Abstract
The AF1q gene, a mixed-lineage leukemia fusion partner, is highly expressed in hematopoietic progenitor cells but has low expression in differentiated cells. We determined the expression of the AF1q gene by reverse transcriptase–polymerase chain reaction in 64 pediatric acute myeloid leukemia (AML) patients treated on Children's Cancer Group clinical trial CCG-2891 and correlated its expression level to clinical characteristics and outcome. AF1q expression in patients varied from 0- to 154-fold compared with normal marrow, and increasing expression level was associated with worsening survival, with a hazard ratio of 1.02 per fold increase in AF1q expression (P = .032). We divided patients into tertile groups based on AF1q expression level. Patients with high AF1q expression (top tertile) had a higher predominance of French-American-British M1 compared to patients with lower 2 tertiles of AF1q expression (43% vs 9%, P = .003). High AF1q expression was associated with poor survival in univariate and multivariate models. Overall survival at 8 years for patients with the high AF1q expression was 19% versus 50% in patients with low AF1q expression, (P = .01). AF1q expression may correlate with clinical outcome in pediatric AML, although it is not clear if AF1q is simply a marker of a more primitive phenotype or contributes directly to leukemogenesis.
Introduction
The normal function of the hematopoietic system depends on precise control of a series of key regulatory genes that govern self renewal, lineage commitment, and differentiation of hematopoietic stem cells and their progeny. In leukemia and lymphoma, these genes are frequently altered by chromosomal translocations, inversions, methylations, or mutations.1-3 Hence, alterations of gene expression levels may result in gain or loss of function.4 Many of these genetic alterations have proven to be useful molecular tools for diagnosis, risk stratification, and minimal residual disease (MRD) monitoring.5-8 In addition, such alterations may also provide potential targets for therapy.9,10
The AF1q gene, located in chromosome 1, band 21, was initially identified as a mixed-lineage leukemia (MLL) fusion partner from an infant acute myelomonocytic leukemia carrying the t(1;11)(q21;q23) translocation.11 There are more than 50 different types of chromosomal translocations involving the MLL gene.12 To date, more than 20 MLL translocation partners have been cloned and characterized.12 In general, the structure of the MLL fusion gene is diverse, and the genes seldom share functional domains, which has made it difficult to develop a unifying theory for how rearranged forms of MLL may contribute to leukemogenesis.13 The MLL partner proteins appear to fall into 2 functional categories: signaling molecules that normally localize to the cytoplasm/cell junctions, or nuclear transcription factors implicated in transcriptional regulation.14 The biologic function of many MLL fusion genes and their impact on leukemogenesis are poorly understood.
The AF1q gene has several unique structural features that differentiate it from other MLL fusion partners. AF1q fuses to the MLL gene in an in-frame fashion with its entire open reading frame (ORF), rather than with a truncated ORF portion as seen in other fusion partners.11,15 There are no well-defined functional domains in the AF1q ORF. The 3′ untranslated region of AF1q contains a series of ATTA-rich sequences, which may contribute to the short half-life of AF1q protein in the intracellular environment.11 AF1q is highly regulated in the human hematopoietic system, and it has been shown that AF1q is highly expressed in CD34-enriched stem cells, and this expression declines by differentiation and is nearly undetectable in peripheral blood.11,16 In addition, AF1q is highly expressed in leukemic cell line NB4, and all-trans retinoic acid (ATRA)–induced differentiation leads to concomitant decline in AF1q expression.17 In CD34-enriched human cord blood stem cells AF1q is coexpressed with other stem cell–related genes such as GATA-2 and STAT5.18
As AF1q has higher expression in immature CD 34+ cells, higher expression of AF1q in myeloid leukemias might identify more “primitive” leukemias with perhaps a worse prognosis. We evaluated the available diagnostic RNA from 64 pediatric AML cases treated on Children's Cancer Group clinical trial CCG-2891 for the expression of AF1q using a quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) assay and studied the association of AF1q expression with clinical characteristics and treatment outcome.
Patients, materials, and methods
Patients and treatment
Newly diagnosed patients with de novo AML registered in Children's Cancer Group (CCG) pediatric AML protocol CCG-2891 were included in this study. Archived RNA from 64 de novo pediatric AML diagnostic marrow samples was available for analysis. The diagnosis of AML was made according to French-American-British (FAB) classification and was confirmed by the CCG-Central Review Committee. All patients had consented to use of diagnostic marrow specimens for biology studies at the time of study entry. This study was approved by the Fred Hutchinson Cancer Research Center Institutional Review Board.
CCG-2891 has been described in detail in other publications.19,20 In brief, CCG-2891 was a prospective randomized trial that accrued 887 de novo AML patients from 1989 to 1995 (excluding Down syndrome AML, myelodysplastic syndrome, secondary AML, or granulocytic sarcoma without marrow involvement). This study randomized pediatric patients with AML at diagnosis to receive 1 of 2 induction regimens that involved 4-day cycles of 5 chemotherapeutic agents (daunomycin, cytarabine, etoposide, 6-thioguannine, and dexamethasone). The following cycles were administered either after 10 days, despite low counts (intensive timing), or after 14 days, depending on the marrow status (standard timing). Induction regimen consisted of a total of 4 cycles in both groups. At the end of induction, patients who achieved remission and had an HLA-matched related donor were eligible to receive an allogeneic bone marrow transplant (allo-BMT). Patients without related donors were randomized to receive either nonmyeloablative chemotherapy or an autologous BMT (auto-BMT).
RNA isolation and cDNA synthesis
Bone marrow samples from patients in the CCG2891 trial were obtained prior to therapy. Total RNA were prepared by using the TRIzol extraction kit (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. The cDNAs were synthesized from 0.5 μg total RNA by using Superscript reverse transcriptase system in 20 μL volume (Invitrogen). The cDNA synthesis was performed under the following conditions recommended by Invitrogen: 42°C for 50 minutes, then 72°C for 15 minutes.
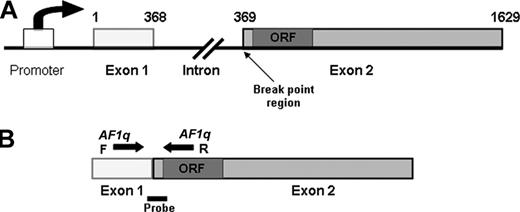
Quantitative PCR primers and fluorescent probe design
The structure of the AF1q gene is shown in Figure 1. The AF1q gene contains 2 exons, and the forward and reverse primers were designed to cross the exon/intron borders in order to avoid genomic DNA amplification. The primer sequences are as follows: AF1qF5′-GCA CTC CCT CCA TCT TTG GA-3′; AF1qR5′-CAG CTC CGA CAG ATC CAG TTC-3′. The size of the PCR amplicon is 133 base pair (bp). The fluorescent probe sequence was: 5′-ACC CTG TGA GTA GCC AGT ACA GTT CCT TTC TTT TC-3′ (Synthegen, LLC; Houston, TX).
AF1q gene structure. (A) AF1q gene contains 2 exons separated by a 9-kb intron. Numbers above the gene represent the nucleotide sequence of the AF1q cDNA. Open reading frame (ORF) is located near the N-terminus of exon 2, proximal to the break-point region. (B) Primers (arrows) and TaqMan probe (solid bar) for the quantitative PCR are represented.
AF1q gene structure. (A) AF1q gene contains 2 exons separated by a 9-kb intron. Numbers above the gene represent the nucleotide sequence of the AF1q cDNA. Open reading frame (ORF) is located near the N-terminus of exon 2, proximal to the break-point region. (B) Primers (arrows) and TaqMan probe (solid bar) for the quantitative PCR are represented.
Quantitative PCR standard curve
The full-length AF1q and β2 microglobulin cDNAs were cloned into the Bluescript plasmid at EcoRI and XbaI sites, respectively. The plasmid DNA that contains both AF1q and β2 microglobulin genes was purified by standard methods (Qiagen, Valencia, CA). A standard curve stock was made that contains 107 molecules/μL of AF1q/β2 plasmid. The standard curves were generated by amplification of a 133-bp AF1q sequence and 80-bp β2 microglobulin sequence from dilution series ranging from 107 to 1 of AF1q/β2 plasmid.
PCR quantitation
The real-time quantitative PCR analysis was performed using the standard TaqMan PCR Core Reagent Kit (Applied Biosystems, Foster City, CA) in a 50-μL reaction on patient's synthesized cDNA. Each quantitative PCR reaction mix contains the cDNA equivalence to 100 ng of total RNA. We performed the quantitative PCR reaction in ABI PRISM 7700 Sequence Detector under the following conditions: stage 1, 50° C for 2 minutes, 95°C for 10 minutes for one cycle; and stage 2, 95°C for 15 seconds, 60°C for 1 minute for a total of 40 cycles. β2 microglobulin gene was used as an internal control to normalize the difference of amount of cDNA in each reaction. AF1q expression level was calculated as the ratio of AF1q to β2 microglobulin amplification and then normalized to normal control bone marrow. Five marrow samples from volunteer donors were used to determine the median control AF1q expression.
Mutational analysis
Analyses of diagnostic marrow specimens of this population for activating mutations of the FLT3 and ras genes were performed and reported previously.21
Statistical methods
Data obtained in CCG-2891 through July 31, 2001, were used to compare patients with various levels of AF1q expression. The significance of observed differences in proportions was tested using the Chi-squared test and Fisher exact test when data were sparse. For continuous data, the Mann-Whitney test was used to compare the medians of distributions.22 Patients lost to follow-up were censored at their last known point of study, with a cutoff of January 31, 2001. Patients who received a hematopoietic cell transplant were censored at the time of transplantation. Actuarial estimates of overall survival (OS) and event-free survival (EFS) from diagnosis, defined as the time from diagnosis to either death (OS) or marrow relapse or death (EFS), were calculated using the Kaplan-Meier method.23 The Kaplan-Meier method also was used to calculate estimates of OS and disease-free survival (DFS) from the end of induction for patients in remission at the end of induction. DFS was defined as the time from the end of induction to relapse or death by any cause. Confidence intervals were calculated using Greenwood estimate of the standard error.24 Differences in OS, EFS, and DFS were tested for significance using the log-rank statistic.25
Factors significant in univariate analysis at P less than .05 were considered for inclusion in multivariate Cox regression models.26 The likelihood ratio test was used to determine whether variables should be added or dropped from the multivariate model. Multivariate analyses included patients with complete covariate data. Statistical corrections for multiple comparisons were not employed.
The positive predictive value (PPV) of high AF1q expression, that is, the likelihood of a person with high AF1q expression dying, was estimated as 1 minus the Kaplan-Meier estimate of 8-year OS for patients with high AF1q expression. The negative predictive value (NPV) of low AF1q expression, that is, the likelihood of a person with low AF1q expression surviving, was estimated as the Kaplan-Meier estimate of 8-year OS for patients with low AF1q expression. PPV and NPV were estimated in a similar manner for nonwhite race, FLT/ITD, and high-risk cytogenetics.
Results
Patient characteristics—study population versus CCG-2891
We initially examined the clinical characteristics of the study population to determine if it was representative of the CCG-2891 de novo AML patients. A comparison of median age, sex, induction regimen, race, and remission induction rate showed no statistically significant difference between our study patients and the rest of the CCG-2891 patients (Table 1). As expected, the diagnostic white blood cell count (WBC) of the archived samples was higher in our study population (45 900/mm3) than the rest of the CCG-2891 population (20 400/mm3) (P = .0004). The study population had a complete remission (CR) rate of 68%, which was lower than the CR rate of 78% for the CCG-2891 study; however, this difference was not statistically significant (P = .21). Actuarial overall survival (OS) at 8 years from on-study and from remission was similar between our study population and the rest of the CCG-2891 patients (P = .636 and .45, respectively).
Patient characteristics in the study population versus CCG 2891 population
. | AF1q study population . | Rest of CCG2891 . | P . |
|---|---|---|---|
| No. | 64 | 823 | NA |
| Median age, y | 8.5 | 7.7 | .532 |
| Median WBC count, per mm3 | 45 900 | 20 400 | .0004 |
| Male, no. (%) | 34 (53.1) | 421 (51.2) | .862 |
| Intensive induction, no. (%) | 40 (62.5) | 499 (60.6) | .871 |
| Race, overall | .289 | ||
| White, no. (%) | 47 (73) | 550 (67) | .343 |
| Nonwhite, no. (%) | 17 (27) | 273 (33) | |
| CR rate, % | 68.3 | 77.7 | .132 |
| 8-year OS from diagnosis, % | 40 | 40 | .636 |
| 8-year OS from remission, % | 48 | 52 | .445 |
. | AF1q study population . | Rest of CCG2891 . | P . |
|---|---|---|---|
| No. | 64 | 823 | NA |
| Median age, y | 8.5 | 7.7 | .532 |
| Median WBC count, per mm3 | 45 900 | 20 400 | .0004 |
| Male, no. (%) | 34 (53.1) | 421 (51.2) | .862 |
| Intensive induction, no. (%) | 40 (62.5) | 499 (60.6) | .871 |
| Race, overall | .289 | ||
| White, no. (%) | 47 (73) | 550 (67) | .343 |
| Nonwhite, no. (%) | 17 (27) | 273 (33) | |
| CR rate, % | 68.3 | 77.7 | .132 |
| 8-year OS from diagnosis, % | 40 | 40 | .636 |
| 8-year OS from remission, % | 48 | 52 | .445 |
NA indicates not applicable.
AF1q expression in normal marrow and in CD34 selected progenitor cells
AF1q expression was determined in 5 marrow specimens obtained from healthy donors by real-time quantitative RT-PCR and corrected for the β2 microglobulin expression levels. AF1q expression ranged from 1.54 × 105 to 2.36 × 105AF1q copies per microgram of total RNA, with a median of 2 × 105 copies per microgram RNA in the normal marrow. We further evaluated the expression level in CD34 selected peripheral blood stem cells. AF1q expression level was 1.8 × 106 copies per microgram of total RNA, approximately 9-fold higher than that in normal marrow.
AF1q expression level varies in pediatric AML
AF1q expression level was determined for the 64 patients. AF1q expression ranged from 0 to3 × 107 copies per microgram RNA and normalized to the median AF1q expression level in the normal control bone marrows. After normalization for control marrow, AF1q expression levels ranged from 0- to 154.2-fold of the expression in the normal bone marrow (median, 4.7-fold). Compared with the normal marrow control, 54 of the 64 (84%) patients in our study population had elevated AF1q expression, and the remaining 10 (16%) either lacked AF1q expression (n = 1) or had expression equal to or lower than healthy volunteer marrow (n = 9). To test whether variation in AF1q expression might be due to the variation in blast percentage, we compared the marrow blast percentage in the lowest tertile of AF1q expression to that of the middle and highest tertile of AF1q expression of the population. Blast percentage of the one third of patients with the lowest AF1q expression (first tertile) was 81.5%, compared with 80% and 86% in patients with the mid-range (second tertile) or high (third tertile) AF1q expression (P = .36).
High AF1q expression is associated with poor outcome in pediatric AML
We studied whether AF1q expression level correlates with clinical outcome in pediatric patients with AML. For survival determination, patients who had received allogeneic stem cell transplantation were censored at the time of transplant. Patients who had AF1q expression at or below the normal range had an overall survival of 60%, versus 40% for those with elevated AF1q expression (P = .41). We subsequently evaluated the AF1q expression as a continuous variable, showing that increasing AF1q expression level was associated with worsening overall survival with a hazard ratio (HR) of 1.02 per each fold increase in AF1q expression (P = .031; Table 2). For example, a 10-fold increase in AF1q expression would increase risk of death by 1.20-fold (or 20%). To further delineate the clinical significance of AF1q expression, we divided the study population into 3 equal groups (tertiles) based on AF1q expression level and asked if clinical outcome differed based on AF1q expression. The first tertile included patients with the lowest AF1q expression (0- to 2.8-fold the normal control, n = 22), the second tertile included patients with mid-range AF1q expression (3- to 8.5-fold the normal control, n = 21), and the third tertile included patients with highest AF1q expression (≥ 9-fold normal control, n = 21). Cox regression analysis revealed that increasing the AF1q expression by tertiles was associated with worse survival, as patients in the second and third tertile had increasing risk of death compared with the patients in the first tertile. Actuarial OS at 8 years from diagnosis for the patients in the first tertile was 59% compared with 37% and 19% (5-year OS) for the patients in the second and third tertiles, respectively, (Table 2, P = .035).
Survival of patients with elevated AF1q expression
AF1q expression by tertiles . | HR versus first tertile . | P . | Overall survival, %* . | P . |
|---|---|---|---|---|
| First tertile | NA | NA | 59 | .035 |
| Second tertile | 1.4 | .483 | 37 | NA |
| Third tertile | 2.9 | .019 | 19 | NA |
AF1q expression by tertiles . | HR versus first tertile . | P . | Overall survival, %* . | P . |
|---|---|---|---|---|
| First tertile | NA | NA | 59 | .035 |
| Second tertile | 1.4 | .483 | 37 | NA |
| Third tertile | 2.9 | .019 | 19 | NA |
AF1q expression as a continuous variable: HR per fold increase in AF1q expression = 1.02, and P = .32. First tertile versus second tertile, P = .476; second tertile versus third tertile, P = .063; first tertile versus third tertile, P = .026. For the first tertile, N = 22; for the second and third, N = 21.
NA indicates not applicable.
Pairwise comparisons for overall survival.
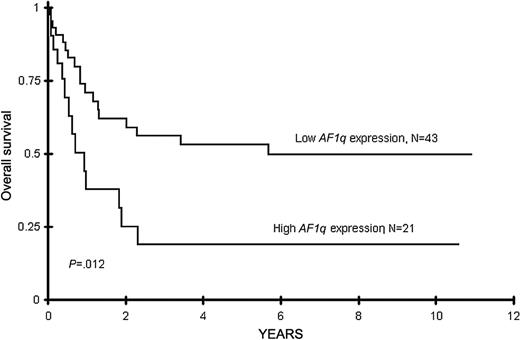
We also compared the clinical outcome of the patients with the highest AF1q expression (ie, third tertile) to the remaining two thirds of the patients with lower AF1q expression. Remission induction rate was similar between the patients with low and high AF1q expression (70% vs 66%, P = .922). Patients with lower AF1q expression (n = 43) had an actuarial OS at 8 years from diagnosis of 50% compared to 19% for the patients with high AF1q expression (n = 21, P = .01) (Figure 2). Actuarial DFS at 8 years from remission in patients with lower AF1q expression (n = 28) was 50% compared with 0% (n = 12) in patients with high AF1q expression (P < .001), as patients with high AF1q expression had a relapse rate of 100% at 8 years compared with 50% in patients with lower AF1q expression (Table 3, P = .002). Thus, increasing AF1q appeared to correspond with poor clinical outcome, and high AF1q expression level distinguished patients with significantly higher relapse rate and worse outcome. In contrast, of the 14 high-AF1q patients who achieved a CR, 4 went on to receive a matched family donor stem cell transplant in first CR. Of these 4 patients, 3 are long-term survivors, with 1 patient dying of nonleukemic causes more than 6 years after transplantation. The remaining 10 patients with high AF1q who were in CR and did not have matched family donors were randomized to autologous stem cell transplantation or continued chemotherapy. All of these patients relapsed and died (6 with early relapse, 2 after autologous stem cell transplantation, and 2 from relapse after completing chemotherapy).
The Kaplan-Meier estimates for overall survival from diagnosis in AML patients with high or low AF1q expression. High AF1q expression is defined as highest tertile and low expression is defined as the lower 2 tertiles of AF1q expression.
The Kaplan-Meier estimates for overall survival from diagnosis in AML patients with high or low AF1q expression. High AF1q expression is defined as highest tertile and low expression is defined as the lower 2 tertiles of AF1q expression.
Patient characteristics in the AF1q study population
. | Low AF1q expression* . | High AF1q expression† . | P . |
|---|---|---|---|
| No. | 43 | 21 | NA |
| Median age, y | 8.6 | 8.3 | .721 |
| Median WBC count per mm3 | 637 000 | 437 000 | .403 |
| No. of males (%) | 21 (49) | 13 (62) | .473 |
| No. of nonwhites (%) | 10 (23) | 7 (33) | .578 |
| Intensive induction, no. (%) | 25 (58) | 15 (71) | .45 |
| CR rate, % | 70 | 65 | .922 |
| FAB classification, no. (%) | .057 | ||
| M0 | 1 (2.3) | 0 (0) | .99 |
| M1 | 4 (9.3) | 9 (42.8) | .003 |
| M2 | 12 (27.9) | 4 (19.0) | .547 |
| M3 | 3 (12) | 1 (4.8) | .99 |
| M4 | 15 (43.8) | 4 (19.0) | .251 |
| M5 | 6 (13.9) | 2 (9.5) | .99 |
| M6 | 2 (4.6) | 0 (0) | .99 |
| M7 | 0 (0) | 1 (4.8) | .328 |
| Cytogenetics, no. (%) | |||
| No. | 25 | 12 | .569 |
| Normal | 8 (32) | 6 (50) | .47 |
| t(15;17) | 2 (8) | 1 (8.3) | .99 |
| t(8;21) | 3 (12) | 1 (8.3) | .99 |
| 11q23 | 6 (24)‡ | 1 (8.3)§ | .389 |
| Del 7 | 0 (0) | 0 (0) | NA |
| Del 5 | 0 (0) | 1 (8.3) | .324 |
| Others | 5 (20) | 1 (8.3) | .641 |
| Mutation status, no. (%) | |||
| No. | 43 | 21 | .082 |
| No mutation | 23 (53.5) | 10 (47.6) | .791 |
| K-ras | 3 (7) | 2 (9.5) | .99 |
| N-ras | 9 (21) | 0 (0) | .025 |
| Flt3/ITD | 5 (11.6) | 5 (23.8) | .275 |
| Flt3/ALM | 2 (4.6) | 3 (14.3) | .320 |
| All FLT3 mutations | 7 (16.2) | 8 (38.1) | .066 |
| Survival/relapse | |||
| 8-year OS from diagnosis, % | 50% | 19% | .01 |
| 8-year DFS from remission, % | 50% | 0% | .001 |
| Relapse rate at 8 years, % | 50% | 100% | .002 |
. | Low AF1q expression* . | High AF1q expression† . | P . |
|---|---|---|---|
| No. | 43 | 21 | NA |
| Median age, y | 8.6 | 8.3 | .721 |
| Median WBC count per mm3 | 637 000 | 437 000 | .403 |
| No. of males (%) | 21 (49) | 13 (62) | .473 |
| No. of nonwhites (%) | 10 (23) | 7 (33) | .578 |
| Intensive induction, no. (%) | 25 (58) | 15 (71) | .45 |
| CR rate, % | 70 | 65 | .922 |
| FAB classification, no. (%) | .057 | ||
| M0 | 1 (2.3) | 0 (0) | .99 |
| M1 | 4 (9.3) | 9 (42.8) | .003 |
| M2 | 12 (27.9) | 4 (19.0) | .547 |
| M3 | 3 (12) | 1 (4.8) | .99 |
| M4 | 15 (43.8) | 4 (19.0) | .251 |
| M5 | 6 (13.9) | 2 (9.5) | .99 |
| M6 | 2 (4.6) | 0 (0) | .99 |
| M7 | 0 (0) | 1 (4.8) | .328 |
| Cytogenetics, no. (%) | |||
| No. | 25 | 12 | .569 |
| Normal | 8 (32) | 6 (50) | .47 |
| t(15;17) | 2 (8) | 1 (8.3) | .99 |
| t(8;21) | 3 (12) | 1 (8.3) | .99 |
| 11q23 | 6 (24)‡ | 1 (8.3)§ | .389 |
| Del 7 | 0 (0) | 0 (0) | NA |
| Del 5 | 0 (0) | 1 (8.3) | .324 |
| Others | 5 (20) | 1 (8.3) | .641 |
| Mutation status, no. (%) | |||
| No. | 43 | 21 | .082 |
| No mutation | 23 (53.5) | 10 (47.6) | .791 |
| K-ras | 3 (7) | 2 (9.5) | .99 |
| N-ras | 9 (21) | 0 (0) | .025 |
| Flt3/ITD | 5 (11.6) | 5 (23.8) | .275 |
| Flt3/ALM | 2 (4.6) | 3 (14.3) | .320 |
| All FLT3 mutations | 7 (16.2) | 8 (38.1) | .066 |
| Survival/relapse | |||
| 8-year OS from diagnosis, % | 50% | 19% | .01 |
| 8-year DFS from remission, % | 50% | 0% | .001 |
| Relapse rate at 8 years, % | 50% | 100% | .002 |
NA indicates not applicable.
Low is defined as lower 2 tertiles of AF1q expression.
High is defined as highest tertile of AF1q expression.
Two t(9;11), 2 del (11)(q23), 1 t(4;11).
t(9;11).
Characteristics of the patients with elevated AF1q expression
We compared the laboratory and clinical characteristics of patients with high (top tertile) and low (lower 2 tertiles) AF1q expressions. The parameters of sex, age, diagnostic WBC, and induction regimen in patients with high AF1q expression were compared to that of patients with lower AF1q expression. Patients with high or low AF1q expression had similar demographics and clinical characteristics (Table 1). Diagnostic WBC was lower in patients with high AF1q expression, but this difference did not achieve statistical significance (43 700 vs 63 700, P = .4, Table 3). Of the 21 patients with high AF1q expression, 9 (43%) had an M1 FAB morphology compared with 4 (9%) of the 43 of the patients with low AF1q expression (P = .003).
Thirty-seven patients had cytogenetic information available, of which 12 had high AF1q expression and the other 25 had low AF1q expression. Of the 12 patients with high AF1q expression, 6 (50%) and 8 of 25 patients (32%) with low AF1q expression had normal karyotype (P = .47). There was no predominance of a particular cytogenetic abnormality in the patients with high AF1q expression. Of the 7 patients with 11q23, 6 had low AF1q expression (P = .389), and there were no t(1;11) translocations. Only 1 patient with high AF1q expression had high-risk cytogenetics (deletion 5) (Table 3).
All 64 patients in this study were previously screened for activating mutations of the FLT3 and ras genes. In the high AF1q expression group, 5 (24%) patients had FLT3 internal tandem duplication (FLT3/ITD), and an additional 3 (14%) patients had activation loop mutation of the FLT3 gene (FLT3/ALM). Activating mutations of the FLT3 gene was seen in 7 of 43 (16%) patients with low AF1q expression (5 FLT3/ITD and 2 FLT3/ALM). Prevalence of FLT3 activating mutations was 38% versus 16% in the high versus low AF1q expression group (P = .066, Table 3). Mutations in the ras gene were present in 9.5% of the patients with high AF1q expression, compared with 28% in the low AF1q expression (P = .118), and specifically, N-ras mutations were not seen in the high AF1q group (21% vs 0%, P = .025, Table 3). Thus, activating mutations of the FLT3 or ras genes were seen in 10 of 21 (48%) of the patients with high AF1q and in 20 of 43 (47%) patients in the patients with low AF1q expression (P = .93).
Prognostic significance of high AF1q expression
Cox regression analysis was used to determine the prognostic significance of high AF1q expression and compare it with that of other known prognostic factors. Specifically, we used univariate analysis to assess the prognostic significance of AF1q expression level (high vs low), induction timing (intensive vs standard timing), diagnostic WBC (< vs > 100 000/mm3), race (nonwhite vs white), ras (activated vs wild type), FLT3/ITD (positive vs negative), FAB (M1 vs other FAB phenotypes), and cytogenetics (high risk vs other). The HR for death for patients with high AF1q expression was 2.4 compared to those with low AF1q expression (P = .015). Induction timing, diagnostic WBC count higher than 100 000, activated ras, and nonwhite race also were univariately associated with adverse overall survival (Table 4). Patients with FLT3/ITD, FAB M1 classification, and high-risk cytogenetics had HR for death of 1.78, 1.24, and 1.53, respectively, although they did not reach statistical significance (Table 4). Multivariate analysis of the univariate predictors of poor outcome (high AF1q, standard timing induction regimen, high diagnostic WBC, race, activated ras, and FLT3/ITD) revealed that high AF1q expression was the single independent prognostic factor for adverse survival from diagnosis (Table 4).
Cox regression analysis of prognostic factors for survival from diagnosis
. | Univariate . | . | . | Multivariate . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | HR . | 95% CI . | P . | HR . | 95% CI . | P . | ||||
| AF1q expression, high versus low | 2.41 | 1.19-4.88 | .015 | 3.05 | 1.31-7.09 | .009 | ||||
| Timing, standard versus intensive | 1.24 | 1.02-1.49 | .03 | 0.88 | 0.39-1.98 | .758 | ||||
| WBC, at least 100 versus fewer than 100, mm3 | 1.29 | 1.02-1.62 | .033 | 1.41 | 0.60-3.35 | .430 | ||||
| Race, nonwhite versus white | 1.30 | 1.07-1.59 | .008 | 0.94 | 0.41-2.19 | .893 | ||||
| ras gene, activated versus wild type | 2.04 | 1.15-3.63 | .015 | 1.76 | 0.67-4.65 | .254 | ||||
| FLT3/ITD, positive versus negative | 1.78 | 0.89-3.54 | .101 | 1.13 | 0.39-3.30 | .820 | ||||
| FAB, M1 versus other FAB | 1.24 | 0.51-3.03 | .636 | 0.74 | 0.27-2.03 | .557 | ||||
| Cytogenetics, high-risk versus other | 1.53 | 0.91-2.59 | .111 | — | — | — | ||||
. | Univariate . | . | . | Multivariate . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | HR . | 95% CI . | P . | HR . | 95% CI . | P . | ||||
| AF1q expression, high versus low | 2.41 | 1.19-4.88 | .015 | 3.05 | 1.31-7.09 | .009 | ||||
| Timing, standard versus intensive | 1.24 | 1.02-1.49 | .03 | 0.88 | 0.39-1.98 | .758 | ||||
| WBC, at least 100 versus fewer than 100, mm3 | 1.29 | 1.02-1.62 | .033 | 1.41 | 0.60-3.35 | .430 | ||||
| Race, nonwhite versus white | 1.30 | 1.07-1.59 | .008 | 0.94 | 0.41-2.19 | .893 | ||||
| ras gene, activated versus wild type | 2.04 | 1.15-3.63 | .015 | 1.76 | 0.67-4.65 | .254 | ||||
| FLT3/ITD, positive versus negative | 1.78 | 0.89-3.54 | .101 | 1.13 | 0.39-3.30 | .820 | ||||
| FAB, M1 versus other FAB | 1.24 | 0.51-3.03 | .636 | 0.74 | 0.27-2.03 | .557 | ||||
| Cytogenetics, high-risk versus other | 1.53 | 0.91-2.59 | .111 | — | — | — | ||||
— indicates we were unable to obtain estimate because cytogenetics had zero variance when added to the multivariate model.
We also used the univariate and multivariate analyses to ascertain the prognostic significance of the aforementioned parameters on death or relapse from the time of achieving remission. Patients with high AF1q expression had a nearly 5-fold higher risk of relapse or death from the time of achieving remission (P = .002). After accounting for induction regimen, diagnostic WBC, race, activated ras, and FLT3/ITD in the multivariate model, the hazard ratio for relapse or death remained elevated at more than 6 (P = .002). In the univariate model induction regimen, race, activated ras, and diagnostic WBC were associated with poor DFS; however, in the multivariate model that included high AF1q expression, all parameters lost clinical significance while high AF1q expression remained a strong predictor of outcome.
Finally, we assessed the predictive values (positive and negative) of high AF1q expression for survival and compared the values with those of other established prognostic factors. In this study, positive predictive value (PPV) for AF1q expression (likelihood of a person with high AF1q expression dying) was 81% compared with negative predictive value (NPV) of 50% (likelihood that a person with low AF1q expression would survive). The PPV and NPV for WBC higher than 100 000, nonwhite race, FLT3/ITD, and high-risk cytogenetics were 65% and 42%, 65% and 43%, 78% and 44%, and 73% and 42%, respectively.
Discussion
In this study we demonstrate significant variation in expression of AF1q gene in pediatric AML and show that AF1q expression level correlates with clinical outcome in this population. Patients with high AF1q expression had a remission induction rate similar to those with lower expression, but suffered a higher relapse rate. The poor outcome associated with high AF1q expression remained after adjustment for other factors associated with poor outcome. As this preliminary study was conducted in a subset of patients from the CCG-2891 study, these results should be interpreted with caution. However, the study population did not significantly differ from the CCG-2891 population except for having a higher white count, which is difficult to avoid in retrospective analyses. The underlying reason for such a difference is likely the higher proportion of samples with high white counts in the reference laboratory, as higher WBC may also explain the somewhat lower (though statistically not significant) CR rate in this study group. Taken together, the data suggest that high AF1q expression may be a new determinant of poor outcome in pediatric AML. This study also suggested that high expression of AF1q was an independent prognostic factor for poor outcome. Patients with high AF1q had lower white counts, had higher proportion of patients who received intensive induction (which should affect the outcome favorably in this population). Despite these 2 favorable factors among patients with high AF1q expression, these patients had a substantially worse outcome. Interestingly, patients with high AF1q expression had a higher percentage of FAB M1 but a lower percentage of M4 subtype compared to patients with low AF1q expression (M1: 42.8% vs 9.3%, P = .003; M4: 43.8% vs 19%, P = .251). The data seemed to support that high AF1q expression might be related to less-differentiated FAB subtypes. However, increased HR in patients with M1 subtype did not reach statistical significance (HR 1.24, P = .636). This was consistent with recent observations reported by CCG2891 investigators that patients with minimally differentiated AML-M0 had no differences in demographic, cytogenetics, and clinical outcome when compared to patients with more-differentiated non-M0 subtypes.27 There was no predominance of high-risk cytogenetics in the high AF1q group, and indeed, more than 90% of the patients with high AF1q had normal or traditionally considered “favorable risk” cytogenetics. Furthermore, in multivariate analysis, which adjusted for known prognostic factors including WBC, race, FLT3/ITD, and activated ras, high AF1q expression was the sole independent prognostic factor in this patient population.
To date, there have been more than 20 MLL fusion partner genes cloned. The functions of most of these genes are unknown, as are the gene expression levels in leukemia cases. This study demonstrates the variation in expression of this MLL fusion partner in leukemia and shows that higher expression level of the AF1q gene is associated with an adverse clinical outcome. AF1q expression is highly regulated in human and murine hematopoietic cell differentiation processes,11,17 and AF1q expression is markedly elevated in undifferentiated progenitor cells.16 This observation was further supported by the fact that AF1q expression was highest in acute promyelocytic leukemic (APL) cell line NB4 entering S phase and declined to nearly undetectable levels after ATRA-induced differentiation.16 Gene expression profile studies have demonstrated direct correlation between AF1q, WT-1, and c-kit gene expression and indirect correlation with CEBPα expression (D.L.S. and W.T., unpublished data, June 2003). These observations suggest that AF1q gene expression may be dysregulated in leukemia and that the level of expression might reflect the differentiation and proliferative state of the leukemia. There are, however, examples of genes that are expressed in immature hematopoietic cells yet not related to clinical significance or FAB classification. For example, c-kit expression predicted a higher CR rate to induction therapy in adult AML but had no effect on OS, DFS, and FAB classification.28 In addition, the expression of ELF-1 and MEF genes are linked to hematopoietic cell differentiation and proliferation, but may be differentially associated with phenotype and outcome, as a study showed that ELF-1 expression appears not related to FAB classification and clinical outcome, whereas MEF expression level is associated with FAB M2/M3 and a favorable prognosis.29
The function of AF1q currently is unknown. However, given that its high expression appears clinically relevant and associated with a significantly poor outcome, studies examining the structure, function, and biologic properties of AF1q may clarify its clinical significance. Although it is possible that high AF1q may play an integral part in leukemogenesis, it may also be that high AF1q expression is simply a surrogate marker for more-primitive, less-differentiated leukemia with high resistance to nonmyeloablative chemotherapy. Clearly further basic research is needed to understand the biology of this gene. In addition, the clinical use of AF1q expression needs to be further evaluated in large clinical trials. Such large studies would have adequate power to appropriately validate its clinical significance in the context of other prognostic markers. Evaluation of the prognostic significance of the AF1q gene expression in large multi-institutional trials are currently under way.
Prepublished online as Blood First Edition Paper, June 24, 2004; DOI 10.1182/blood-2003-12-4347.
W.T. and S.M. contributed equally to this work.
Supported by National Institutes of Health training grant T32 (W.T.); Lauri Strauss Leukemia Foundation (W.T.); the National Institutes of Health K23 (D.L.S.); and the National Institutes of Health CA 18029 (J.P.R.), FIJC-99/INT (S.M.), and CHRC NIA 62-4532 (S.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal