Abstract
Cellular inactivation through killer immunoglobulin-like receptors (KIRs) may allow neoplastic cells to evade host natural killer (NK) cell–mediated immunity. Recently, alloreactive NK cells were shown to mediate antileukemic effects against acute myelogenous leukemia (AML) after mismatched transplantation, when KIR ligand incompatibility existed in the direction of graft-versus-host disease (GVHD). Therefore, we investigated whether solid tumor cells would have similar enhanced susceptibility to allogeneic KIR-incompatible NK cells compared with their KIR-matched autologous or allogeneic counterparts. NK populations enriched and cloned from the blood of cancer patients or healthy donors homozygous for HLA-C alleles in group 1 (C-G1) or group 2 (C-G2) were tested in vitro for cytotoxicity against Epstein-Barr virus–transformed lymphoblastic cell lines (EBV-LCLs), renal cell carcinoma (RCC), and melanoma (MEL) cells with or without a matching KIR-inhibitory HLA-C ligand. Allogeneic NK cells were more cytotoxic to tumor targets mismatched for KIR ligands than their KIR ligand–matched counterparts. Bulk NK populations (CD3–/CD2+/CD56+) expanded 104-fold from patients homozygous for C-G1 or C-G2 had enhanced cytotoxicity against KIR ligand–mismatched tumor cells but only minimal cytotoxicity against KIR ligand–matched targets. Further, NK cell lines from C-G1 or C-G2 homozygous cancer patients or healthy donors expanded but failed to kill autologous or KIR-matched MEL and RCC cells yet had significant cytotoxicity (more than 50% lysis at 20:1 effector-target [E/T] ratio) against allogeneic KIR-mismatched tumor lines. These data suggest immunotherapeutic strategies that use KIR-incompatible allogeneic NK cells might have superior antineoplastic effects against solid tumors compared with approaches using autologous NK cells.
Introduction
Natural killer (NK) cells have antigen-independent tumor cytotoxicity and have been shown in murine models to control and prevent tumor growth and dissemination.1,2 However, the exact role that NK cells play in the control of cancer in humans remains a matter of controversy.
NK cells do not rearrange genes coding for specific antigen receptors; rather, their recognition of targets is regulated through a balance of activating or inhibitory signals.3 It is necessary to inactivate NK cells to prevent their destruction of normal host tissues.4 Therefore, even in the presence of an activating ligand, inhibitory ligands expressed on cells may deliver overriding signals that culminate in a net suppression of NK cell function. Recently, a growing number of NK cell inhibitory and activating receptors were characterized.5-8 NK cells can recognize major histocompatibility (MHC) class I and class I–like molecules through killer immunoglobulin–like receptors (KIRs) expressed on their surfaces. MHC class I ligation of KIRs on normal and malignant tissues suppresses NK cell function. MHC class I molecules fall into groups that serve as ligands for specific KIR, resulting in the inhibition of NK cell–mediated cytotoxicity. Polymorphisms in amino acids residing at positions 77 and 80 of HLA-C dictate specificity for its target KIR.9-11 KIR2DL1 recognizes group 2 HLA-C molecules that have an asparagine at position 77 and a lysine at position 80, whereas KIR2DL2 and KIR2DL3 recognize group 1 HLA-C molecules that have a serine at position 77 and an asparagine at position 80.
The inactivation of NK cells by self-HLA molecules might be a mechanism permitting malignant host cells to evade NK cell–mediated immunity. Because tumor KIR ligands are always matched to NK cell KIR, autologous NK cells would be inhibited by MHC class I–expressing tumors, even in the presence of activating ligands. This may in part explain the failure of adoptively transfused autologous NK12 or lymphokine-activated killer (LAK) cells to mediate antitumor effects against most metastatic solid tumors.13 Only tumor cells that have lost MHC class I expression or have a dominant-activating ligand are predicted to be susceptible to such populations.14
Ruggeri et al15,16 and others17 have shown allogeneic NK cells can mediate antileukemic effects against AML after allogeneic haploidentical and partially mismatched unrelated hematopoietic cell transplantation when KIR/KIR ligand incompatibility exists in the graft-versus-host (GVH) direction (defined as an MHC class I KIR ligand that is absent in the recipient but present in the donor). In this setting, donor NK cells expand that are not inhibited by ligands expressed on recipient leukemia cells, substantially reducing the risk for disease relapse compared with those who receive KIR-compatible transplants. This beneficial effect does not appear to occur against acute lymphoblastic leukemia (ALL), perhaps because these populations lack the activating ligands required to trigger NK cytolytic activity.
HLA-matched allogeneic hematopoietic cell transplantation has recently been reported to induce T cell–mediated graft-versus-tumor effects in select solid tumors.18-22 However, HLA-mismatched transplantation in this setting has not yet been explored. The aim of this study was to evaluate in vitro the effects of KIR ligand–mismatched NK cell populations against solid tumor cells. We sought to test the hypothesis that allogeneic NK cell populations, which sometimes expand after HLA-mismatched transplantation and which previously had been shown to kill Epstein-Barr virus–transformed lymphoblastoid cell lines (EBV-LCLs) and acute myelogenous leukemia/chronic myelogenous leukemia (AML/CML) cells in vitro through KIR-ligand incompatibility, would have enhanced antitumor cytotoxicity against renal cell carcinoma (RCC) and melanoma (MEL) cells with KIR incompatibility compared with autologous or allogeneic KIR–matched NK cells.
Materials and methods
Generation of cell lines
Epstein-Barr virus–transformed lymphoblastoid cell lines (EBV-LCLs) were established by culturing peripheral blood mononuclear cells (PBMCs) in the presence of 100 μg/mL cyclosporin A (CSA; Sandoz Pharmaceuticals, Washington, DC) with EBV supernatant harvested from the cell line B95-8 (American Type Culture Collection [ATCC], Manassas, VA). The human melanoma (MEL) cell lines St-MEL and R-MEL and the human renal cell carcinoma (RCC) cell line Wh-RCC were established from biopsies of metastatic lesions and a renal tumor primary specimen (kindly provided by Dr S. Rosenberg, National Cancer Institute [NCI]). The human RCC cell line SJ-RCC was established from a nephrectomy sample procured at the NCI and maintained in our laboratory. EBV-LCL, MEL, and RCC lines were maintained in RPMI 1640 with 10% fetal calf serum (FCS).
MHC class I typing was performed at the National Institutes of Health HLA laboratory using polymerase chain reaction with sequence-specific primers (PCR-SSPs). Briefly, genomic DNA was first extracted from peripheral blood lymphocytes or tumor samples using primers and PCR conditions described by Bunce et al.23 The final PCR product was visualized on a 2% agarose gel stained with ethidium bromide, with results determined by the presence or absence of an appropriately sized band. For the purpose of this study, tumor lines were specifically selected to be homozygous for HLA-C alleles that would be predicted to ligate a single HLA-C–reactive KIR (either KIR2DL2/3 or KIR2DL1; Table 1).
HLA data on NK and tumor cells used in experiments
. | NK cells . | Tumor cells . | . | . | HLA class I . | . | . | HLA-C group homozygous . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | . | RCC . | MEL . | LCL . | A . | B . | Cw . | . | ||||
| R | - | - | + | + | 02, 23 | 15, 35 | 02, 04 | 2 | ||||
| St | + | - | + | + | 02, 29 | 07, 08 | 0701, 0702 | 1 | ||||
| Wh | - | + | - | - | 03, 01 | 08, 44 | 05, (-)† | 2 | ||||
| SJ | - | + | - | + | 11, 11 | 51, 55 | 01, 0303 | 1 | ||||
| Volle | - | - | - | + | 02, 03 | 07, 51 | 01, 0702 | 1 | ||||
| KR0350 | + | - | - | + | 02, 68 | 40, 44 | 0304, 0704 | 1 | ||||
| SKEM* | + | - | - | - | 11, 11 | 51, 55 | 01, 0303 | 1 | ||||
| MORRC | + | - | - | - | 02, 11 | 35, 40 | 02, 04 | 2 | ||||
. | NK cells . | Tumor cells . | . | . | HLA class I . | . | . | HLA-C group homozygous . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | . | RCC . | MEL . | LCL . | A . | B . | Cw . | . | ||||
| R | - | - | + | + | 02, 23 | 15, 35 | 02, 04 | 2 | ||||
| St | + | - | + | + | 02, 29 | 07, 08 | 0701, 0702 | 1 | ||||
| Wh | - | + | - | - | 03, 01 | 08, 44 | 05, (-)† | 2 | ||||
| SJ | - | + | - | + | 11, 11 | 51, 55 | 01, 0303 | 1 | ||||
| Volle | - | - | - | + | 02, 03 | 07, 51 | 01, 0702 | 1 | ||||
| KR0350 | + | - | - | + | 02, 68 | 40, 44 | 0304, 0704 | 1 | ||||
| SKEM* | + | - | - | - | 11, 11 | 51, 55 | 01, 0303 | 1 | ||||
| MORRC | + | - | - | - | 02, 11 | 35, 40 | 02, 04 | 2 | ||||
HLA-C group patients are grouped according to HLA-C alleles and corresponding KIR ligands.
SKEM is an HLA-identical sibling donor for RCC patient SJ.
Wh-RCC lost an HLA-Cw locus.
NK cell culture media
Bulk NK cells and NK cells isolated after limiting-dilution cloning were grown in Cellgro SCGM serum-free media (CellGenix, Gaithersburg, MD) containing 10% human AB serum, 50 U /mL penicillin, 50 μg/mL streptomycin, and 500 IU/mL interleukin-2 (IL-2) (unless otherwise specified).
Flow cytometry
Cell surface phenotype was determined by flow cytometry using the following antihuman monoclonal antibodies (mAbs): anti–CD16-phycoerythrin (PE) (clone B73.1 mouse immunoglobulin G1 [IgG1]; Becton Dickinson, San Jose, CA), anti–CD158a-PE (clone EB6, mouse IgG1; Immunotech, Marseilles, France), anti–CD158b-PE (clone GL183, mouse IgG1; Immunotech), anti–CD56-Cy-chrome (clone MY31, mouse IgG1; Becton Dickinson), anti–CD3-fluorescein isothiocyanate (FITC) (clone SK7, mouse IgG1; Becton Dickinson), anti–LFA-1/CD11a-PE (clone HI111, mouse IgG1; PharMingen, San Diego, CA), and antihuman HLA-A, -B, -C–PE (clone G46-2.6, mouse IgG1; PharMingen). Cells were also stained with their corresponding isotype-matched control mAbs (PharMingen). All samples were analyzed using EPICS XL-MCL (Beckman Coulter, Miami, FL). EXPO 32 ADC software (Beckman Coulter) was used for data acquisition and analysis.
Evaluation of NKG2 ligands on tumor cell lines
Unconjugated antibodies to NKG2L, MICA (M363), MICB (M364), ULBP1 (M295), ULBP2 (M311) and ULBP3 (M551) were a kind gift from David Cosman (Amgen, Seattle, WA). Secondary antibody staining was accomplished with AlexaFluor488-conjugated goat antimouse immunoglobulin (Molecular Probes, Eugene, OR). Specific staining was revealed through the use of MAB272, an I-309 reactive IgG1 isotype control antibody (R&D Systems, Minneapolis, MN). Staining was evaluated using a FACScan instrument (Becton Dickinson).
Isolation of NK cells and expansion of NK cell lines
PBMCs obtained from healthy donors or cancer patients with HLA-C alleles homozygous in group 1 (C-G1; ligates KIR2DL2/3) or group 2 (C-G2; ligates KIR2DL1) were enriched for NK cell populations by negative depletion using immunomagnetic beads (Dynal NK cell isolation kit; Dynal Biotech, Lake Success, NY) according to the manufacturer's recommended conditions. Briefly, PBMCs were incubated with an antibody mix containing anti-CD3, anti-CD14, anti-CD36, anti-CDw123, and anti-HLA class 2 DR/DP for 10 minutes at 4° C with gentle tilting and rotation. Cells were washed with phosphate buffer saline (PBS)/0.1% human AB serum and centrifuged for 8 minutes at 500g. The supernatant was discarded, and the cell pellet was resuspended in PBS/0.1% human AB serum and incubated at 4° C for 10 minutes after the addition of depletion Dynabeads. The cell suspension was placed in a magnetic particle concentrator (Dynal MPC; Dynal Biotech) for 2 minutes, the supernatant was removed, and the depletion bead step was repeated. NK cell purity was determined by flow cytometry. One portion of the enriched NK cells was used to generate relatively homogeneous NK cell lines by limiting dilution at a density of 1 to 10 cells/well in a 96-well U-bottom plate containing 500 IU/mL of IL-2 and 104 irradiated (100 Gy) allogeneic EBV-LCLs per well as feeder cells. Half the culture medium was replaced weekly with new NK cell media containing fresh IL-2. The remaining portion of enriched NK cells was expanded as bulk NK populations in NK cell media containing 500 IU/mL of IL-2 and irradiated (100 Gy) allogeneic EBV-LCL feeder cells (1000:1 EBV-LCL/NK cell ratio). Two weeks after expansion, bulk NK cells were stained with anti–CD158b-FITC and anti–CD56 Cy-chrome and were then sorted using a MoFlo flow Cytometer (DAKO-Cytomation, Fort Collins, CO) to isolate CD56+/CD158b+ populations that were reexpanded for 2 weeks using EBV-LCL feeders and NK cell media as described. In most experiments, allogeneic EBV-LCLs with HLA-C alleles homozygous in a group opposite the NK cells were used as feeder cells.
Optimizing NK cell expansion
In bulk NK cell expansions, EBV-LCLs heterogeneous or homogeneous for C-G1 or C-G2 were used as feeders, with the proportions of expanded CD158a+ or CD158b+ NK cells determined by flow cytometry. Fold expansion was measured by staining bulk NK cells with trypan blue, then dividing the total number of viable cells after expansion by the number of viable cells before expansion.
Cytotoxicity assay
NK cell cytotoxicity was assessed using standard chromium release assays, as described previously.24 Briefly, 106 target cells were incubated with 100 μCi (3.7 MBq)
The percentage of specific lysis was calculated using the formula: 100 × (count per minute [cpm] released from test sample – cpm spontaneous release)/(cpm maximum release – cpm spontaneous release). All values shown represent the average of duplicates or triplicates ± 1 SD. Some targets were preincubated with 20 μg/mL of the monoclonal antibody W6/32 (anti-pan class 1 MHC monoclonal antibody; DAKO) to assess the impact of MHC class I blockade on NK cell cytotoxicity.
Results
Impact of EBV-LCL feeders and HLA class 1 expression on in vitro NK cell expansion
Preferential proliferation of NK cells occurs when EBV-LCLs are cultured with PBMCs.25,26 Therefore, we first investigated whether EBV-LCL feeder cells could be used to enhance the expansion of bulk NK cell populations. NK cells (1 × 103) enriched from PBMCs by negative depletion (typically 90% or more CD3– and CD56+) were cultured in stem cell growth medium (SCGM) containing 500 IU of IL-2, alone or with 1 × 106 irradiated EBV-LCLs. After 14 days, NK cells containing EBV-LCL feeders expanded more than 104-fold compared with NK cell populations cultured in IL-2 containing SCGM media alone, where approximately a 102-fold net expansion was typically observed (data not shown). No significant differences in NK cell expansion were observed when using EBV-LCLs with HLA-C alleles that were matched compared with mismatched for NK cell HLA-C groups. However, a significantly higher percentage of KIR 2DL2/3-positive NK cells was expanded from an HLA-C group 1 homozygous donor when EBV-LCL feeders were used that either lacked or had heterozygous expression for HLA-C alleles known to ligate KIR2DL2/3 (Table 2). Varying the IL-2 concentration from 5 to 500 IU/mL did not influence NK cell expansion when EBV-LCLs were used as feeders.
Expansion of bulk NK cells: donor HLA-C group 1 homozygous
Locus and concentration . | Expansion fold . | CD3-CD56+ or CD3-CD16+, % . | CD3-CD56+CD158b+, % . |
|---|---|---|---|
| HLA-Cw locus of feeder cells | |||
| Homozygous for C-G2 | (1.8 ± 1.6) × 104 | 79.5 ± 5.2 | 41 ± 9.1† |
| Homozygous for C-G1 (autologous) | (1.32 ± 0.87) × 104 | 72.5 ± 11.6 | 24 ± 8.8† |
| Heterozygous for C-G2 and C-G1 | (1.8 ± 0.45) × 104 | 78.6 ± 4.5 | 33 ± 9.5 |
| IL-2 concentration, IU/mL* | |||
| 100 | 2.6 × 104 | 76.6 | 35.5 |
| 50 | 2.5 × 104 | 75.7 | 27.2 |
| 5 | 6.8 × 103 | 68.2 | 21.8 |
Locus and concentration . | Expansion fold . | CD3-CD56+ or CD3-CD16+, % . | CD3-CD56+CD158b+, % . |
|---|---|---|---|
| HLA-Cw locus of feeder cells | |||
| Homozygous for C-G2 | (1.8 ± 1.6) × 104 | 79.5 ± 5.2 | 41 ± 9.1† |
| Homozygous for C-G1 (autologous) | (1.32 ± 0.87) × 104 | 72.5 ± 11.6 | 24 ± 8.8† |
| Heterozygous for C-G2 and C-G1 | (1.8 ± 0.45) × 104 | 78.6 ± 4.5 | 33 ± 9.5 |
| IL-2 concentration, IU/mL* | |||
| 100 | 2.6 × 104 | 76.6 | 35.5 |
| 50 | 2.5 × 104 | 75.7 | 27.2 |
| 5 | 6.8 × 103 | 68.2 | 21.8 |
NK cells were enriched by negative selection from PBMCs of a healthy donor (KR0350) homozygous for HLA-C group 1 (C-G1), then expanded in vitro using irradiated EBV-LCL feeders: (1) HLA-C group 2 (C-G2) EBV-LCLs; (2) autologous EBV-LCLs; (3) HLA-C group is heterozygous EBV-LCLs. Two weeks later, expanded NK cells were counted, and phenotype analyses were performed. The difference in CD3-CD56+CD158b+ population between homozygous for C-G2 feeder cells and autologous feeder cells was significant (P = .0085). Values represent averages ± SD from 3 independent experiments.
Values represent averages from 2 independent experiments.
P = .0085.
Susceptibility of RCC and melanoma cells to NK cells with or without KIR ligand incompatibility: bulk NK cultures
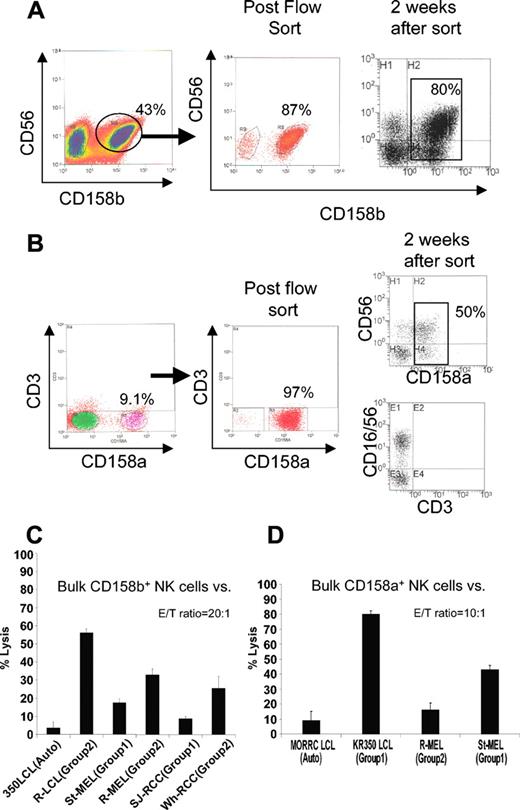
We first assessed in vitro whether melanoma and RCC cells would be more susceptible to killing by bulk allogeneic NK populations when the tumor lacked an HLA-C allele for a specific KIR expressed on NK cell effectors. NK cells enriched from PBMCs by negative selection from a donor homozygous for HLA-C group 1 (C-G1) were expanded in vitro using irradiated EBV-LCLs as feeder cells. Two weeks later, NK cells expressing CD158b (KIR 2DL2/3) were isolated by flow sorting and were then expanded for 2 more weeks to remove remaining bound antibodies (Figure 1A).
Isolation of CD158b+ (KIR 2DL2/3) and CD158a+ (KIR 2DL1) NK populations. (A) NK cells were enriched by negative selection from PBMCs of a healthy donor (KR0350) homozygous for HLA-C group 1 (C-G1), then expanded in vitro using irradiated EBV-LCL feeders. Two weeks later, CD3–/CD56+/CD158b+ NK cells were isolated by flow sorting, then expanded for another 2 weeks to remove any remaining surface-bound antibodies. (B) In a similar fashion, CD158a+ NK cell populations were isolated by flow sorting from healthy donor MORRC homozygous for HLA-C group 2 (C-G2). Percentages of CD3–/CD56+/CD1586b+ cells are shown. (C) At a 20:1 E/T ratio, NK cells enriched for CD158b (80% positive) lysed a significantly higher percentage of KIR-incompatible EBV-LCL, melanoma, and RCC cells homozygous for HLA-C group 2 (KIR incompatible) compared with HLA-C group 1 homozygous KIR-matched tumor targets. (D) These cells lysed a significantly higher percentage of EBV-LCL and melanoma cells homozygous for HLA-C group 1 (KIR incompatible) compared with the HLA-C group 2 homozygous KIR-matched targets. Mean values +SD of 3 samples are shown.
Isolation of CD158b+ (KIR 2DL2/3) and CD158a+ (KIR 2DL1) NK populations. (A) NK cells were enriched by negative selection from PBMCs of a healthy donor (KR0350) homozygous for HLA-C group 1 (C-G1), then expanded in vitro using irradiated EBV-LCL feeders. Two weeks later, CD3–/CD56+/CD158b+ NK cells were isolated by flow sorting, then expanded for another 2 weeks to remove any remaining surface-bound antibodies. (B) In a similar fashion, CD158a+ NK cell populations were isolated by flow sorting from healthy donor MORRC homozygous for HLA-C group 2 (C-G2). Percentages of CD3–/CD56+/CD1586b+ cells are shown. (C) At a 20:1 E/T ratio, NK cells enriched for CD158b (80% positive) lysed a significantly higher percentage of KIR-incompatible EBV-LCL, melanoma, and RCC cells homozygous for HLA-C group 2 (KIR incompatible) compared with HLA-C group 1 homozygous KIR-matched tumor targets. (D) These cells lysed a significantly higher percentage of EBV-LCL and melanoma cells homozygous for HLA-C group 1 (KIR incompatible) compared with the HLA-C group 2 homozygous KIR-matched targets. Mean values +SD of 3 samples are shown.
KIR ligand compatible (eg, homozygous for C-G1) or KIR ligand incompatible (eg, homozygous for C-G2) tumor targets were then tested in vitro for differences in susceptibility to the cytotoxic effects of CD158b-enriched (approximately 80% positive) bulk NK cells.
A significantly greater percentage of KIR incompatible, C-G2 homozygous, melanoma, and RCC lines were lysed by CD158b-enriched NK cells than by KIR compatible C-G1 homozygous tumor counterparts (Figure 1B). A similar pattern of cytotoxicity was observed against KIR ligand–compatible (autologous) compared with KIR ligand–incompatible EBV-LCL targets. Likewise, CD158a-enriched NK cells lysed a higher percentage of KIR incompatible, C-G1 homozygous EBV-LCL and melanoma cells compared with C-G2 homozygous KIR-matched targets (Figure 1B,D).
NK lines
We next isolated NK cell lines from healthy donors or patients with cancer that specifically killed KIR-incompatible EBV-LCL populations and then evaluated their cytotoxic effects against KIR ligand–matched compared with–mismatched melanoma or RCC targets. PBMCs from healthy donors or cancer patients homozygous for C-G1 or C-G2 were enriched for NK cell populations by negative depletion, then cloned by limiting dilution using allogeneic KIR ligand–mismatched EBV-LCLs as feeder cells. NK lines were phenotyped using fluorescence-activated cell sorter (FACS) and were tested for cytotoxicity against KIR-matched (autologous) or KIR-mismatched EBV-LCL targets.
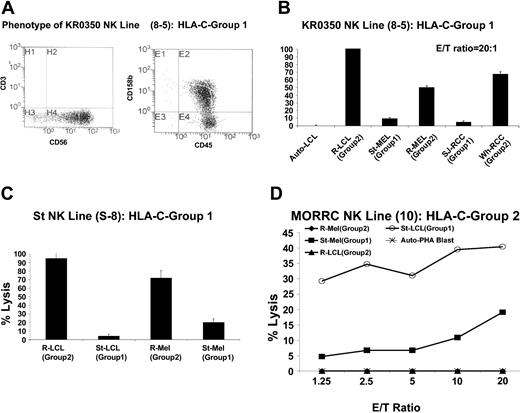
Thirty-four CD3–/CD56+ NK lines were expanded from a healthy donor homozygous for C-G1 (KR0350); 9 lysed EBV-LCL cells homozygous for C-G2 (35%-100% lysis at an effector-target [E/T] ratio of 20:1), with minimal to no lysis of KIR-matched (autologous) EBV-LCLs (data not shown). Line 8-5 expanded sufficiently for further experiments (Figure 2A).
RCC and melanoma cells are more susceptible to the cytotoxic effects of KIR-incompatible NK clones than KIR-matched lines. NK cells were enriched and then cloned by limiting dilution from the PBMCs of cancer patient St (homozygous for HLA-C group 2) or from healthy donors homozygous for HLA-C group 1. (A) NK cell line 8-5, expanded from a C-G1 homozygous healthy donor (KR0350), killed a significantly higher percentage of C-G2 homozygous KIR-incompatible EBV-LCL, melanoma, and RCC cells than KIR-matched autologous EBV-LCL or KIR-matched allogeneic C-G1 homozygous melanoma and RCC cells (B). (C) NK line S-8, expanded from a C-G1 homozygous patient with widely metastatic melanoma (St), killed a significantly higher percentage of KIR-mismatched C-G2 homozygous EBV-LCLs and melanoma cells than autologous KIR-matched EBV-LCL and melanoma cells, in which minimal cytotoxicity was observed. Mean values +SD of 3 samples are shown. (D) MORRC NK line 10, expanded from a C-G2 homozygous healthy donor (MORRC), was cytotoxic to KIR-incompatible C-G1 homozygous but not KIR-matched C-G2 homozygous targets.
RCC and melanoma cells are more susceptible to the cytotoxic effects of KIR-incompatible NK clones than KIR-matched lines. NK cells were enriched and then cloned by limiting dilution from the PBMCs of cancer patient St (homozygous for HLA-C group 2) or from healthy donors homozygous for HLA-C group 1. (A) NK cell line 8-5, expanded from a C-G1 homozygous healthy donor (KR0350), killed a significantly higher percentage of C-G2 homozygous KIR-incompatible EBV-LCL, melanoma, and RCC cells than KIR-matched autologous EBV-LCL or KIR-matched allogeneic C-G1 homozygous melanoma and RCC cells (B). (C) NK line S-8, expanded from a C-G1 homozygous patient with widely metastatic melanoma (St), killed a significantly higher percentage of KIR-mismatched C-G2 homozygous EBV-LCLs and melanoma cells than autologous KIR-matched EBV-LCL and melanoma cells, in which minimal cytotoxicity was observed. Mean values +SD of 3 samples are shown. (D) MORRC NK line 10, expanded from a C-G2 homozygous healthy donor (MORRC), was cytotoxic to KIR-incompatible C-G1 homozygous but not KIR-matched C-G2 homozygous targets.
Although this line had no cytotoxicity against autologous EBV-LCLs, it lysed nearly 100% of EBV-LCLs homozygous for C-G2, consistent with killing through KIR incompatibility. At a 20:1 E/T ratio, a significantly higher percentage of KIR ligand–mismatched melanoma and RCC cells (homozygous for C-G2) were killed than KIR ligand–matched (homozygous for C-G1) tumor targets (Figure 2B). Similarly, enriched NK cells were expanded after limiting dilution from the PBMCs of a patient (St) with widely metastatic melanoma who was homozygous for C-G1; NK line S-8 from this patient killed a significantly higher percentage of KIR ligand–mismatched EBV-LCLs (95%) and melanoma cells (72%) than KIR ligand–matched autologous targets (5% and 19% of EBV-LCL and melanoma cells respectively; Figure 2C).
NK lines could also be expanded from a healthy donor homozygous for C-G2 with enhanced cytotoxicity against KIR ligand–mismatched allogeneic EBV-LCLs and melanoma cells homozygous for C-G1 compared with KIR ligand–matched allogeneic EBV-LCLs and melanoma cells homozygous for C-G2 (Figure 2D).
Impact of MHC class I expression on KIR-matched and -mismatched NK cell targets
MHC class I blockade has been shown to enhance NK cell cytotoxicity against KIR ligand–matched tumors in vitro. Therefore, we investigated the cytotoxic potential of allogeneic KIR ligand–mismatched NK cells against RCC and melanoma cells compared with KIR ligand–matched NK cells targeting tumors in which MHC class I had been blocked with the monoclonal antibody W6/32.
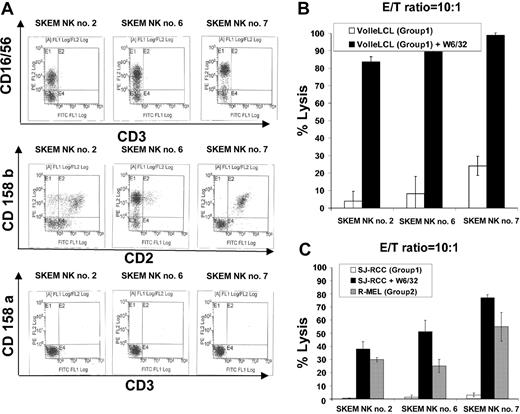
NK cells enriched from the PBMCs of a healthy donor, SKEM (C-G1 homozygous; HLA-identical sibling to RCC patient SJ) were expanded after limiting dilution and screened for cytotoxicity against KIR-matched (C-G1 homozygous) versus KIR-mismatched (C-G2 homozygous) targets. Three NK lines (SKEM NK lines 2, 6, and 7; Figure 3A) with significantly greater cytotoxicity against C-G2–mismatched versus C-G1–matched EBV-LCLs were identified.
Impact of MHC class I blockade on NK cells targeting KIR ligand–matched compared with KIR ligand–mismatched tumor targets. (A) NK cell lines 2, 6, and 7 were isolated by limiting dilution from the PBMCs of a healthy donor, SKEM (C-G1 homozygous: HLA-identical sibling to RCC patient SJ). (B) All 3 lines lysed a minority of C-G1 homozygous KIR-matched VOLLE EBV-LCLs. Preincubation of VOLLE EBV-LCLs with W6/32 significantly enhanced the cytotoxic effects of all 3 lines, consistent with MHC class I molecules inhibiting NK cell function. (C) NK lines 2, 6, and 7 were significantly more cytotoxic to KIR ligand–mismatched R-MEL cells (C-G2 homozygous) compared with KIR-matched (C-G1 homozygous) SJ cells, in which virtually no cytotoxicity was observed. Preincubation of SJ RCC cells with W6/32 significantly increased the cytotoxic effects of all 3 lines against this KIR-matched RCC target. Mean values +SD of 3 samples are shown.
Impact of MHC class I blockade on NK cells targeting KIR ligand–matched compared with KIR ligand–mismatched tumor targets. (A) NK cell lines 2, 6, and 7 were isolated by limiting dilution from the PBMCs of a healthy donor, SKEM (C-G1 homozygous: HLA-identical sibling to RCC patient SJ). (B) All 3 lines lysed a minority of C-G1 homozygous KIR-matched VOLLE EBV-LCLs. Preincubation of VOLLE EBV-LCLs with W6/32 significantly enhanced the cytotoxic effects of all 3 lines, consistent with MHC class I molecules inhibiting NK cell function. (C) NK lines 2, 6, and 7 were significantly more cytotoxic to KIR ligand–mismatched R-MEL cells (C-G2 homozygous) compared with KIR-matched (C-G1 homozygous) SJ cells, in which virtually no cytotoxicity was observed. Preincubation of SJ RCC cells with W6/32 significantly increased the cytotoxic effects of all 3 lines against this KIR-matched RCC target. Mean values +SD of 3 samples are shown.
At a 10:1 E/T ratio, NK lines lysed 4% to 25% of Volle EBV-LCLs (matched for C-G1); the cytotoxicity of all 3 lines increased significantly (greater than 80%) when Volle EBV-LCLs were precultured with the monoclonal antibody W6/32 (pan class 1 MHC antibody), consistent with MHC class I molecules on the target cells functioning as NK cell KIR ligands (Figure 3B). SKEM NK lines were significantly more cytotoxic against KIR ligand–mismatched, C-G2 homozygous R-melanoma cells than against KIR ligand–matched, C-G1 homozygous SJ RCC cells, in which virtually no cytotoxicity was observed. Preculturing SJ RCC cells with the pan MHC class I monoclonal antibody W6/32 substantially increased the cytotoxic effects of all 3 NK cell lines against this KIR-matched tumor target.
Impact of gamma irradiation on NK cell function
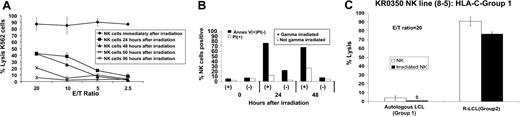
Strair et al39 recently demonstrated that metastatic RCC may regress in humans after the adoptive infusion of irradiated HLA-mismatched allogeneic PBMCs. Our in vitro data suggest adoptively infused KIR-incompatible allogeneic NK cells might also have beneficial antitumor effects in vivo. Therefore, the impact of gamma irradiation on NK cell–mediated tumor cytotoxicity was further investigated. Bulk NK cells expanded from PBMCs of healthy donors (as previously described) were gamma irradiated (50 Gy) and then tested for cytotoxicity against NK cell–sensitive K562 cells 0, 24, 48, 60, and 96 hours after irradiation (Figure 4A).
Impact of γ-irradiation on NK cell function. Bulk NK cells expanded from the PBMCs of healthy donor KR0350 were γ-irradiated (50 Gy) and tested for cytotoxicity against K562 cells (A) 0, 24, 48, 60, and 96 hours after irradiation. (B) Although irradiation induced NK apoptosis (annexin V–positive) and necrosis (PI-positive), persistent NK cell cytotoxicity was observed up to 48 hours after γ-irradiation. (C) C-G1 homozygous NK cell lines 8-5 from donor KR0350 maintained a high level of cytotoxicity against KIR-incompatible R-EBV-LCL cells 24 hours after 50 Gy γ-irradiation. Mean values +SD of 3 samples are shown.
Impact of γ-irradiation on NK cell function. Bulk NK cells expanded from the PBMCs of healthy donor KR0350 were γ-irradiated (50 Gy) and tested for cytotoxicity against K562 cells (A) 0, 24, 48, 60, and 96 hours after irradiation. (B) Although irradiation induced NK apoptosis (annexin V–positive) and necrosis (PI-positive), persistent NK cell cytotoxicity was observed up to 48 hours after γ-irradiation. (C) C-G1 homozygous NK cell lines 8-5 from donor KR0350 maintained a high level of cytotoxicity against KIR-incompatible R-EBV-LCL cells 24 hours after 50 Gy γ-irradiation. Mean values +SD of 3 samples are shown.
Although most NK cells quickly became annexin V- or propidium iodide (PI)–positive (Figure 4B) by FACS (consistent with radiation-induced apoptosis and death), significant cytotoxicity against K562 cells could still be measured 48 hours after irradiation. NK cell line 8-5 from healthy donor KR0350 maintained its ability to kill KIR-incompatible EBV-LCL cells 24 hours after irradiation, with only a slight decrease in its cytotoxic capacity compared with a nonirradiated control (Figure 4C).
Discussion
Ruggeri et al15,16 and others17 have shown that the risk for AML relapse in patients receiving KIR-incompatible haploidentical or partially mismatched unrelated allogeneic transplants is significantly reduced when a KIR epitope mismatch in the direction of GVH exists. In this population, NK cells can be expanded after transplantation that kill AML and CML cells in vitro through KIR incompatibility. Furthermore, these alloreactive NK cell populations can mediate curative antileukemia effects against AML when adoptively infused into leukemia-bearing NOD/SCID mice.16 Equally important, alloreactive NK cell populations appear to mediate these graft-versus-leukemia effects without causing graft-versus-host disease (GVHD).16
Here we demonstrate that similar NK cell–mediated effects occur in vitro against neoplastic cells of epithelial origin. Specifically, we show that melanoma and RCC cells are more susceptible to the cytotoxic effects of allogeneic NK cells expressing KIR that cannot be ligated by the tumor's inhibitory HLA-C alleles because of HLA mismatching. In this study, we generated NK cell lines from donors or patients who were homozygous for group 1 or group 2 HLA-C alleles, in whom the frequency of NK cells with alloreactivity against targets lacking the same HLA-C allele group was high. Furthermore, we used EBV-LCL, melanoma, and RCC tumor targets that were homozygous for group 1 or group 2 HLA-C alleles, so that the impact of KIR mismatching on NK cell cytotoxicity could be easily evaluated.
Although they varied in frequency, NK cells that selectively killed HLA-C group–mismatched EBV-LCLs could be isolated from all donors studied. In most cases, NK cells with enhanced cytotoxicity against KIR-incompatible EBV-LCLs also killed a significantly higher percentage of melanoma and RCC cells that lacked a matching inhibitory HLA-C KIR ligand compared with KIR ligand–matched tumor cells. The differences in the level of cytotoxicity were often striking, with substantially higher (more than 60%) lysis of KIR ligand–mismatched tumor targets compared with KIR-matched targets in which little to no lysis occurred. This NK cell phenomenon was not restricted to healthy donors; NK cell lines from cancer patients homozygous for C-G1 or C-G2 were identified that failed kill autologous tumor cells while having significantly greater cytotoxicity against allogeneic KIR ligand–mismatched tumor lines. In this study, we restricted KIR incompatibility to the HLA-C locus. Although we did not evaluate the impact of KIR mismatching in HLA-B or -A loci, one might speculate from our findings that such NK cell populations might have enhanced cytotoxicity similar to that shown to occur against AML and EBV-LCL cells.
The balance between activating and inhibitory signals ultimately determines whether target cells will be susceptible to NK cell–mediated lysis. In contrast to myelogenous leukemia (AML and CML), Ruggeri et al15 found only a minority of ALL cells were killed in vitro by KIR-mismatched NK populations. This in vitro finding correlated well with their clinical data showing no beneficial impact of NK cell alloreactivity on ALL relapse. The presence of LFA-1 on AML cells and its absence on NK cell–resistant ALL cells suggested this adhesion molecule might function as a ligand for NK cell–activating receptors. The activating ligands on tumor cells used in these experiments that were necessary to initiate alloreactive NK cell killing are unknown but are being investigated. Flow cytometric analysis showed all our RCC and melanoma lines to be LFA-1 negative (data not shown). However, our tumor lines had variable densities of MHC class I chain–related antigens (MICA or MICB)27,28 or UL16-binding proteins29-32 (ULBPs; Table 3), known ligands for the NK cell–activating receptor NKG2D.
Expression of NKG2D ligands measured by flow cytometry
NKG2D ligands . | Tumor cell lines, % . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | St-MEL . | R-MEL . | SJ-RCC . | Wh-RCC . | |||
| MIC A/B | 8 | 13 | 17 | 10 | |||
| ULBP1 | 0 | 0 | 52 | 0 | |||
| ULBP2 | 0 | 0 | 52 | 0 | |||
| ULBP3 | 80 | 0 | 98 | 9 | |||
NKG2D ligands . | Tumor cell lines, % . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | St-MEL . | R-MEL . | SJ-RCC . | Wh-RCC . | |||
| MIC A/B | 8 | 13 | 17 | 10 | |||
| ULBP1 | 0 | 0 | 52 | 0 | |||
| ULBP2 | 0 | 0 | 52 | 0 | |||
| ULBP3 | 80 | 0 | 98 | 9 | |||
Expression of NKG2D ligands; stress-inducible surface glycoprotein MIC A/B, UL16-binding protein (ULBP), ULBP 1, ULBP 2, and ULBP 3 were measured using flow cytometry.
Previous studies have shown that leukemia, lymphoma,33 and melanoma lines34,35 with these ligands can be specifically lysed by NKG2D–expressing NK cells. It remains to be determined whether the level of expression of different activating ligands might be used as a surrogate to identify those malignancies with susceptibility to KIR-incompatible NK cells.36
Efforts to boost host NK cell–mediated immunity against solid tumors through IL-2 administration or the adoptive infusion of LAK cells have been largely unsuccessful.37,38 KIR-mediated inhibition of NK cell function by tumor HLA class 1 or class 1–like ligands may be a major factor limiting autologous-based NK cell therapies. Murine and human studies have shown that alloreactive NK cells do not induce a GVH reaction.16 These findings, combined with our data showing that KIR-incompatible NK cells have enhanced tumor cytotoxicity compared with KIR-matched or autologous NK populations, could be used as the rationale to investigate their antineoplastic potential in humans. Using irradiated EBV-LCLs as feeders, we successfully expanded bulk NK populations 104-fold, 102-fold higher than the maximum expansion previously reported achievable when PBMCs were used as feeders. Therefore, it appears feasible that 1010 or more allogeneic NK cells could be expanded ex vivo for adoptive infusion from a starting source of only 10 to 25 mL of peripheral blood. Such populations would likely have to be irradiated before their adoptive infusion in the allogeneic setting to prevent long-term engraftment and possible transfusion-associated GVHD as a consequence of T-lymphocyte contamination. Not surprisingly, irradiation induced NK cell apoptosis and, ultimately, cell death. However, our in vitro data showed that some level of NK cell cytotoxic function was maintained for up to 48 hours after exposure to 50 Gy. Indeed, γ-irradiated, partially mismatched allogeneic PBMC infusions have recently been reported to induce disease regression in some patients with metastatic RCC.39 Our data suggest that adoptively infused and irradiated allogeneic NK cells with KIR incompatibility could have similar or even enhanced antitumor effects compared with unmanipulated allogeneic PBMC infusions containing a relatively small proportion of NK cells.
A potential limitation of our study was the use of RCC and melanoma cell lines, which potentially could represent a minor subset of the entire tumor cell population. Low numbers of tumor cells and poor tumor cell purity from contamination of metastatic lesions with nonmalignant cells (stromal, endothelial, hematopoietic) are technical factors that limited the use of fresh tumor samples in these experiments. Although studies are ongoing, preliminary data on fresh and early-passage tumor cells isolated from large metastatic lesions have shown similarly enhanced susceptibility to KIR-incompatible NK cells as was observed with established tumor lines (data not shown).
Finally, allogeneic hematopoietic stem cell transplantation has recently been shown to induce graft-versus-tumor effects in some patients with advanced cytokine-refractory metastatic RCC.40 Because HLA-matched siblings were used as donors, alloreactive NK cells would not be expected to expand. Our data showing enhanced susceptibility of RCC and melanoma cells to NK cells with KIR incompatibility suggest the NK cell–mediated antitumor effects seen against AML after KIR-incompatible mismatched allotransplantation might be induced in a similar fashion against select solid tumors. Transplantation trials for metastatic RCC using unrelated donors are already in progress.41 Based on our findings, trials that evaluate the use of single HLA-mismatched unrelated donors with KIR incompatibility (in the direction of the GVH) should be considered. This would also provide an opportunity to investigate the impact of posttransplantation alloreactive NK cell infusions, which have been previously shown to facilitate engraftment, reduce the risk for GVHD, and eradicate leukemia in murine models of mismatched transplantation.16
Prepublished online as Blood First Edition Paper, March 11, 2004; DOI 10.1182/blood-2003-12-4438.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Steven A. Rosenberg (NCI), Kenichi Hanada (NCI), and Giuseppe Sconocchia (National Heart, Lung, and Blood Institute) for their valuable intellectual input and advice.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal