1 during ES cell differentiation and in vivo. (A) Western blot analysis of Notch1 expression during EB differentiation. Roughly equal levels of total protein (as determined by serial dilution and Coomassie staining) from lysates of ES cells (plated in the absence of feeder cells) or EBs at the indicated ages (in days) were used to compare endogenous Notch1 protein levels using an anti-Notch1 antibody (AN1). Since the antibody is part of the intracellular domain of Notch1, it detects an approximately 120-kDa band, the transmembrane intracellular (TMIC) portion of Notch1 that is formed during processing of the full-length protein during transport to the cell surface. Analysis was also performed on Notch1-deficient ES cells as a negative control. WT indicates wild type. (B) FACS analysis of Notch1 expression in ES cells and developing embryoid bodies (EBs) at various days of differentiation. ES or EB cells were treated to form a single-cell suspension and stained with a rabbit antibody to the extracellular domain of Notch1, followed by antirabbit biotin secondary and finally streptavidin phycoerythrin. Profiles indicated by dotted lines represent cells stained without primary antibody. (C) FACS analysis of Notch1 expression in a Notch1-deficient ES cell line, treated identically to wild-type lines analyzed in panel B. (D) FACS analysis of Notch1 expression in the early mouse embryo. Approximately 8.25-dpc wild-type embryos were dissociated to single cells and pooled for analysis similar to EB analysis. The profile indicated by dotted lines represents cells stained without primary antibody. Percentage of cells in the M2 window is shown. (E) Staining for an activated epitope of Notch1 in a section of an approximately 7.5-dpc wild-type embryo with an antibody to the N-terminus of the intracellular domain of Notch1 (NICD). Nuclear staining is indicated by Bis-benzimide (green) and NICD is indicated in red. Yellow cells represent specific nuclear NICD staining. Note positive NICD detection in the mesodermal layer of the embryo proper (arrowhead) but absence of detection in the presumptive blood island aggregates in the extraembryonic region (arrow). Similar staining patterns were observed in multiple sections from 3 different wild-type embryos at approximately 7.5 dpc. Image was visualized using Olympus 100 ×/1.25 oil objective lenses. Magnification, × 100.
Expression of Notch1 during ES cell differentiation and in vivo. (A) Western blot analysis of Notch1 expression during EB differentiation. Roughly equal levels of total protein (as determined by serial dilution and Coomassie staining) from lysates of ES cells (plated in the absence of feeder cells) or EBs at the indicated ages (in days) were used to compare endogenous Notch1 protein levels using an anti-Notch1 antibody (AN1). Since the antibody is part of the intracellular domain of Notch1, it detects an approximately 120-kDa band, the transmembrane intracellular (TMIC) portion of Notch1 that is formed during processing of the full-length protein during transport to the cell surface. Analysis was also performed on Notch1-deficient ES cells as a negative control. WT indicates wild type. (B) FACS analysis of Notch1 expression in ES cells and developing embryoid bodies (EBs) at various days of differentiation. ES or EB cells were treated to form a single-cell suspension and stained with a rabbit antibody to the extracellular domain of Notch1, followed by antirabbit biotin secondary and finally streptavidin phycoerythrin. Profiles indicated by dotted lines represent cells stained without primary antibody. (C) FACS analysis of Notch1 expression in a Notch1-deficient ES cell line, treated identically to wild-type lines analyzed in panel B. (D) FACS analysis of Notch1 expression in the early mouse embryo. Approximately 8.25-dpc wild-type embryos were dissociated to single cells and pooled for analysis similar to EB analysis. The profile indicated by dotted lines represents cells stained without primary antibody. Percentage of cells in the M2 window is shown. (E) Staining for an activated epitope of Notch1 in a section of an approximately 7.5-dpc wild-type embryo with an antibody to the N-terminus of the intracellular domain of Notch1 (NICD). Nuclear staining is indicated by Bis-benzimide (green) and NICD is indicated in red. Yellow cells represent specific nuclear NICD staining. Note positive NICD detection in the mesodermal layer of the embryo proper (arrowhead) but absence of detection in the presumptive blood island aggregates in the extraembryonic region (arrow). Similar staining patterns were observed in multiple sections from 3 different wild-type embryos at approximately 7.5 dpc. Image was visualized using Olympus 100 ×/1.25 oil objective lenses. Magnification, × 100.
Finally, we examined the accumulation of activated Notch1 with an antibody specific to the N-terminus of Notch1 intracellular domain (NICD, formed upon cleavage of Notch1 at the transmembrane region by γ-secretase) in early embryo sections. Activated Notch1 epitope was not detected in blood islands in the extraembryonic mesoderm of embryos at 7.5 dpc, a time point when primitive erythroid progenitors are detected in this region (Figure 3E, arrow).1 Consistent with reported mRNA expression of Notch1 at this stage II3 though, we were able to detect a subset of mesodermal cells in the embryo proper in which Notch1 appeared to be activated (Figure 3E, arrowhead).
Early definitive hematopoietic progenitors were not perturbed in the absence of Notch1 in vivo or in differentiating ES cells
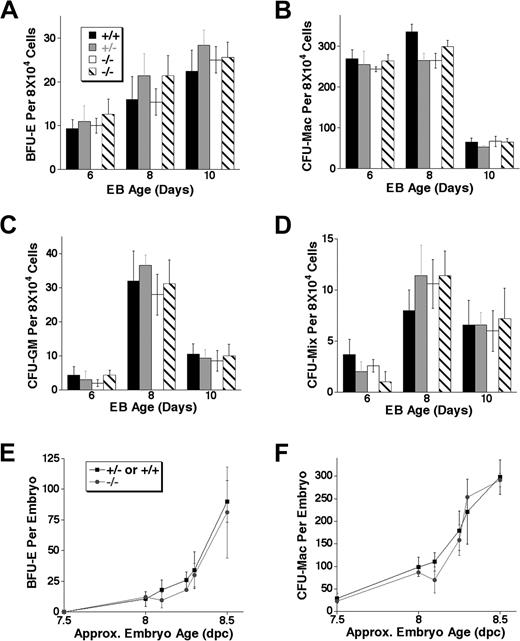
To determine whether the lack of Notch1 affected early definitive hematopoiesis, we first examined the ability of Notch1-deficient EBs to give rise to definitive erythroid and myeloid CFU progenitors. This early definitive progenitor activity can be detected in the ES cell differentiation system in a wave that begins after the onset of primitive erythropoiesis, corresponding to the sequential onset of primitive and definitive CFU activity seen in vivo in the early yolk sac.1 Cells from EBs differentiated for 6 to 10 days were replated in methylcellulose medium in the presence of the appropriate cytokines to detect various definitive colony-forming progenitors, including definitive erythroid (BFU-E, burst-forming unit–erythroid, Figure 4A), myeloid (CFU-Mac, colony-forming unit–macrophage, Figure 4B; and CFU-GM, colony-forming unit–granulocyte/monocyte, Figure 4C), and mixed definitive (CFU-mix, containing erythroid and other myeloid cells, Figure 4D). We found that there was no significant difference in the total number of definitive progenitor colonies derived from Notch1–/– EBs relative to wild-type and Notch1+/– controls at any time point during EB differentiation. Furthermore, colony size and morphology were not noticeably altered in the absence of Notch1 in any of the lineages examined (not shown). These results indicated that though primitive erythropoiesis was altered in the absence of Notch1 during ES cell differentiation, definitive progenitors developed normally without Notch1 in this system.
Notch1-deficient (Notch1in32 allele) EBs and early embryos display normal definitive colony-forming unit (CFU) progenitor activity. (A-D) EBs differentiated in liquid culture for 6, 8, or 10 days were trypsinized to obtain single-cell suspensions and replated to assay for the following definitive colony-forming progenitors: (A) burst-forming unit–erythroid (BFU-E), (B) colony-forming unit–macrophage (CFU-Mac) (C) colony-forming unit–granulocyte/monocyte (CFU-GM), or for (D) colony-forming unit–mix (CFU-Mix). Values represent the average of 3 replicate platings and error bars indicate SD. Similar results were obtained in independent experiments from EBs differentiated for 10 or 12 days. Wild type (+/+), Notch1 heterozygous (+/–), and 2 independently derived Notch1 null (–/–) ES cell lines were examined. (E-F) Notch1-deficient embryos or littermate controls were dissected at various stages of development. The entire embryo or the dissected yolk sac was dispersed to a single-cell suspension and assayed in methylcellulose with cytokines for the following definitive colony-forming progenitors: (E) definitive erythroid (BFU-E) and (F) macrophage (CFU-Mac). Each time point represents the average number of CFUs from Notch1-deficient (–/–) or control (+/– or +/+) littermate embryos whose approximate chronologic age was based on morphology and/or somite numbers. Error bars represent SD. Similar results not shown in this graph were obtained in additional experiments performed at various embryonic stages.
Notch1-deficient (Notch1in32 allele) EBs and early embryos display normal definitive colony-forming unit (CFU) progenitor activity. (A-D) EBs differentiated in liquid culture for 6, 8, or 10 days were trypsinized to obtain single-cell suspensions and replated to assay for the following definitive colony-forming progenitors: (A) burst-forming unit–erythroid (BFU-E), (B) colony-forming unit–macrophage (CFU-Mac) (C) colony-forming unit–granulocyte/monocyte (CFU-GM), or for (D) colony-forming unit–mix (CFU-Mix). Values represent the average of 3 replicate platings and error bars indicate SD. Similar results were obtained in independent experiments from EBs differentiated for 10 or 12 days. Wild type (+/+), Notch1 heterozygous (+/–), and 2 independently derived Notch1 null (–/–) ES cell lines were examined. (E-F) Notch1-deficient embryos or littermate controls were dissected at various stages of development. The entire embryo or the dissected yolk sac was dispersed to a single-cell suspension and assayed in methylcellulose with cytokines for the following definitive colony-forming progenitors: (E) definitive erythroid (BFU-E) and (F) macrophage (CFU-Mac). Each time point represents the average number of CFUs from Notch1-deficient (–/–) or control (+/– or +/+) littermate embryos whose approximate chronologic age was based on morphology and/or somite numbers. Error bars represent SD. Similar results not shown in this graph were obtained in additional experiments performed at various embryonic stages.
Notch1-deficient embryos exhibit severe vascular defects by 9.5 dpc and are absorbed shortly thereafter.53,57,58 Since the first definitive erythroid and myeloid progenitors can be detected in the yolk sac before 8.5 dpc,1 we examined the onset of extraembryonic definitive hematopoietic progenitors in Notch1-deficient embryos. The presence of colony-forming unit (CFU) progenitors in the yolk sac of Notch1-deficient and +/– and +/+ littermate CD1 embryos (Notch1Δ1 allele58 ) was assayed at various stages of development from 7.5 dpc onward. In these early stages of differentiation, the numbers of definitive erythroid (BFU-E, Figure 4E) and early myeloid (CFU-macrophage, Figure 4F) progenitors obtained from Notch1-deficient yolk sacs and control littermate yolk sacs were similar. Furthermore, the size and morphology of colonies observed in Notch1-deficient embryos were not different from littermate controls (not shown). These results suggested that Notch1 did not play a critical role in the onset of definitive hematopoiesis in the yolk sac and were consistent with the results obtained from the differentiation of ES cells lacking Notch1.
LacZ-tagged (ROSA26) Notch1-deficient ES cells contributed to definitive hematopoietic CFUs in the early yolk sac and fetal liver in chimeric mice, but failed to contribute to long-term, definitive hematopoiesis
The development of Notch1-deficient embryos is arrested shortly after 9 dpc, most likely due to severe widespread defects in vascular remodeling.53 Since HSCs capable of reconstituting multilineage hematopoiesis in conditioned newborn mice and adult mice are not detected until approximately 9 dpc and 10 dpc, respectively, experiments determining the long-term definitive HSC potential of cells from Notch1-deficient embryo-derived tissues are likely to be complicated by the vascular and other developmental defects. Thus, in order to determine whether Notch1 is essential for long-term definitive hematopoiesis in later stage embryos and whether this requirement is cell autonomous within hematopoietic precursors, we generated chimeric mice containing both wild-type cells and Notch1-deficient cells marked by the Rosaβgeo26 (ROSA26) gene. We derived marked (ROSA26) ES cells (wild type, heterozygous, or homozygous at the Notch1 locus). These ES lines were then injected into wild-type blastocysts to determine their potential to contribute to hematopoietic lineages in chimeric embryos.
At various stages of development, the yolk sac or fetal liver of chimeric embryos or bone marrow from postnatal mice was removed and cells from these organs were assayed for CFU potential, followed by staining with X-gal, to determine the fraction of colonies that were ES derived (Figure 5). The remaining embryo or individually dissected organs were fixed and stained whole mount with X-gal to determine overall contribution of LacZ+ cells. Only embryos with a high density of blue-staining (LacZ+) cells in most regions were used for hematopoietic analysis. Using 2 different Notch1+/+; ROSA26+ control ES lines, we found the contribution of LacZ+ (ES derived) definitive colonies at all stages of development examined to be 25% or greater (Table 1). Like these wild-type control ES cells, a relatively high percentage contribution of Notch1-deficient ES cells to the definitive CFU progenitor population in the early yolk sac (9.5-10.5 dpc) was observed in high-percentage chimeras (Table 1). This was consistent with the observation that early definitive hematopoiesis was not severely impaired in the yolk sac of Notch1-deficient embryos. At later stages in the yolk sac (11.5-12.5 dpc) and fetal liver (13.5-14.5 dpc), while LacZ+ Notch1-deficient cells were still detected among definitive hematopoietic CFUs, their contribution was comparably reduced (Table 1). At 11.5 dpc or later, the percentage of LacZ+ (Notch1 deficient, ES derived) colonies was less than 15% in all individual chimeras examined, and averaged 4% (Table 1). The size and type of colonies derived from LacZ+ cells were similar to those obtained from LacZ– (Notch+/+, embryo derived) cells (not shown). From 15.5 dpc to postnatal development, Notch1-deficient cells were no longer detected in any CFU progenitors of the fetal liver or bone marrow (Table 1), despite the fact that Notch1-deficient ES cells still contributed in high percentages to other organs in these chimeras (B.K.H., unpublished data, 2004). Similar results were obtained with 3 independent Notch1–/– ES clones. By contrast, no reduction in marked CFU progenitors was detected in chimeras generated with control LacZ+ (ROSA26); Notch1+/+ ES cells. Overall, these results suggested that while Notch1 was not essential for the development of the early wave of definitive progenitors, it was required, cell autonomously, for long-term contribution to definitive hematopoiesis, consistent with a critical role for Notch1 in the development of the definitive HSC.
Contribution of Notch1-deficient and wild-type cells to definitive hematopoiesis in chimeric mice
Chimera age . | Organ assayed . | Notch1 genotype . | No. of chimeras analyzed . | No. of different ES lines . | Percent LacZ+ CFU . | Range . |
|---|---|---|---|---|---|---|
| 9.5 to 10.5 | YS | +/+ | 1 | 1 | 29 | |
| -/- | 6 | 3 | 35 ± 19 | 15-66 | ||
| 11.5 to 12.5 | YS | +/+ | 1 | 1 | 33 | |
| -/- | 8 | 2 | 4 ± 5 | 0-14 | ||
| 13.5 to 14.5 | FL | +/+ | 1 | 1 | 51 | |
| -/- | 2 | 1 | 4.5 | 4-5 | ||
| 15.5 to postnatal | FL/BM | +/+ | 5 | 2 | 40 ± 17 | 25-68 |
| -/- | 10 | 3 | 0 | 0 |
Chimera age . | Organ assayed . | Notch1 genotype . | No. of chimeras analyzed . | No. of different ES lines . | Percent LacZ+ CFU . | Range . |
|---|---|---|---|---|---|---|
| 9.5 to 10.5 | YS | +/+ | 1 | 1 | 29 | |
| -/- | 6 | 3 | 35 ± 19 | 15-66 | ||
| 11.5 to 12.5 | YS | +/+ | 1 | 1 | 33 | |
| -/- | 8 | 2 | 4 ± 5 | 0-14 | ||
| 13.5 to 14.5 | FL | +/+ | 1 | 1 | 51 | |
| -/- | 2 | 1 | 4.5 | 4-5 | ||
| 15.5 to postnatal | FL/BM | +/+ | 5 | 2 | 40 ± 17 | 25-68 |
| -/- | 10 | 3 | 0 | 0 |
Contribution of Notch1-deficient (Notch1Δ1 allele) ES cells (-/-) and control wild-type (+/+) ES cells to definitive hematopoietic CFU progenitors in yolk sac (YS), fetal liver (FL), and bone marrow (BM) of high-percentage chimeras at various stages of development. Colonies were stained with X-gal stain to determine the percentages of overall colonies obtained from ES-derived (LacZ+) and embryo-derived (LacZ-) progenitors. We did not observe any bias in the distribution of erythroid, myeloid (macrophage or monocyte/granulocyte), or mixed colonies in the LacZ+ and LacZ- populations (not shown). For each age range, the number of chimera analyzed and the number of different ES lines used to generate these chimeras are indicated, as well as the average, standard deviation, and range of percent contribution of LacZ+ CFUs.
Discussion
EBs from Notch1-deficient ES cell lines or EBs in which all Notch signaling is blocked with γ-secretase inhibitors produced significantly more primitive erythroid progenitors relative to their wild-type and heterozygous counterparts or DMSO-treated controls, whereas a similar difference in primitive erythroid progenitor potential was not detected between Notch1-deficient embryos and wild-type or heterozygous littermates. We found that Notch1 protein was continuously and extensively expressed in ES cells and in EB cells throughout differentiation, whereas it was previously shown that in the developing embryo Notch1 mRNA expression was initiated between embryonic day 6.5 and 7.517,23 and remained temporally and spatially modulated throughout development. Furthermore, whereas we can detect activation-specific Notch1 epitopes during the time in development in which primitive erythroid progenitors emerge within blood islands (approximately 7.0 dpc), these epitopes are detected only in embryonic mesoderm and not in presumptive extraembryonic blood islands. Thus, one explanation is that the absence of Notch1 does not affect the differentiation of primitive erythroid CFU progenitors in the yolk sac in vivo simply because Notch1 is not active in this tissue at the critical time. However, we cannot rule out the possibility that the fate of the primitive erythroid progenitors is determined in the embryonic mesoderm prior to migration to extraembryonic sites.70 It was recently shown that quantitative differences in primitive erythroid progenitors detected between differentiated Runx1+/+ and Runx1+/– ES cells were not observed between Runx1+/+ and Runx1+/– embryos.71 It was suggested that subtle differences may not be detectable due to the rapid kinetics of the early embryonic waves of hematopoiesis in vivo. Thus, while a subtle role for Notch1 in quantitatively regulating primitive erythroid progenitor formation cannot be ruled out, our in vitro and in vivo analyses demonstrate that Notch1 is not an essential factor in primitive erythroid progenitor development.
In addition to being dispensable for primitive erythropoiesis, we show that Notch1 is not required in vitro or in vivo for the wave of early, transient definitive hematopoiesis preceding development of the HSC. We have demonstrated that Notch1-deficient cells are capable of producing early definitive progenitors based on several criteria. First, the timing of definitive progenitor emergence is similar in Notch1-deficient and wild-type mice yolk sacs and in EBs. The temporal sequence we observe is similar to that reported previously in another wild-type mouse strain1 and EBs from other wild-type ES lines.60 Second, by Wright-Giemsa stain (not shown), we confirmed that these colonies contain cell types representative of definitive hematopoiesis (for example, BFU-Es, containing enucleated erythrocytes). Finally, we have found that Notch1-deficient cells still contribute to a small percentage of CFU progenitors detected in chimeric mice in the 13.5- to 14.5-dpc fetal liver; thus, these progenitors cannot belong to the primitive lineage since primitive progenitor CFU activity is localized solely within the yolk sac during a short window of development (approximately 7 to 8.5 dpc).1
The definitive HSC has the ability to give rise to long-term multilineage hematopoiesis in vivo. To determine whether Notch1 activity is required for development of the definitive HSC, we used a powerful chimera approach in which LacZ-tagged Notch1–/– ES cells are allowed to codevelop with wild-type cells, thereby allowing for the assessment of the potential of Notch1–/– ES cells to contribute to long-term hematopoiesis. We definitively show that Notch1 plays a direct and critical role in regulating the developmental potential of an HSC precursor. These results extend the previous finding of Kumano et al47 by showing that this defect in HSC development in the absence of Notch1 is a cell-intrinsic event.
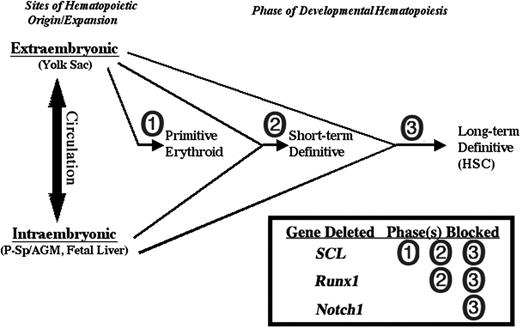
Mice lacking the critical hematopoietic transcription factor SCL have defects in primitive and definitive hematopoietic progenitors,72 and are thus distinct from Notch1-deficient mice in which primitive erythropoiesis and early definitive hematopoiesis is spared. Superficially, mice lacking the transcription factor Runx1 have a phenotype similar to that reported for Notch1-deficient mice in that definitive HSCs are absent.46,73 However, while Runx1-deficient embryos and ES cells fail to give rise to definitive hematopoietic progenitors of any kind, even in the early yolk sac,46,59 here we demonstrate that Notch1-deficient cells can contribute to early definitive CFU progenitors. This distinct aspect of a Notch1 requirement in hematopoietic development suggests a novel stage in the differentiation of the definitive HSC from its precursor, one that distinguishes the self-renewing HSC from earlier definitive hematopoietic progenitors that have only a limited lifespan (Figure 6). Further studies to address the exact identity of the precursor stage at which Notch1 is necessary will require the ability to efficiently and specifically delete Notch1 activity at various steps of development in the early hematopoietic versus endothelial compartments. Such studies will yield novel insight into the origin and nature of the hematopoietic stem cell.
Distinct Notch1 requirement in developmental hematopoiesis. Notch1 is required only for the development of the long-term definitive hematopoietic stem cell compartment while dispensable for earlier, short-term definitive hematopoietic progenitors that are detected in embryoid bodies from in vitro ES cell differentiation and the early yolk sac/fetal liver in vivo. In contrast, the critical hematopoietic transcription factor SCL is required for the development of all hematopoietic progenitors, primitive and definitive,72 and the transcription factor Runx1 is required for all definitive, but not primitive, progenitors.46,59
Distinct Notch1 requirement in developmental hematopoiesis. Notch1 is required only for the development of the long-term definitive hematopoietic stem cell compartment while dispensable for earlier, short-term definitive hematopoietic progenitors that are detected in embryoid bodies from in vitro ES cell differentiation and the early yolk sac/fetal liver in vivo. In contrast, the critical hematopoietic transcription factor SCL is required for the development of all hematopoietic progenitors, primitive and definitive,72 and the transcription factor Runx1 is required for all definitive, but not primitive, progenitors.46,59
Prepublished online as Blood First Edition Paper July 13, 2004; DOI 10.1182/blood-2004-03-1224.
Supported by National Institutes of Health (NIH) grants GM55479 and HD044056, and the Washington University/Pfizer Biomedical Research program (R.K.); NIH grant CA75315, and grant 9940116N from the American Heart Association (G.D.L.). G.D.L. is an Established Investigator of the American Heart Association. B.K.H. was supported in part by the NIH Medical Scientist Training Program.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank E. Ross and J. Mudd of the Washington University School of Medicine (WUSM) ES Core facility; K. McGrath, J. Palis, K. Choi, and P. Faloon for technical advice; Mia Wallace (WUSM Mouse Genetics Core facility) and Yu Mei Wu for blastocyst injections; M. La Ragina for MAP testing; C.L. Hsieh for karyotyping ES lines; and members of the Longmore and Kopan labs for critical discussion and advice. We also thank Grace Wang, a summer student in the lab.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal