Abstract
Inflammatory stimulation of endothelial cells by tumor necrosis factor α (TNF-α) involves activation of nuclear factor κB (NF-κB) and p38 mitogen-activated protein (MAP) kinase signaling pathways. A reliable analysis of the gene expression program elicited by TNF-α and its assignment to distinct signaling pathways is not available. A sophisticated analysis of oligonucleotide microarrays covering more than 13 000 genes allowed definition of the TNF-α-regulated endothelial gene expression profile and novel TNF-α-induced genes. Virtually all TNF-α-inducible genes were dependent on IκB kinase 2 (IKK2)/NF-κB activation, whereas a minor number was additionally modulated by p38. Furthermore, genes suppressed by IKK2/NF-κB were newly identified. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) and flow cytometry confirmed reliability of data. Thus, these results define a list of primary candidates for targeted modulation of endothelial functions during inflammation. (Blood. 2004;103:3365-3373)
Introduction
The vascular endothelium is critically involved in the regulation of inflammation. Its activation through extracellular stimuli triggers intracellular signal transduction cascades and coordinately elicits gene expression programs resulting in the expression of adhesion molecules, cytokines, and chemokines, which finally guide leuko cytes to sites of inflammatory challenge.1 Tumor necrosis factor α (TNF-α), one of the most important promoters of inflammation, induces activation of at least 2 major signaling pathways involved in endothelial activation (ie, the IκB kinase/nuclear factor κB [IKK/NF-κB] pathway and the p38 mitogen-activated protein [MAP] kinase cascade). In resting cells, NF-κB is inactive because of binding to inhibitor of κB (IκB) proteins, thus preventing DNA binding. Upon endothelial activation, the critical step in NF-κB activation is the phosphorylation of IκB by a signalosome complex consisting of 2 catalytic subunits, the IκB kinases IKK1 (IKKα) and IKK2 (IKKβ), and a scaffolding subunit, NF-κB essential modulator (NEMO) (IKKγ).2-4 IκB phosphorylation and degradation allow NF-κB to translocate to the nucleus where it binds to specific DNA motifs in the promoter region of target genes. Mice with targeted deletion of IKK2 or NEMO are deficient for cytokine-induced IκB degradation and nuclear NF-κB-DNA binding activity.5,6 Accordingly, stable expression of dominant-negative mutants of IKK2 in human endothelial cells completely blocks cytokine-induced expression of NF-κB-dependent target genes.7
In addition to the NF-κB pathway, MAP kinase signaling cascades are pivotal to stress and inflammatory responses. One of these cascades results in activation of the p38 MAP kinase whose pharmacologic inhibition blocks production of proinflammatory mediators such as TNF-α and interleukin 1 (IL-1). Furthermore, p38 has been shown to be involved in the expression of endothelial adhesion molecules such as vascular cell adhesion molecule 1 (VCAM-1) and chemokines.8,9 It is now becoming evident that p38 is an important target for novel pharmacologic compounds that are expected to disrupt inflammation.10 The relative contribution of NF-κB pathways and p38 pathways to TNF-α-dependent gene expression has not systematically been analyzed in endothelial cells. Anti-TNF-α-based strategies for treatment of inflammatory disorders such as rheumatoid arthritis, Crohn disease, and psoriasis and clinical trials studying pharmacologic inhibitors interfering with NF-κB pathways and p38 signaling pathways show promising anti-inflammatory efficacy.10-14 An accurate assignment of TNF-α-induced gene expression patterns to distinct signaling pathways is desirable for a deeper understanding of basic pathomechanisms of inflammation as well as for identification of novel target molecules allowing innovative treatment strategies.
Employing oligonucleotide microarray analysis of more than 13 000 genes, we defined the TNF-α-induced expression profile of primary human endothelial cells validated by extensive statistical analysis. Surprisingly, virtually all TNF-α-regulated genes appeared to be controlled by IKK2/NF-κB when analyzing cells with impaired NF-κB signaling. In contrast, the expression of a lower number of genes was additionally modulated by the p38 MAP kinase pathway. We furthermore identified genes that are negatively regulated by TNF-α via activation of NF-κB. A sufficient number of individual experiments and strict statistical criteria resulted in a high rate of confirmation at the mRNA and protein levels by real-time reverse transcriptase-polymerase chain reaction (RT-PCR) and flow cytometry. Our study provides the first microarray analysis that allows, due to statistically strict evaluation, reliable conclusions on both TNF-α-regulated gene expression and its dependence on distinct intracellular signaling pathways in endothelial cells.
Materials and methods
Cytokines and reagents
Human recombinant TNF-α was obtained from R&D Systems (Wiesbaden, Germany) and inhibitor SB202190 from Calbiochem (Bad Soden, Germany). All other reagents were purchased from Sigma-Aldrich (Deisenhofen, Germany) unless otherwise specified.
Cells and cell culture
Primary endothelial cells derived from human umbilical veins (HUVECs) were obtained from Clonetics (via Cell Systems, St Katharinen, Germany) and cultured as described elsewhere.7 HUVECs were used between passages 3 and 4 and exposed to TNF-α and/or SB202190 as indicated. The ΦNX amphotropic retrovirus producer cells were kindly provided by G. Nolan (Stanford, CA) and cultured as described elsewhere.7
Immunoprecipitation, immune-complex kinase assay, Western blotting
Preparation of whole-cell extracts was performed as described earlier.7 After removal of cell debris, HUVEC lysates were incubated with 1 μg/mL rabbit antiserum against mitogen-activated protein kinase-activated protein kinase-3/chromosome 3p kinase (MK-3/3pK), a p38 effector kinase. Thereafter, immune complexes were precipitated with protein A agarose (Roche Molecular Biochemicals, Mannheim, Germany). Samples were then incubated with heat shock protein 27 (Hsp27) as substrate for MK-3/3pK in the presence of 100 μM unlabeled adenosine triphosphate (ATP), 5 μCi (0.185 MBq) γ32[P]-ATP, and kinase buffer for 15 minutes at 30°C. Samples were subsequently subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blotted, and visualized by autoradiography or detected by phosphoimaging. Western blot analysis with a polyclonal antiserum against MK-3/3pK was performed to confirm equal loading of proteins.
Retroviral infection
The pCFG5-IEGZ retroviral vector containing an insert of kinase-dead IKK2 (dnIKK2), which is coupled to the expression of a green fluorescent protein (GFP)-zeocin resistance fusion gene through an internal ribosomal entry site, was used for infection of HUVECs as described.7 The efficacy of gene expression was controlled by flow cytometrically monitoring GFP expression, which ranged between 82% and 87% at day 9 after infection.
Flow cytometry
For detection of cell surface molecules on endothelial cells, nonspecific binding was blocked with 1% bovine serum albumin in phosphate-buffered saline (PBS), and immunostaining was performed with mouse monoclonal antibodies against VCAM-1, E-selectin (both obtained from Dianova, Hamburg, Germany), intercellular adhesion molecule-1 (ICAM-1; Immunotech, Marseille, France), CD69, CD137 (Becton Dickinson, Heidelberg, Germany), or corresponding isotype controls and fluorescein isothiocyanate (FITC)-conjugated second-stage reagents (Dianova). Retrovirally infected cells were fixed with 4% paraformaldehyde after stimulation and then successively stained with primary antibodies, biotin-SP-conjugated goat antimouse immunoglobulin G (IgG; Dianova) and streptavidin-Cy-chrome (Becton Dickinson). Fluorescence was determined using a FACScan (Becton Dickinson). Cells expressing dnIKK2 after retroviral infection were identified by detecting expression of GFP in the fluorescence channel 1 (FL1).
An intracellular flow cytometry staining procedure was employed for detection of chemokines and cytokines as described earlier.15 Cells were incubated with monoclonal antibodies against fractalkine, IL-6 (both obtained from R&D Systems), IL-8, or monocyte chemoattractant protein-1 (MCP-1; both obtained from Becton Dickinson), which had been diluted in permeabilization buffer (0.1% saponin/1% fetal calf serum/PBS). Thereafter, cells were stained with FITC-conjugated second-stage reagents or, in the case of retrovirally infected cells, with biotin-SP-conjugated goat antimouse IgG (Dianova) and streptavidin-Cy-chrome (Becton Dickinson), respectively.
DNA microarray hybridization
Total cellular RNA was isolated from 5 independent experiments with wild-type HUVECs that were exposed for 5 hours to medium as control, 10 μM SB202190 or 2 ng/mL TNF-α, in the absence or presence of 10 μM SB202190, respectively (RNeasy kit; Qiagen, Hilden, Germany). Four additional independent experiments were performed with HUVECs that had been infected with empty retroviral expression vector (n = 4) or a vector containing a dominant-negative mutant of IKK2 (n = 4). These cells were exposed to medium or stimulated with 2 ng/mL TNF-α resulting in 16 independent gene expression analyses. Thus, the degrees of freedom in these sets of experiments were 16 and 10, respectively.16
Samples for microarray hybridization was prepared according to the manufacturer's instructions (Affymetrix, Santa Clara, CA).
Ten micrograms of fragmented cRNA was, together with control cRNAs and grid alignment oligonucleotides, hybridized for 16 hours under constant rotation at 45°C to Affymetrix Human Genome 133 A Gene Chip arrays (Affymetrix). Transcripts are represented on the chip as probe sets consisting of about 11 probe pairs of 25 base pairs-long oligonucleotides, one set of perfect match sequences (PM) and one of mismatch sequences (MM); the latter contains a base substitution in the middle of the probe sequence. Representing transcripts by multiple oligonucleotides ensures technical replication within the array.17 Arrays were washed and stained using the Gene Chip Fluidics Station 400 (Affymetrix). Fluorescent signals were detected by the HP G2500A Gene Array Scanner (Affymetrix).
Statistical analysis of microarray data
Data were processed by MicroArray Suite (MAS) Software 5.0 (Affymetrix). Signals were scaled to a target intensity of 100 and log transformed. The algorithm applied in the MAS 5.0 software assigned individual average background values to each cell intensity and subtracted it from single raw cell intensities. To consider nonspecific binding and systematic errors of intensity ratios at low intensity levels,17,18 all probe sets with insignificant differences between PM and MM were removed by a further algorithm that computed discrimination values for each probe pair. Single raw values were calculated for each probe set (ie, transcript from the median of discrimination values and their significance was assessed by using a one-sided Wilcoxon rank test). Finally, the whole algorithm resulted in the determination of reasonable array zone-related background thresholds and the assignment of detection calls, “absent” or “present,” to each transcript.
While the MAS 5.0 software only allows comparison of 2 individual data sets, we further studied the Affymetrix intensity files with the Expressionist Suite software package (GeneData, Basel, Switzerland) for a more sophisticated statistical analysis. A chain of statistical methods is applied as a mining tool to identify genes significantly regulated in multiple independent experiments.19 The software package includes the Expressionist Refiner v3.0.4, which permits a global chip quality control with detection and masking of outliers and array defects, fluorescence gradient correction, and variance regularization. For final identification of significant gene expression differences we used Expressionist Analyst v4.0.5. First, data were normalized to a logarithmic mean of 280 in all experiments. In order to evaluate how the 20 data sets from experiments with wild-type HUVECs and the 16 data sets from experiments with retrovirally infected HUVECs were grouped together, we performed a principal component analysis. We filtered for genes with valid values (ie, an expression over background or a detection call “on”) in more than 50% of the respective experimental group. Finally, n-fold changes were determined and screened for their significance by applying a t test. We retained only genes with a fold change of at least 2.5 or no more than -2.5 and a P value of less than .05. To consider “on/off” phenomena in gene expression we defined genes that were present in each of the 5 individual experiments of the stimulation (or inhibition) groups and absent in all 5 experiments of the control group (ie, genes with an “on-off” ratio of 5:0, as significantly regulated). In the experimental settings with wild-type HUVECs we furthermore considered genes with on-off ratios of 0:5, 5:1, 1:5, 4:0, and 0:4, respectively, and in the experiments with vector-infected HUVECs we chose 4:0, 0:4, 4:1, 1:4, 3:0, and 0:3 ratios as significantly regulated. To differentiate on/off phenomena occurring around the background threshold from significant on/off phenomena we used normalized Analyst raw data and performed n-fold determinations and student t test calculations without applying the valid value filter. Being aware of the low significance of intensity ratios at low intensity levels we only defined regulations with a high fold change of at least 5 and a P value of less than .05 as relevant, since “on” then means an expression level high over background. Providing exact n-fold values would not be appropriate, since “off” means expression at background level. We therefore categorized on/off events in grades of expression over background as the following: + indicates n-fold range 5 to 10; ++, n-fold range 10 to 25; +++, n-fold range 25 to 50; and ++++, n-fold more than 50. Indicated gene accession numbers were derived from the GenBank database.
Real-time RT-PCR
For real-time RT-PCR, RNA from every experimental group was analyzed in duplicate. cDNA was synthesized from 4 μg of total RNA using SuperScript II RNase H-reverse transcriptase (Invitrogen, Carlsbad, CA). Primers were designed using the Primer Express software package (Applied Biosystems, Foster City, CA) and obtained from MWG Biotech (Ebersberg, Germany). The primers used for PCR analysis were as follows: bone morphogenetic protein 2 (BMP2) forward, 5′-TGCTAGTAACTTTTGGCCATGATG-3′ and reverse, 5′-GTTTCCGCTGTTTGTGTTTGG-3′; VCAM-1 forward, 5′-ACATGGAATTCGAACCCAAACA-3′ and reverse, 5′-GGCTGACCAAGACGGTTGTATC-3′; ICAM-1 forward, 5′-ACCTCCCCACCCACATACATTT-3′ and reverse, 5′-GGCATAGCTTGGGCATATTCC-3′; CD137 forward, 5′-TCCGCAGATCATCTCCTTCTTTC-3′ and reverse, 5′-TTTCTGCCCCGTTTAACAACAG-3′; cycloxygenase 2 (Cox2) forward, 5′-GGGTTGCTGGTGGTAGGAATG-3′ and reverse, 5′-AGCATAAAGCGTTTGCGGTACTC-3′; TNF-related apoptosis-inducing ligand (TRAIL) forward, 5′-GTCTCTCTGTGTGGCTGTAACTTACG-3′ and reverse, 5′-AAACAAGCAATGCCACTTTTGG-3′; cellular inhibitors of apoptosis protein 1 (c-IAP-1) forward, 5′-CAAGTGGTTTCCAAGGTGTGAGTAC-3′ and reverse, 5′-GATGTGGATAGCAGCTGTTCAAGTAG-3′; IMAGE 4711494 forward, 5′-CAAGAAGGGTTTTTGTGACTGAATC-3′ and reverse, 5′-TCCTTGTTTTGCTCCAACACTAATC-3′; coxsackie virus and adenovirus receptor (CAR) forward, 5′-GCTAAGGTAGCTGCCCCTAATCTAAG-3′ and reverse, 5′-CGGAGAGCACAGATGAGACATATG-3′. Real-time RT-PCR was performed using the Quanti-Tect SYBR Green PCR kit (Qiagen) under conditions as described earlier.20 Data were acquired with the ABI PRISM 7900 (Applied Biosystems). Gene expression was normalized with respect to endogenous housekeeping control genes, glyceraldehyde phosphate dehydrogenase (GAPDH) and interferon-stimulated gene factor (ISGF), which were determined not to have significantly changed in the different experiments. The relative expression of respective genes was calculated by using the comparative threshold cycle (CT) method as described.21
Results
TNF-α-induced gene expression in primary endothelial cells
To assess the gene expression profile of cytokine-stimulated primary endothelial cells, HUVECs were exposed to 2 ng/mLTNF-α for 5 hours or left untreated. This early time point was chosen to minimize indirect autocrine or paracrine effects. Total RNA was isolated from HUVECs obtained from 5 independent experiments and processed for DNA microarray hybridization. Efficacy of endothelial activation was controlled by flow cytometric analysis of E-selectin and ICAM-1 expression (Figure 1). Restricting the inclusion criteria to at least a 2.5-fold increase of gene expression or a considerable on/off switch (for details refer to “Materials and methods”) we identified 58 genes that were induced upon exposure to TNF-α. Most of the genes could be assigned to functional groups such as chemokines/cytokines, cell surface receptors, components of the coagulation system, inflammatory response genes, signal and transcription factors, apoptosis- or cell proliferation-related genes, metabolism components, or cell transporters/channels (Figure 2A). Our analysis added 23 new molecules to the list of TNF-α-inducible endothelial genes that had not been described earlier. These novel genes include activin β, CD69, CD137, Nef-associated factor (Naf1), Jun B proto-oncogene, nuclear receptor interacting protein-1 (NRIP-1), and TRAIL (Table 1; the complete and detailed list of genes is available on the Blood website; see the Supplemental Tables link at the top of the online article).
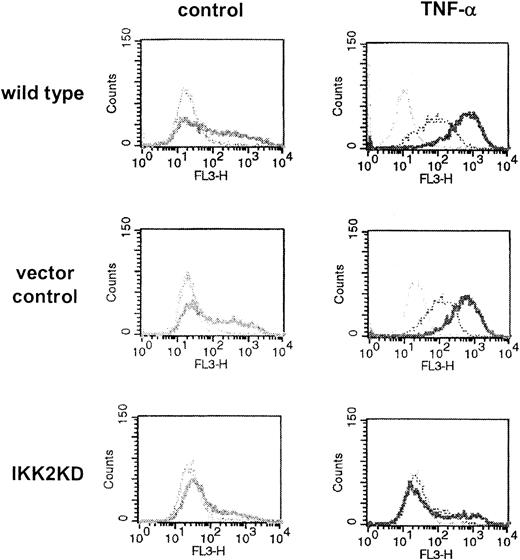
TNF-α-regulated expression of adhesion molecules ICAM-1 and E-selectin dependence on the IKK2/NF-κB pathway. To control for successful activation of the endothelial cells later to be processed for microarray analysis as well as for functionality of the dominant-negative IKK2 mutant, flow cytometry was performed in parallel experiments. Briefly, wild-type primary endothelial cells (HUVECs, top) or cells infected with parental vector as control (middle) or with retrovirus carrying a dominant-negative mutant of IKK2 (bottom) were exposed to medium or 2 ng/mL TNF-α. Cells were immunostained for ICAM-1 (bold profiles), E-selectin (dotted profiles), or incubated with IgG isotype control (thin profiles). Successful infection of HUVECs with the retrovirus was confirmed by coexpression of the GFP gene.
TNF-α-regulated expression of adhesion molecules ICAM-1 and E-selectin dependence on the IKK2/NF-κB pathway. To control for successful activation of the endothelial cells later to be processed for microarray analysis as well as for functionality of the dominant-negative IKK2 mutant, flow cytometry was performed in parallel experiments. Briefly, wild-type primary endothelial cells (HUVECs, top) or cells infected with parental vector as control (middle) or with retrovirus carrying a dominant-negative mutant of IKK2 (bottom) were exposed to medium or 2 ng/mL TNF-α. Cells were immunostained for ICAM-1 (bold profiles), E-selectin (dotted profiles), or incubated with IgG isotype control (thin profiles). Successful infection of HUVECs with the retrovirus was confirmed by coexpression of the GFP gene.
Assignment of TNF-α-regulated genes to functional groups. Endothelial genes that are induced at least 2.5-fold or switched on (A) or repressed at least 2.5-fold or switched off (B) by TNF-α are assigned to functional groups.
Assignment of TNF-α-regulated genes to functional groups. Endothelial genes that are induced at least 2.5-fold or switched on (A) or repressed at least 2.5-fold or switched off (B) by TNF-α are assigned to functional groups.
TNF-α-mediated up-regulation of selected IKK2/NF-κB- and p38 MAP kinase-dependent genes in endothelial cells (HUVECs)
Genes up-regulated by TNF-α . | Accession no. . | Fold-change or grade of expression over background in wild-type HUVECsa . | P . | Induction dependent on IKK2/NF-κB, % . | Induction dependent on p38 MAP kinase, % . |
|---|---|---|---|---|---|
| Chemokines/cytokines | |||||
| IL-8 | AF043337.1 | 17.3 | < .001 | 100.0 | 53.1 |
| Bone morphogenetic protein 2 | AA583044 | 4.8 | < .001 | 77.4 | 50.9 |
| Inhibin beta Ab | M13436.1 | 3.1 | < .001 | 100.0 | 44.7 |
| CXCL6c | NM_002993.1 | 2.7 | < .001 | 100.0 | No |
| Fraktalkined | NM_002996.1 | +++ | < .05 | 100 | No |
| Cell-surface receptors | |||||
| VCAM-1 | NM_001078.1 | 38.1 | < .001 | 100.0 | 52.7 |
| E-selectin | NM_000450.1 | 35.7 | < .001 | 100.0 | No |
| ICAM-1 | NM_000201.1 | 22.1 | < .001 | 100.0 | No |
| CD137 antigene | NM_001561.2 | 3.0 | < .001 | ND | No |
| CD69 antigen | L07555.1 | ++ | < .05 | 100.0 | No |
| Coagulation | |||||
| Coagulation factor IIIf | NM_001993.2 | ++ | < .05 | ND | 100.0 |
| Inflammation/immune response | |||||
| Cyclo-oxygenase 2g | NM_000963.1 | 2.9 | < .005 | ND | 100.0 |
| Signaling/transcription | |||||
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alphah | AI078167 | 7.4 | < .001 | 100.0 | No |
| Jagged 1 | U77914.1 | 5.0 | < .001 | 100.0 | No |
| Nef-associated factor 1i | NM_006058.1 | 4.7 | < .001 | 100.0 | No |
| Ephrin-A1j | NM_004428.1 | 4.5 | < .001 | 78.9 | No |
| Jun B proto-oncogene | NM_002229.1 | 3.2 | < .001 | No | No |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2k | BC002844.1 | 3.0 | < .05492 | 100.0 | No |
| Nuclear receptor interacting protein 1l | A1824012 | 2.9 | < .001 | 100 | 44.0 |
| Apoptosis/cell proliferation | |||||
| TNF-α-induced protein 3m | NM_006290.1 | 13.0 | < .001 | 100.0 | No |
| TRAILn | NM_003810.1 | 4.0 | < .005 | ND | 46.4 |
| c-IAP-2o | U37546.1 | ++ | < .05 | 100.0 | No |
| Metabolism | |||||
| Guanylate binding protein 1, interferon-inducible, 67 kDa | NM_002053.1 | 3.6 | < .001 | 100.0 | 49.3 |
| Transporters/channels | |||||
| Transporter 1, ATP-binding cassette, subfamily B | NM_000593.2 | + | < .05 | 100.0 | No |
| Others | |||||
| HIV type 1 enhancer binding protein 2p | AL023584 | 4.0 | < .001 | 66.0 | No |
Genes up-regulated by TNF-α . | Accession no. . | Fold-change or grade of expression over background in wild-type HUVECsa . | P . | Induction dependent on IKK2/NF-κB, % . | Induction dependent on p38 MAP kinase, % . |
|---|---|---|---|---|---|
| Chemokines/cytokines | |||||
| IL-8 | AF043337.1 | 17.3 | < .001 | 100.0 | 53.1 |
| Bone morphogenetic protein 2 | AA583044 | 4.8 | < .001 | 77.4 | 50.9 |
| Inhibin beta Ab | M13436.1 | 3.1 | < .001 | 100.0 | 44.7 |
| CXCL6c | NM_002993.1 | 2.7 | < .001 | 100.0 | No |
| Fraktalkined | NM_002996.1 | +++ | < .05 | 100 | No |
| Cell-surface receptors | |||||
| VCAM-1 | NM_001078.1 | 38.1 | < .001 | 100.0 | 52.7 |
| E-selectin | NM_000450.1 | 35.7 | < .001 | 100.0 | No |
| ICAM-1 | NM_000201.1 | 22.1 | < .001 | 100.0 | No |
| CD137 antigene | NM_001561.2 | 3.0 | < .001 | ND | No |
| CD69 antigen | L07555.1 | ++ | < .05 | 100.0 | No |
| Coagulation | |||||
| Coagulation factor IIIf | NM_001993.2 | ++ | < .05 | ND | 100.0 |
| Inflammation/immune response | |||||
| Cyclo-oxygenase 2g | NM_000963.1 | 2.9 | < .005 | ND | 100.0 |
| Signaling/transcription | |||||
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alphah | AI078167 | 7.4 | < .001 | 100.0 | No |
| Jagged 1 | U77914.1 | 5.0 | < .001 | 100.0 | No |
| Nef-associated factor 1i | NM_006058.1 | 4.7 | < .001 | 100.0 | No |
| Ephrin-A1j | NM_004428.1 | 4.5 | < .001 | 78.9 | No |
| Jun B proto-oncogene | NM_002229.1 | 3.2 | < .001 | No | No |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2k | BC002844.1 | 3.0 | < .05492 | 100.0 | No |
| Nuclear receptor interacting protein 1l | A1824012 | 2.9 | < .001 | 100 | 44.0 |
| Apoptosis/cell proliferation | |||||
| TNF-α-induced protein 3m | NM_006290.1 | 13.0 | < .001 | 100.0 | No |
| TRAILn | NM_003810.1 | 4.0 | < .005 | ND | 46.4 |
| c-IAP-2o | U37546.1 | ++ | < .05 | 100.0 | No |
| Metabolism | |||||
| Guanylate binding protein 1, interferon-inducible, 67 kDa | NM_002053.1 | 3.6 | < .001 | 100.0 | 49.3 |
| Transporters/channels | |||||
| Transporter 1, ATP-binding cassette, subfamily B | NM_000593.2 | + | < .05 | 100.0 | No |
| Others | |||||
| HIV type 1 enhancer binding protein 2p | AL023584 | 4.0 | < .001 | 66.0 | No |
Data shown in bold represent observations that have so far not been reported in literature.
ND indicates not detectable, since our strict statistical criteria do not appraise the respective gene transcripts in empty expression vector-infected HUVECs as TNF-α inducible; and +++ indicates an expression over background with an n-fold range of 25-50; ++, n-fold range of 10 to 25; and +, n-fold range of 5 to 10.
For details, see “Materials and methods.”
Activin beta-A chain.
GCP-2.
CX3CL1.
TNF receptor superfamily, member 9.
Thromboplastin, tissue factor, CD142.
Cox2, prostaglandin-endoperoxidase synthase 2.
IκBα.
Naf1.
TNF-α-induced protein 4.
p49/p100.
NRIP-1.
A20.
Tumor necrosis factor (ligand) superfamily; member 10.
Baculoviral IAP repeat-containing 3.
HIVEP2.
Identification of IKK2-dependent genes
To examine the impact of NF-κB on TNF-α-induced gene expression we infected HUVECs with a retroviral vector bearing an insert of dnIKK2 or empty parental expression vector as control. Gene expression profiles obtained after exposure to TNF-α were studied in 4 independent experiments. Efficient inhibition of the IKK2/NF-κB pathway was controlled by flow cytometric analysis of TNF-α-induced surface expression of E-selectin and ICAM-1 as described earlier7 (Figure 1). Intracellular signaling pathways not related to NF-κB were not affected (eg, interferon-γ-induced expression of ICAM-1; data not shown).7 In unstimulated cells, blockade of IKK2 did not exert a relevant alteration in gene induction of at least 2.5-fold. Upon exposure to TNF-α, all 58 genes except one (Jun B proto-oncogene) identified in wild-type HUVECs were also up-regulated in vector-infected HUVECs; however, 11 of these did not meet our strict inclusion criteria including tissue factor and Cox2. TNF-α-induced gene expression was completely inhibited in cells expressing dnIKK2 (Table 1). In 21 of these genes IKK2/NF-κB dependence could be demonstrated here for the first time (the complete and detailed list of genes is provided in the Supplemental Tables).
Identification of TNF-α-inducible p38-dependent genes
For identification of TNF-α-inducible genes that are regulated in a p38 MAP kinase-dependent manner, HUVECs were stimulated with 2 ng/mL TNF-α for 5 hours in the absence or presence of 10 μM SB202190. Efficient blockade of p38 activity was confirmed by measuring the activity of a p38 effector kinase, MK-3/3pK, by immune complex kinase assay and by demonstrating inhibition of VCAM-1 surface expression that is known to be p38 dependent (Figure 3). Incubation of HUVECs with SB202190 in the absence of TNF-α did not result in at least a 2.5-fold induction of gene expression. Selecting only genes that were inhibited for at least 40% with a significance of P less than .05 in 5 independent experiments analysis revealed that pretreatment of cells with SB202190 significantly reduced the TNF-α-mediated up-regulation of 11 genes and completely blocked transcriptional induction of another 2 genes (Table 1; the complete and detailed list of genes is provided in the Supplemental Tables). Two levels of inhibition, about 50% and 100%, respectively, became apparent, which may indicate different mechanisms of p38-dependent gene regulation. For 8 of the genes, a dependence on p38 MAP kinase is demonstrated here for the first time.
Activation of endothelial p38 MAP kinase by TNF-α is efficiently inhibited by SB202190. To confirm efficient inhibition of p38 activity in endothelial cells to be studied by microarray profiling, immune complex kinase assays (A) and flow cytometry (B) were performed on cells obtained from parallel experiments. (A) HUVECs were exposed to medium or TNF-α for 30 or 300 minutes in the absence or presence of 10 μM SB202190, which had been added to the culture medium 30 minutes prior to stimulation. After cell lysis, MK-3/3pK, a kinase acting downstream of p38, was immunoprecipitated and assayed for activity using Hsp27 as substrate. Protein loads were controlled by Western blot using an antiserum against MK-3/3pK. (B) Cells were stimulated with TNF-α for 5 hours in the absence or presence of 10 μM SB202190 as indicated and subsequently studied by flow cytometry for surface expression of an established p38-dependent gene, VCAM-1 (thick lines). Isotype controls are additionally shown (thin lines).
Activation of endothelial p38 MAP kinase by TNF-α is efficiently inhibited by SB202190. To confirm efficient inhibition of p38 activity in endothelial cells to be studied by microarray profiling, immune complex kinase assays (A) and flow cytometry (B) were performed on cells obtained from parallel experiments. (A) HUVECs were exposed to medium or TNF-α for 30 or 300 minutes in the absence or presence of 10 μM SB202190, which had been added to the culture medium 30 minutes prior to stimulation. After cell lysis, MK-3/3pK, a kinase acting downstream of p38, was immunoprecipitated and assayed for activity using Hsp27 as substrate. Protein loads were controlled by Western blot using an antiserum against MK-3/3pK. (B) Cells were stimulated with TNF-α for 5 hours in the absence or presence of 10 μM SB202190 as indicated and subsequently studied by flow cytometry for surface expression of an established p38-dependent gene, VCAM-1 (thick lines). Isotype controls are additionally shown (thin lines).
TNF-α-mediated down-regulation of gene expression
TNF-α stimulation of HUVECs resulted in significant down-regulation or complete transcriptional block of 18 genes (Figure 2B; Table 2; the complete and detailed list of genes is provided in the Supplemental Tables). Similar results were obtained with retroviral control vector-infected cells where 7 of these genes met our strict inclusion criteria. Analyzing cells expressing dominant-negative IKK2, we realized that TNF-α-mediated down-regulation of these genes was almost completely dependent on the IKK2/NF-κB pathway. The only gene that was switched off upon TNF-α treatment in an IKK2-independent manner was DKFZp761P1010, which codes for a hypothetical protein with yet unknown function. In addition, blockade of the IKK2/NF-κB pathway repressed basal expression of 4 genes in a range of 2.5- to 2.7-fold (ie, thyroid-stimulating hormone, adaptor-related protein complex 4, glutaminylpeptide cyclotransferase, and clone 24479 mRNA; data not shown). None of the 18 genes repressed by TNF-α inhibition could be abrogated by blockade of p38 MAP kinase. SB202190 blocked the basal expression of 4 genes: regulator of G-protein signaling 3, RGC32 protein, hypothetical protein FLJ23056, and protein kinase NM18423 (data not shown).
TNF-α-mediated down-regulation of selected genes in endothelial cells
Genes down-regulated by TNF-α . | Accession no. . | Fold-change or grade of expression over background in wild-type HUVECs* . | P . | Inhibition dependent on IKK2/NF-κB, % . | Inhibition dependent on p38 MAP kinase, % . |
|---|---|---|---|---|---|
| Cell-surface receptors | |||||
| Coxsackie virus and adenovirus receptor† | NM_001338.1 | −2.8 | < .001 | 100.0 | No |
| Signaling/transcription | |||||
| Inhibitor of DNA binding 1, dominant-negative helix-loop-helix protein | D13889 | −3.6 | < .001 | ND | No |
| SRY-box 18‡ | NM_018419.1 | −3.2 | < .001 | 100 | No |
| CBP/p300-interacting transactivator 2§ | AF109161.1 | −2.9 | < .001 | ND | |
| v-yes-1 Yamaguchi sarcoma viral-related oncogene homolog | AI356412 | −2.8 | < .005 | 100.0 | No |
| Inflammation/immune response | |||||
| DnaJ homolog, subfamily B; member 4∥ | BG252490 | −4.0 | < .001 | 100.0 | No |
| Apoptosis/cell proliferation/cell cycle | |||||
| Cyclin E2 | NM_004702.1 | −2.7 | < .001 | ND | No |
| Cyclin A1 | NM_003914.1 | −2.5 | < .001 | ND | No |
Genes down-regulated by TNF-α . | Accession no. . | Fold-change or grade of expression over background in wild-type HUVECs* . | P . | Inhibition dependent on IKK2/NF-κB, % . | Inhibition dependent on p38 MAP kinase, % . |
|---|---|---|---|---|---|
| Cell-surface receptors | |||||
| Coxsackie virus and adenovirus receptor† | NM_001338.1 | −2.8 | < .001 | 100.0 | No |
| Signaling/transcription | |||||
| Inhibitor of DNA binding 1, dominant-negative helix-loop-helix protein | D13889 | −3.6 | < .001 | ND | No |
| SRY-box 18‡ | NM_018419.1 | −3.2 | < .001 | 100 | No |
| CBP/p300-interacting transactivator 2§ | AF109161.1 | −2.9 | < .001 | ND | |
| v-yes-1 Yamaguchi sarcoma viral-related oncogene homolog | AI356412 | −2.8 | < .005 | 100.0 | No |
| Inflammation/immune response | |||||
| DnaJ homolog, subfamily B; member 4∥ | BG252490 | −4.0 | < .001 | 100.0 | No |
| Apoptosis/cell proliferation/cell cycle | |||||
| Cyclin E2 | NM_004702.1 | −2.7 | < .001 | ND | No |
| Cyclin A1 | NM_003914.1 | −2.5 | < .001 | ND | No |
All data shown represent observations that have so far not been reported in literature.
ND indicates not detectable, since our strict statistical criteria do not appraise the respective gene transcripts in empty expression vector-infected HUVECs as TNF-α inducible.
For details, see “Materials and methods.”
CAR.
Sex-determining region Y.
MSG-related protein 1; MRG1 protein.
HSP40.
Confirmation of microarray data by real-time RT-PCR and flow cytometry
Quantitative real-time RT-PCR of 8 exemplary genes (BMP2, VCAM-1, ICAM-1, CD137, Cox2, TRAIL, c-IAP-2, IMAGE 4711) confirmed expression patterns obtained by microarray analysis (Table 3). Validation of microarray data by RT-PCR has earlier been reported to be only rarely successful in cases with an induction of no more than 4.0-fold20 ; nevertheless, our analysis even verified regulation of such genes (Cox2, CD137).
Confirmation of microarray data by real-time RT-PCR: TNF-α—mediated differential gene expression in HUVECs
Genes . | Accession no. . | Fold-change in wild-type HUVECs after exposure to TNF-α, analyzed by real-time RT-PCR . | IKK2/NF-kB—dependent regulation, analyzed by real-time RT-PCR . | p38 MAP kinase—dependent regulation, analyzed by real-time RT-PCR . | Confirmation of complete microarray data set by real-time RT-PCR . |
|---|---|---|---|---|---|
| Bone morphogenetic protein 2* | AA583044 | 6.0 | Yes | Yes | Yes |
| VCAM-1 | NM_001078.1 | 132.5 | Yes | Yes | Yes |
| ICAM-1 | NM_000201.1 | 65.3 | Yes | No | Yes |
| CD137† | NM_001561.2 | 32.9 | Yes | No | IKK2/NF-kB dependence not detected by microarray |
| Cyclo-oxygenase 2‡ | NM_000963.1 | 4.1 | ND | Yes | Yes |
| TRAIL§ | NM_003810.1 | 4.9 | Yes | Yes | Yes |
| c-IAP-2∥ | U37546.1 | 84.5 | Yes | No | Yes |
| Clone IMAGE:4711494, mRNA | BF575213 | 16.0 | Yes | No | Yes |
| Coxsackie virus and adenovirus receptor¶ | NM_001338.1 | −30.9 | Yes | No | Yes |
Genes . | Accession no. . | Fold-change in wild-type HUVECs after exposure to TNF-α, analyzed by real-time RT-PCR . | IKK2/NF-kB—dependent regulation, analyzed by real-time RT-PCR . | p38 MAP kinase—dependent regulation, analyzed by real-time RT-PCR . | Confirmation of complete microarray data set by real-time RT-PCR . |
|---|---|---|---|---|---|
| Bone morphogenetic protein 2* | AA583044 | 6.0 | Yes | Yes | Yes |
| VCAM-1 | NM_001078.1 | 132.5 | Yes | Yes | Yes |
| ICAM-1 | NM_000201.1 | 65.3 | Yes | No | Yes |
| CD137† | NM_001561.2 | 32.9 | Yes | No | IKK2/NF-kB dependence not detected by microarray |
| Cyclo-oxygenase 2‡ | NM_000963.1 | 4.1 | ND | Yes | Yes |
| TRAIL§ | NM_003810.1 | 4.9 | Yes | Yes | Yes |
| c-IAP-2∥ | U37546.1 | 84.5 | Yes | No | Yes |
| Clone IMAGE:4711494, mRNA | BF575213 | 16.0 | Yes | No | Yes |
| Coxsackie virus and adenovirus receptor¶ | NM_001338.1 | −30.9 | Yes | No | Yes |
ND indicates not detectable, since strict statistical criteria do not appraise the gene transcript in empty expression vector—infected HUVECs as TNF-α induced.
BMP2.
TNF receptor superfamily member 9.
Cox2, prostaglandin-endoperoxidase synthase 2.
Tumor necrosis factor (ligand) superfamily member 10.
Baculoviral IAP repeat—containing 3.
CAR.
Dependence on the IKK2/NF-κB pathway could be verified in all cases. Due to our inclusion criteria, TRAIL has not been considered as TNF-α inducible in the experimental set of empty expression vector-infected endothelial cells; IKK2/NF-κB dependence of TRAIL could therefore not be tested. Nevertheless, we found a 2.24-fold induction of TRAIL by TNF-α in vector cells that was significantly inhibited in cells expressing dominant-negative IKK2 (data not shown); these observations were confirmed by real-time RT-PCR. Regarding the p38 pathway, real-time RT-PCR also completely confirmed microarray data (Table 3).
Intracellular and surface flow cytometry analyses were performed to verify gene expression at the protein level. TNF-α inducibility was exemplarily confirmed for surface receptors CD69 and CD137 and chemokines fractalkine, IL-8 (Figure 4), and MCP-1 (data not shown). Expression of E-selectin, ICAM-1, and VCAM-1 as analyzed in our setting of quality control was in complete accordance with the microarray data (Figures 1 and 3B; Tables 1-2). In addition, we verified that endothelial expression of CD69 occurs in an IKK2/NF-κB-dependent manner (Figure 5A). Divergent results were only obtained in one case: consistent with microarray data, flow cytometry revealed an up-regulation of CD137 in wild-type but not in vector-infected endothelial cells after TNF-α stimulation (Figures 4 and 5B). Employing real-time RT-PCR, we detected IKK2/NF-κB-dependent induction of CD137 in vector-infected cells but only after rather high numbers of PCR cycles. In this case real-time RT-PCR appeared to be more sensitive than the microarray, however, without consequences with respect to protein expression.
Confirmation of TNF-α-regulated endothelial gene expression profiles by flow cytometry. Expression of selected genes found to be TNF-α-regulated by microarray analysis was surveyed by flow cytometry. Endothelial cells were stimulated with TNF-α for 16 hours and processed for surface (CD69, CD137) or intracellular flow cytometry (fractalkine, IL-8, IL-6), respectively. Specific profiles are shown by thick lines and isotype control profiles appear as thin lines.
Confirmation of TNF-α-regulated endothelial gene expression profiles by flow cytometry. Expression of selected genes found to be TNF-α-regulated by microarray analysis was surveyed by flow cytometry. Endothelial cells were stimulated with TNF-α for 16 hours and processed for surface (CD69, CD137) or intracellular flow cytometry (fractalkine, IL-8, IL-6), respectively. Specific profiles are shown by thick lines and isotype control profiles appear as thin lines.
TNF-α-regulated expression of CD69 and CD137 after inhibition of the IKK2/NF-κB signaling pathway. Endothelial cells were infected with parental retrovirus or retrovirus containing dominant-negative IKK2 and subsequently stimulated with 2 ng/mL TNF-α for 16 hours. (A) Flow cytometry of CD69 surface expression revealed a significant induction by TNF-α in cells infected with the parental virus; this can be completely blocked in cells expressing dominant-negative IKK2. (B) In contrast to wild-type HUVECs, retrovirally infected HUVECs do not up-regulate CD137 upon exposure to TNF-α. Specific profiles are shown by thick lines and isotype control profiles appear as thin lines.
TNF-α-regulated expression of CD69 and CD137 after inhibition of the IKK2/NF-κB signaling pathway. Endothelial cells were infected with parental retrovirus or retrovirus containing dominant-negative IKK2 and subsequently stimulated with 2 ng/mL TNF-α for 16 hours. (A) Flow cytometry of CD69 surface expression revealed a significant induction by TNF-α in cells infected with the parental virus; this can be completely blocked in cells expressing dominant-negative IKK2. (B) In contrast to wild-type HUVECs, retrovirally infected HUVECs do not up-regulate CD137 upon exposure to TNF-α. Specific profiles are shown by thick lines and isotype control profiles appear as thin lines.
One gene that we would have expected to be up-regulated by TNF-α in endothelial cells was IL-6. To our surprise, however, microarray analysis did not reveal an up-regulation. To assess reliability of microarray data we studied IL-6 expression by intracellular flow cytometry. HUVECs expressed IL-6 already under basal conditions, and stimulation by TNF-α did indeed not further up-regulate its level of expression (Figure 4).
Finally, we controlled the expression of a gene that is down-regulated by TNF-α. Real-time RT-PCR confirmed that the CAR is repressed upon stimulation with TNF-α (Table 3). Overall, control experiments at the mRNA and protein levels revealed a high degree of congruence with the microarray data impressively confirming the reliability and validity of our experimental approach.
Discussion Employing oligonucleotide microarray technology we systemati cally analyzed the gene expression program of primary endothelial cells as it is elicited by TNF-α. The validity of such approaches strongly depends on the quality of the experimental design.16 Affymetrix U133A oligonucleotide microarrays covering at least 13 000 characterized human full-length genes and Expressionist software allowed a comprehensive and reliable acquisition of expression data that was assured by multiple oligonucleotides for a particular gene ensuring technical replication,17 accurate normalization and background subtraction, and thorough analysis with strict statistical evaluation.18
Most earlier reports studying TNF-α-induced gene expression were hampered by a lack of sufficient technical and experimental replications (usually n ≤ 2),22-28 which, however, are essential for estimation of variability and statistical validity.16 Control experiments of these studies at the protein level only included genes already known to be induced by TNF-α in endothelial cells such as IL-8 or MCP-1.25,28 The highest standard of quality is provided in a recent study on human aortic endothelial cells, which included sufficient experimental replicates allowing a validated statistical analysis.29 However, the microarray filters used in this study comprised only 2 technical replicates per gene, and arrays did not contain mismatch sequences for individual genes to control nonspecific background binding. This may explain why, depending on the amplification procedure employed, lists of up to 1150 differentially regulated genes were obtained in that study. The authors reported a maximal validation of 67% of differential regulated genes by quantitative real-time PCR that is significantly lower compared with the rate of confirmation obtained in the present study.29 Global gene expression patterns of endothelial cells were not related to distinct intracellular signaling pathways in any of the previous reports.
In our study we only considered genes as regulated by TNF-α and assigned the regulation to distinct intracellular signaling pathways when confirmation could be achieved on the basis of statistical analysis of 5 independent experiments. TNF-α regulation of these genes was furthermore confirmed in another 4 independent experiments using cells infected with parental control retrovirus. Our study thus provides not simply a phenotypical screening but rather a statistically validated definition of TNF-α-induced endothelial genes. Microarray data were confirmed to almost 90% at the mRNA and protein level, which reflects the quality of study design and analysis.
In the present report, we identified 58 out of 13 000 genes to be significantly up-regulated by TNF-α in endothelium. Most of the genes can be subsumed to a group with primarily inflammatory functions (eg, chemokines and adhesion molecules). Other genes identified here act antiapoptotic or promote survival and cell growth (A20, c-IAP-2, superoxide dismutase 2). A third group is characterized by proapoptotic and antiproliferative properties (TRAIL). A number of genes play a role in feedback regulation of the NF-κB pathway (IκBα, A20, nuclear factor of κ light polypeptide gene enhancer in B cells 2, and jagged 1). The potential roles of the latter as down-regulators of NF-κB activation in endothelial cells have not been studied yet.
A considerable number of the genes have not previously been identified to be regulated by TNF-α. For others, endothelial expression and regulation by TNF-α is demonstrated for the first time (among these are inhibin βA, Naf1, Jun B, NRIP-1, TRAIL, CD69, and CD137). Flow cytometric analysis of the latter revealed that also minor but still significant fold changes of gene expression observed by the microarray technology can be confirmed at the protein level.
A major goal of our study was to analyze the role of 2 principal proinflammatory signaling cascades (ie, the NF-κB and p38 MAP kinase pathways) for TNF-α stimulation. Blockade of the NF-κB pathway was achieved by retroviral expression of a dominant-negative mutant of IKK2.7 In vector-infected control cells TNF-α induction of all 58 genes found in wild-type cells could be confirmed (except Jun B proto-oncogene); 11 genes merely did not fully meet our strict inclusion criteria. Slightly reduced levels of gene induction in vector-infected cells may be due to preactivation caused by overexpression of selection marker genes of the retroviral construct.30,31 However, the general pattern of gene expression was not altered, excluding major effects of the retrovirus itself.
A somehow surprising observation of our study was that virtually all genes that are up-regulated or switched on by TNF-α required activation of IKK2. In some of these genes NF-κB dependence has not previously been described (among others BMP2, inhibin βA, CXCL6, Naf1, ephrin A1, NRIP-1, CD69, and jagged 1). In 3T3-L1 adipocytes, inhibition of the IKK2/NF-κB pathway suppressed only about 60% to 70% of the genes normally induced by TNF-α.24 In HeLa cells, 14 of 16 genes were identified by quantitative real-time PCR that were inducible by TNF-α in an NF-κB-dependent manner, but this report lacks a general analysis of NF-κB-dependent gene expression.26 Our observation of total dependence of TNF-α-induced gene expression patterns on the IKK2/NF-κB pathway may therefore be a particular property of endothelial cells. It may explain the fact that pharmacologic inhibition of NF-κB predominantly targets proinflammatory events despite ubiquitous abundance of this transcription factor system.
Interestingly, a minor number of 13 genes appears to underlie significant but not essential additional control by the p38 MAP kinase pathway. For 9 of these genes, p38-dependent regulation was identified here for the first time (eg, TRAIL, BMP2, and activin βA). The mechanism of p38-dependent regulation of gene expression is not completely understood, but increasing evidence suggests specific posttranscriptional events that result in stabilization of mRNA. Upon treatment with extracellular stimuli such as cytokines, the stability of mRNAs containing adenosine uridine (AU)-rich elements (AREs) can be increased in a p38-dependent and ARE-controlled manner.32,33 A search in the ARED 2.0 database34 revealed that 3 genes that we identified as p38 dependent (ie, NRIP-1, guanylate binding protein-1, and chromosome 8 open reading frame 4) contain ARE motifs; for these a similar mechanism may be operative. However, the expression of a number of other TNF-α-inducible genes that also contain ARE is apparently not inhibited by blockade of p38. Furthermore, p38 may modulate gene expression at the level of transactivation by interacting with components of the enhanceosome, such as the transcriptional coactivator CBP/p300.15 Recently, mitogen- and stress-activated protein kinase-1 (MSK1), which acts downstream of p38 kinase, has been identified to phosphorylate the NF-κB/Rel family member p65 as well as components of the chromatin environment, thereby interfering with NF-κB-dependent gene transcription.35
A surprising result of our study was the identification of a substantial number of genes that were significantly down-regulated by TNF-α. Results in vector- and IKK2-mutant-infected cells showed that down-regulation also requires activation of the IKK2/NF-κB pathway, whereas no role can be assigned to p38 MAP kinase signaling. Down-regulated genes include the CAR, transactivating factors, and kinases promoting cell cycle progression. It thus appears conceivable that pharmacologic interference with TNF-α or NF-κB might result in undesirable inhibition of gene depression and lead to adverse effects, such as increased susceptibility for coxsackie virus or adenovirus infections36 or uncontrolled cell cycle progression.
Overall, the results obtained in this study are 3-fold. First, we here provide a systematic and statistically validated definition of TNF-α-induced gene expression in endothelial cells. Second, regulation of individual genes by TNF-α was assigned to distinct intracellular signaling pathways. Virtually all TNF-α-regulated genes were dependent on the IKK2/NF-κB pathway, whereas p38 MAP kinase modulated the expression of a minor number of genes. Third, we identified novel target genes that are up-regulated upon exposure to TNF-α in an IKK2/NF-κB- and/or p38-dependent manner. A statistically supported definition of gene expression patterns in distinct target cells, such as that presented here for endothelial cells, is a prerequisite for the development of more specific treatment strategies in inflammatory diseases.
Prepublished online as Blood First Edition Paper, January 8, 2004; DOI 10.1182/blood-2003-09-3296.
Supported by grants from the Interdisciplinary Center of Clinical Research University of Muensterto (J.R.) and from the Deutsche Forschungsgemeinschaft (DFG; grant Go 811-1/3; M.G. and S.L.).
D.V. and M.G. contributed equally to this work.
The online version of the article includes a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal