Abstract
NALP proteins are recently identified members of the CATERPILLER (CARD, transcription enhancer, R(purine)-binding, pyrin, lots of LRR) family of proteins, thought to function in apoptotic and inflammatory signaling pathways. Mutations in the CIAS1 gene, which encodes a member of the NALP (NACHT-, LRR-, and PYD-containing proteins) family, the cryopyrin/NALP3/PYPAF1 protein, expressed primarily in phagocytic cells, were recently found to be associated with a spectrum of autoinflammatory disorders. These include chronic infantile neurologic cutaneous and articular (CINCA) syndrome (also known as neonatal-onset multisystem inflammatory disease [NOMID]), Muckle-Wells syndrome (MWS), and familial cold urticaria (FCU). We describe herein 7 new mutations in 13 unrelated patients with CINCA syndrome and identify mutational hotspots in CIAS1 on the basis of all mutations described to date. We also provide evidence of genotype/phenotype correlations. A 3-dimensional model of the nucleotide-binding domain (NBD) of cryopyrin suggested that this molecule is structurally and functionally similar to members of the AAA+ protein family of ATPases. According to this model, most of the mutations known to affect residues of the NBD are clustered on one side of this domain in a region predicted to participate in intermolecular contacts, suggesting that this model is likely to be biologically relevant and that defects in nucleotide binding, nucleotide hydrolysis, or protein oligomerization may lead to the functional dysregulation of cryopyrin in the MWS, FCU, and CINCA/NOMID disorders. (Blood. 2004;103:2809-2815)
Introduction
CIAS1 gene encodes cryopyrin/NALP3/PYPAF1,1,2 a member of the recently discovered NALP/PYPAF subfamily of the CATERPILLER protein family.3,4 Little is known about the structure and function of the proteins of this subfamily. Each member of the NALP/PYPAF (PYRIN-containing Apaf-1-like protein) family contains an amino-terminal pyrin domain (PYD), a central NACHT domain including a nucleoside triphosphate (NTP)-binding site, and carboxy terminal leucine-rich repeats (LRRs) (for a review, see Tschopp et al5 ). PYD contains 6 antiparallel α-helices that form a compact bundle similar in structure to the CARD death and death-effector domains.6 As a member of the death domain-fold superfamily, PYD probably mediates homotypic interactions between PYD-containing proteins, resulting in the formation of a complex involved in signal transduction. LRRs are 20- to 29-residue-sequence motifs present in multiple proteins with diverse functions. In various members of the CATERPILLER family, as in the NOD (nucleotide-binding oligomerization domain) subfamily, LRRs may act as intracellular sensors of bacterial invasion capable of initiating an inflammatory response. LRRs may thus play a role in detecting pathogen-derived molecules and possibly endogenous nonforeign “alarm signals” such as mammalian DNA and heat-shock proteins, ultimately leading to the induction of inflammatory responses.7,8 The NALP1 LRRs may exert their effects by means of inhibition because their removal in NALP1 makes the protein constitutively active.9 The NACHT domain of NALP contains 7 distinct motifs, including an adenosine triphosphate/guanosine triphosphate (ATP/GTP)-specific P-loop and an Mg2+-binding site typical of nucleoside triphosphatases (NTPases) (Walker A and B motifs, respectively).3 NACHT domains appear related to the nucleotide-binding domain (NBD) of the AP-ATPases family10 or the NB-ARC (nucleotide-binding domain shared by APAF-1, certain gene products, and Ced4) family,11 sharing specific features with similarly positioned motifs.3 Some members of the AP-ATPases family, such as the human APAF-1, are involved in programmed cell death and inflammatory signaling pathways, functions that require nucleotide binding and protein oligomerization mediated through the NBD.12 By analogy, the NACHT domain of NALP may be involved in protein oligomerization.
The role of cryopyrin/NALP3/PYPAF1 is unclear. Its expression is restricted to immune cells and chondrocytes.13 Cryopyrin has been reported to interact with the protein ASC (apoptosis-associated speckle-like protein containing a CARD), a PYD-CARD binding partner of procaspase-1, although this interaction has been called into question.2,14,15 The binding of procaspase-1 induces the processing of pro-interleukin-1 (pro-IL-1) to generate its active form, IL-1, and the activation of nuclear factor kappa B (NF-κB). These findings suggest that cryopyrin is involved in the regulation of apoptosis, the inflammatory signaling pathway, or both. Clear evidence that cryopyrin plays a key role in inflammation was recently provided in vivo by the association of CIAS1 mutations with autoinflammatory diseases. The syndromes familial cold urticaria (FCU) (Mendelian Inheritance in Man [MIM] number, 120 100), Muckle-Wells syndrome (MWS) (MIM, 191 900), and chronic infantile neurologic cutaneous and articular syndrome/neonatal-onset multisystem inflammatory disease (CINCA/NOMID) (MIM, 607 115) are 3 autosomal-dominant disorders resulting from CIAS1 missense mutations.1,13 All involve recurrent inflammatory episodes generally associated with fever, arthralgia, and urticaria. These features are brought on by exposure to cold in FCU, the mildest of these conditions. In the MWS, transient arthritis, neurosensory deafness, and amyloidosis are frequently associated with these manifestations. Patients with CINCA syndrome display the most severe phenotype, with neonatal onset, chronic polymorphonuclear (PMN) meningitis leading to progressive neurologic impairment, and recurrent flare-ups of joint inflammation. Joint involvement varies in severity from mild flare-ups to severe arthropathy and radiologic modifications. Progressive visual impairment and perceptive deafness may also be observed as patients age.16 CINCA syndrome may be fatal. All the mutations identified to date in these 3 disorders are missense mutations within exon 3 of the CIAS1 gene. The mutations associated with a particular condition1,13,17-20 do not appear to be clustered. We analyzed the clinical and molecular features of 13 patients with newly diagnosed CINCA syndrome. We combined these data with those previously obtained for patients with CINCA, MWS, and FCU, which enabled us to identify hotspots of mutation preferentially associated with particular disease expressions. We investigated the molecular consequences of these missense mutations in CIAS1 and in the structure-function relationships of the protein by generating a model of the 3-dimensional (3D) structure of the NBD of the cryopyrin NACHT domain.
Patients and methods
Patients
Twenty-two unrelated patients with suspected CINCA syndrome were analyzed in the genetic study. We studied possible genotype/phenotype correlations in patients with the CIAS1 gene mutation within this cohort and in previously reported patients with autoinflammatory disorders associated with CIAS1 mutations. For FCU, diagnostic criteria were recurrent intermittent episodes of fever, rash, conjunctivitis, articular manifestations primarily after general exposure to cold, and absence of deafness and amyloidosis. Patients with MWS were characterized by similar recurrent episodes of inflammation not triggered by cold and, in some, by progressive deafness and amyloidosis. Diagnostic criteria for CINCA/NOMID syndrome were episodic fever, early-onset urticarial skin rash associated with chronic meningitis, and, in some patients, severe and deforming arthropathies. (Table 1). Patients with NOMID/CINCA syndrome differed considerably in terms of the severity of the condition. Therefore, we assigned patients to 2 groups, the first with transient joints flare-ups only and the second with permanent and deforming arthropathies. Physicians experienced in the diagnosis of CINCA/NOMID syndrome carefully examined each patient, and informed, written consent for participation in this study according to the Declaration of Helsinki was obtained from the patients or their parents. This study was approved by the INSERM review board.
Clinical data and CIAS1 mutations identified in the 14 tested patients with CINCA syndrome
. | . | . | . | Constant and deforming arthropathies . | CIASI Mutation . | . | Frequency of CIAS1 mutations in controls . | CIAS1 mutations in parents . | |
|---|---|---|---|---|---|---|---|---|---|
| Patient . | Age, y* . | Neurologic involvement . | Joint involvement . | . | Nucleotides . | Amino acids . | . | . | |
| 11 | 3 | NA | AG | No | 779G>T | R260L | 0/74 | — | |
| 14 | 45 | + | TA | No | 1043C>T | T348M | 0/98 | ND | |
| 15 | 22 | + | TA | No | 1896G>T | L632F | 0/98 | — | |
| 21 | 8 | +++ | TA | No | 1062G>T | E354D | 0/98 | — | |
| 12 | 5 | NA | PA | Yes | 779G>C | R260P | 0/74 | — | |
| 8 | 13 | ++ | PA | Yes | 907G>A | D303N | 0/74 | ND | |
| 13 | 13 | + | TA/PA | Yes | 908A>G | D303G | 0/112 | — | |
| 17 | 8 | ++ | TA/PA | Yes | 926T>C | F309S | 0/112 | — | |
| 9 | 22 | + | PA | Yes | 1213A>C | T405P | 0/90 | ND | |
| 18 | 5 | + | TA/PA | Yes | 1307C>T | T4361 | 0/78 | ND | |
| 10 | 3 | ++ | PA | Yes | 1709A>G | Y570C | 0/98 | — | |
| 16 | 1 | ++ | PA | Yes | 1709A>G | Y570C | 0/98 | — | |
| 19 | 22 | +++ | PA | Yes | 1709 A>G | Y570C | 0/98 | ND | |
. | . | . | . | Constant and deforming arthropathies . | CIASI Mutation . | . | Frequency of CIAS1 mutations in controls . | CIAS1 mutations in parents . | |
|---|---|---|---|---|---|---|---|---|---|
| Patient . | Age, y* . | Neurologic involvement . | Joint involvement . | . | Nucleotides . | Amino acids . | . | . | |
| 11 | 3 | NA | AG | No | 779G>T | R260L | 0/74 | — | |
| 14 | 45 | + | TA | No | 1043C>T | T348M | 0/98 | ND | |
| 15 | 22 | + | TA | No | 1896G>T | L632F | 0/98 | — | |
| 21 | 8 | +++ | TA | No | 1062G>T | E354D | 0/98 | — | |
| 12 | 5 | NA | PA | Yes | 779G>C | R260P | 0/74 | — | |
| 8 | 13 | ++ | PA | Yes | 907G>A | D303N | 0/74 | ND | |
| 13 | 13 | + | TA/PA | Yes | 908A>G | D303G | 0/112 | — | |
| 17 | 8 | ++ | TA/PA | Yes | 926T>C | F309S | 0/112 | — | |
| 9 | 22 | + | PA | Yes | 1213A>C | T405P | 0/90 | ND | |
| 18 | 5 | + | TA/PA | Yes | 1307C>T | T4361 | 0/78 | ND | |
| 10 | 3 | ++ | PA | Yes | 1709A>G | Y570C | 0/98 | — | |
| 16 | 1 | ++ | PA | Yes | 1709A>G | Y570C | 0/98 | — | |
| 19 | 22 | +++ | PA | Yes | 1709 A>G | Y570C | 0/98 | ND | |
NA indicates not assessed; +, chronic meningitis; ++, chronic meningitis, mental retardation; +++, chronic meningitis, mental retardation, epilepsy, or cerebral atrophy; TA, transient arthritis; AG, arthralgia; PA, persistent arthritis; ND, not done; and—, no mutation.
Current age.
Mutation detection
Genomic DNA was extracted from whole blood using standard procedures. We searched for mutations in genomic DNA using exons with flanking intron sequences and bidirectional fluorescence sequencing, as previously described.13 We tested a panel of control DNA samples for the presence of the CIAS1 mutations identified in each patient by mutation sequencing analysis (for CIAS1, GenBank accession number AF427617).
Sequence analysis and modeling of 3D structure
We used a battery of sequence analysis/structure prediction methods, including similarity searches within the Protein Data Bank (PDB) using PSI-BLAST (position-specific iterative-basic local alignment search tool)21 with a protein-specific score matrix (PSSM) derived from the NACHT family of domains as well as threading procedures (3D-PSSM22 ; FUGUE [http://www.cryst.bioc.cam.ac.uk/~fugue/prfsearch.html]23 ). Resultant alignments were manually checked for accuracy, refined, and extended by hydrophobic cluster analysis (HCA),24,25 which makes it possible to consider the 1D sequence alignment in a structural context because the hydrophobic clusters delineated with this approach are generally consistent with the regular secondary structures.26,27 Secondary structures predicted using this approach are consistent with those predicted by the PSI-PRED (http://www.psipred.net)28 and PHD (http://www.embl-heidelberg.de/predictprotein/predictprotein.html)29 programs (the PSI-PRED prediction is reported above the CIAS1 sequence on Figure 2A). This careful analysis was accompanied by visual inspection of the experimental 3D structures. In particular, the 3D superimposition of the structures, together with associated multiple alignments, made it possible to distinguish core sequences from more variable sequences (Figure 2A). It also led to the identification of positions invariably occupied by hydrophobic amino acids (buried positions) required for conservation of the typical fold of NACHT family (Figure 2A). This method for modeling in conditions of low levels of sequence identity has already been used successfully on many different protein targets (eg, see Paoletti et al30 and Callebaut et al31 ).
Sequence alignment and mutation mapping of CIAS1 NBD. (A) Alignment of the CIAS1 sequence with 4 NBD sequences of known 3D structures corresponding to proteins of the AAA+ ATPases superfamily. p97: d1 AAA domain of membrane fusion ATPase p97 (PDB identifier, 1e32[chain A])36 nsf: D2 hexamerization domain of N-ethylmaleimide-sensitive factor (PDB 1d2n [chain A])34,35 ; Cdc6p (PDB 1fnn [chain A])33 ; and RuvB (PDB 1hqc [chain A]).40 Alignment was generated by threading and PSI-BLAST procedures and was carefully refined and extended using the sensitive hydrophobic cluster analysis (HCA) (“Patients and methods”). Vertical arrows indicate the positions of the CIAS1 mutations. Amino acids of nsf involved in intersubunit contacts35 are boxed in red. Identical amino acids are shaded in black, whereas similar amino acids are shaded in gray with white and black letters for hydrophobic (or amino acids that can substitute for them) and nonhydrophobic amino acids, respectively. Green circles indicate the positions most frequently occupied by hydrophobic amino acids in the NACHT family of domains. These residues mainly correspond to amino acids that, in the aligned NBDs, are buried within the considered structures and that can serve as anchors for the alignment procedure. Moreover, positions of the observed regular secondary structures (underlined and labeled under the sequence (S indicates β-strand; H, helix) in most cases match those of the predicted secondary structures of CIAS1 with respect to its sequence (E indicates extended β-strand; H, helix; C, coil). No accurate structural alignment could be obtained for helix H5 (indicated in brackets). However, the N-termini of the corresponding sequences could be aligned, highlighting the conservation of 2 hydrophobic amino acids. The main original features of the CIAS1 NBD fold with respect to the NBD core structures shown in (A) are (1) the presence of another helix, H3C, after helix H3B (nsf labeling; in this respect, the predicted structure of CIAS1 is thought to be similar to that of Cdc6p,33 in which a longer helix H3C is also present between helix H3B and strand S3 [yellow]) and (2) a large loop linking strand S3 to helix H4. This loop is 7 amino acids longer than the corresponding loop in nsf,34,35 but it also contains an isoleucine-glycine-proline (IGP) sequence, which in the nsf structure forms a tight turn involved in hexamer interactions.34,35 (B) Mapping of mutations on the 3D model of CIAS1 NBD. Two orthogonal views are shown in ribbon representation. The model was constructed based of the alignment shown in panel A. Strands and helices are labeled and colored as in panel A. Positions of mutations are shown and labeled, as are positions of the Walker A T231 (blue on helix H2) and Walker B D300 (orange in strand S3) motifs and as are positions of ATP and of the magnesium ion, as in the nsf structure.35 Note that the conformation of the loop linking strand S3 to helix H4B is hypothetical. Cα-trace of the α-helical domain following NBD in AAA+ ATPases (nsf D235 ) is shown on right to illustrate its position with respect to the NDB. According to HCA (data not shown), T405 can be tentatively located in the C-terminal end of an extended structure, after 2 α-helices, lying near the ATP-binding site (gray circle).
Sequence alignment and mutation mapping of CIAS1 NBD. (A) Alignment of the CIAS1 sequence with 4 NBD sequences of known 3D structures corresponding to proteins of the AAA+ ATPases superfamily. p97: d1 AAA domain of membrane fusion ATPase p97 (PDB identifier, 1e32[chain A])36 nsf: D2 hexamerization domain of N-ethylmaleimide-sensitive factor (PDB 1d2n [chain A])34,35 ; Cdc6p (PDB 1fnn [chain A])33 ; and RuvB (PDB 1hqc [chain A]).40 Alignment was generated by threading and PSI-BLAST procedures and was carefully refined and extended using the sensitive hydrophobic cluster analysis (HCA) (“Patients and methods”). Vertical arrows indicate the positions of the CIAS1 mutations. Amino acids of nsf involved in intersubunit contacts35 are boxed in red. Identical amino acids are shaded in black, whereas similar amino acids are shaded in gray with white and black letters for hydrophobic (or amino acids that can substitute for them) and nonhydrophobic amino acids, respectively. Green circles indicate the positions most frequently occupied by hydrophobic amino acids in the NACHT family of domains. These residues mainly correspond to amino acids that, in the aligned NBDs, are buried within the considered structures and that can serve as anchors for the alignment procedure. Moreover, positions of the observed regular secondary structures (underlined and labeled under the sequence (S indicates β-strand; H, helix) in most cases match those of the predicted secondary structures of CIAS1 with respect to its sequence (E indicates extended β-strand; H, helix; C, coil). No accurate structural alignment could be obtained for helix H5 (indicated in brackets). However, the N-termini of the corresponding sequences could be aligned, highlighting the conservation of 2 hydrophobic amino acids. The main original features of the CIAS1 NBD fold with respect to the NBD core structures shown in (A) are (1) the presence of another helix, H3C, after helix H3B (nsf labeling; in this respect, the predicted structure of CIAS1 is thought to be similar to that of Cdc6p,33 in which a longer helix H3C is also present between helix H3B and strand S3 [yellow]) and (2) a large loop linking strand S3 to helix H4. This loop is 7 amino acids longer than the corresponding loop in nsf,34,35 but it also contains an isoleucine-glycine-proline (IGP) sequence, which in the nsf structure forms a tight turn involved in hexamer interactions.34,35 (B) Mapping of mutations on the 3D model of CIAS1 NBD. Two orthogonal views are shown in ribbon representation. The model was constructed based of the alignment shown in panel A. Strands and helices are labeled and colored as in panel A. Positions of mutations are shown and labeled, as are positions of the Walker A T231 (blue on helix H2) and Walker B D300 (orange in strand S3) motifs and as are positions of ATP and of the magnesium ion, as in the nsf structure.35 Note that the conformation of the loop linking strand S3 to helix H4B is hypothetical. Cα-trace of the α-helical domain following NBD in AAA+ ATPases (nsf D235 ) is shown on right to illustrate its position with respect to the NDB. According to HCA (data not shown), T405 can be tentatively located in the C-terminal end of an extended structure, after 2 α-helices, lying near the ATP-binding site (gray circle).
We assessed alignments by calculating Z scores (differences between the observed scores and the mean scores of a distribution of scores calculated from the alignment of one sequence with 1000 randomized versions of the other). Z-score values are expressed in standard deviation units of the random distribution. Mean Z-score values calculated for the alignment of CIAS1 with the 4 sequences shown in Figure 2A were 6.0 (maximum, 7.1) and 7.5 (maximum, 8.4) for identity and similarity (Blosum 62 matrix) scores, respectively, whereas the mean identity level was 11.6%. These values were similar to those calculated from alignments of the NBDs of known 3D structures (eg, identity and similarity Z-score values for the alignment 1fnn/1hqc [12.5% identity] are 6.1 and 7.1, respectively).
Results
Novel CIAS1 mutations identified in patients with CINCA syndrome
We have previously reported 7 different missense mutations in the CIAS1 gene associated with CINCA syndrome in 7 unrelated families.13 Since this first description, 4 additional CINCA syndrome-associated mutations have been reported.20 CIAS1 mutations have also been reported in patients with MWS and FCU.1,17-19 We studied 22 additional patients and identified CIAS1 gene mutations in 13 with CINCA/NOMID syndrome (Table 1). Nine of these patients had particularly severe disease, with persistent arthropathy associated with radiologically evident bone deformities (Table 1). None of these patients had a family history of the disease.
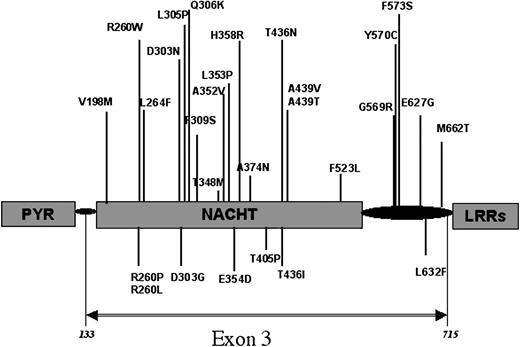
Both strands of the CIAS1 coding sequence and all the exon/intron flanking sequences were screened by direct sequencing of polymerase chain reaction (PCR) fragments, as previously described.13 We also searched for mutations in DNA of the parents' patients, when available, and in a panel of control samples (Table 1). All the mutations identified in this cohort of patients, as in the previously reported ones, consisted of missense mutations located in exon 3 of CIAS1 (Table 1 and Figure 1). Seven of the 13 mutations identified in CIAS1 are new. In 3 patients, the mutation affected a residue not previously identified as involved in CIAS1-associated diseases—E354D in patient 21 (P21), T405P in P9, and L632F in P15 (Table 1; Figure 1). In another 4 patients, the mutations affected residues previously reported to be mutated in CIAS1-associated syndromes (CINCA syndrome, MWS, or FCU) but with a different substitution: R260L was identified in P11 and R260P in P12, whereas R260W was previously reported in 5 families with either MWS or FCU (Figure 1; Table 2) as independent events occurring in each family. T436I was identified in P18, whereas T436N was previously observed in a family in which 3 members were affected by CINCA syndrome.13 D303G was found in P13, but a different transition at the same residue, D303N, was previously observed in 2 members of a family and 1 sporadic occurrence of CINCA/MWS overlapping phenotype and in 1 patient with CINCA syndrome.13,18,38 Finally, 6 mutations identified in patients from this study had been previously reported: D303N (P8), T438M (P14) in 3 different families with MWS18 and in 1 sporadic occurrence of CINCA syndrome,39 F309S (P17) in 1 patient with CINCA/NOMID syndrome of severe phenotype,13 and Y570C in 3 patients (P10, P16, P19) 2 other patients with severe CINCA/NOMID syndrome20,39 (Table 1). None of the mutations were found in controls, as in patients' parents, when tested (Table 1). Overall, these findings showed that regardless of disease severity, all CIAS1 mutations identified to date were missense mutations. In addition, though they occurred de novo, these mutations were present in a number of patients, indicating the probable existence of mutation hotspots. We therefore investigated the distribution of these mutations according to disease severity and according to whether mutation hot spots designated critical functional residues on a predicted 3D structure of the cryopyrin NACHT domain.
Locations of the mutations in CIAS1 encoding protein. All the mutations identified to date in CIAS1 that cause FCU, MWS, or CINCA/NOMID syndromes are located in exon 3, which encodes the NACHT domain and its flanking regions. Mutations previously reported are indicated above the protein structure, whereas new mutations identified in this study appear below the protein structure.
Locations of the mutations in CIAS1 encoding protein. All the mutations identified to date in CIAS1 that cause FCU, MWS, or CINCA/NOMID syndromes are located in exon 3, which encodes the NACHT domain and its flanking regions. Mutations previously reported are indicated above the protein structure, whereas new mutations identified in this study appear below the protein structure.
Distribution of all the CIASI missense mutations identified to date, relative to the spectrum of CIASI-associated illnesses
Disorder . | Mutation . | Family or patient . | Reference . |
|---|---|---|---|
| FCU | V198M‡ | 2F | Hoffman et al,1 Dode et al18 |
| R260W§ | 2F | Dode et al18 | |
| L305P | 1P | Aganna et al19 | |
| L353P | 1P | Hoffman et al17 | |
| A439V | 1F | Hoffman et al1 | |
| E627G | 1F | Hoffman et al1 | |
| MWS | V198M‡ | 1F | Aganna et al19 |
| R260W§ | 3F and 1P | Dode et al,18 Aganna et al19 | |
| A352V | 1F | Hoffman et al1 | |
| T348M∥ | 2F and 1P | Dode et al18 | |
| A439T | 1F | Dode et al18 | |
| G569R | 1F | Dode et al18 | |
| CINCA* | D303N¶ | 1F and 1P | Feldmann et al,13 Dode et al,18 Granel et al38 |
| Q306K | 1P | Feldmann et al13 | |
| T348M∥ | 2P | Rosen-Wolff et al39 and current study | |
| E354D | 1P | Current study | |
| H358R | 1P | Feldmann et al13 | |
| T436N | 1F | Feldmann et al13 | |
| L632N | 1P | Current study | |
| M662T | 1P | Feldmann et al13 | |
| R260L | 1P | Current study | |
| CINCA† | R260P | 1P | Current study |
| L264F | 1P | Aksentijevich et al20 | |
| D303N¶ | 2P | Aksentijevich et al20 and current study | |
| D303G | 1P | Current study | |
| F309S | 2P | Feldmann et al13 and current study | |
| A374N | 1P | Aksentijevich et al20 | |
| T405P | 1P | Current study | |
| T436I | 1P | Current study | |
| F523L | 2P | Aksentijevich et al20 | |
| Y570C | 5P | Hoffman et al,1 Aksentijevich et al,20 Rosen-Wolff et al,39 and current study | |
| F573S | 1P | Feldmann et al13 |
Disorder . | Mutation . | Family or patient . | Reference . |
|---|---|---|---|
| FCU | V198M‡ | 2F | Hoffman et al,1 Dode et al18 |
| R260W§ | 2F | Dode et al18 | |
| L305P | 1P | Aganna et al19 | |
| L353P | 1P | Hoffman et al17 | |
| A439V | 1F | Hoffman et al1 | |
| E627G | 1F | Hoffman et al1 | |
| MWS | V198M‡ | 1F | Aganna et al19 |
| R260W§ | 3F and 1P | Dode et al,18 Aganna et al19 | |
| A352V | 1F | Hoffman et al1 | |
| T348M∥ | 2F and 1P | Dode et al18 | |
| A439T | 1F | Dode et al18 | |
| G569R | 1F | Dode et al18 | |
| CINCA* | D303N¶ | 1F and 1P | Feldmann et al,13 Dode et al,18 Granel et al38 |
| Q306K | 1P | Feldmann et al13 | |
| T348M∥ | 2P | Rosen-Wolff et al39 and current study | |
| E354D | 1P | Current study | |
| H358R | 1P | Feldmann et al13 | |
| T436N | 1F | Feldmann et al13 | |
| L632N | 1P | Current study | |
| M662T | 1P | Feldmann et al13 | |
| R260L | 1P | Current study | |
| CINCA† | R260P | 1P | Current study |
| L264F | 1P | Aksentijevich et al20 | |
| D303N¶ | 2P | Aksentijevich et al20 and current study | |
| D303G | 1P | Current study | |
| F309S | 2P | Feldmann et al13 and current study | |
| A374N | 1P | Aksentijevich et al20 | |
| T405P | 1P | Current study | |
| T436I | 1P | Current study | |
| F523L | 2P | Aksentijevich et al20 | |
| Y570C | 5P | Hoffman et al,1 Aksentijevich et al,20 Rosen-Wolff et al,39 and current study | |
| F573S | 1P | Feldmann et al13 |
Boldface indicates independent mutations identified in several patients. F indicates family; and P, patient.
Reference citations relate to previous descriptions of the corresponding mutations.
CINCA with chronic meningitis and transient joint flares.
CINCA with chronic meningitis and permanent, deforming arthropathies.
Similar mutations observed in patients from different groups.
Distribution of the CIAS1 mutation as a function of disease severity
We evaluated a possible genotype/phenotype correlation within the spectrum of CIAS1 mutations found in this and other studies by classifying cases according to diagnosis and severity of disease presentation. Patients with FCU and CIAS1 mutations were reported to have episodes of fever, rash, and articular manifestations primarily after natural or experimental generalized cold exposure, without deafness or amyloidosis. In the other groups, patients had similar recurrent episodes of symptoms but without cold triggering and often with deafness and amyloidosis. Patients with MWS were differentiated from patients with CINCA syndrome on the basis of chronic meningitis that occurred in patients with CINCA syndrome but not in those with MWS. Finally, within the group of patients with CINCA syndrome, severity of disease expression was estimated on the basis of the severity of neurologic symptoms and the development of arthropathy, as assessed by radiography (Tables 1,2). Using these diagnostic criteria, several of the identified mutations were found to be associated with the same phenotype. For instance, extremely severe expression of CINCA syndrome was associated in 5 patients with Y570C mutations (3 in our group [P10, P16, P19] and 2 previously reported).20,39 Detailed medical histories were available for 4 of these patients. Each had severe arthropathy before 1 year of age resulting in metaphyseal enlargement and severe contractures. Severe neurologic symptoms with mental retardation were observed in all, associated with epilepsy in 1 patient (P19) and with cerebral atrophia and hydrocephaly in 3 patients (P16, P19, and a patient in Rosen-Wolff et al's study39 ). Prematurity and dysmaturity were observed in 3 patients (P10, P16, P19). They all failed to thrive (weight, less than -2 SD), had growth failure (height, less than -2 SD), and dysmorphy. P19 died at 22 years of age. The F309S mutation was found in 2 patients with severe articular and neurologic diseases, 1 of whom died in early adulthood. The F523L mutation was also found in 2 other patients with severe CINCA syndrome, as reported by Aksentijevich et al.20 Some mutations may be common to groups of patients contiguous in terms of severity. For instance, R260W and V198M were found in several families with MWS or FCU,1,18 and T348M occurred in 3 families with severe expression of MWS and in 2 patients18,39 with mild expression of CINCA syndrome involving acute episodes of arthritis and mild neurologic problems consisting of sporadic headache caused by chronic meningitis as confirmed by examination of the cerebrospinal fluid (CSF). The D303N mutation was found in patients with severe or moderate expression of CINCA syndrome. One patient with the D303N mutation was initially reported to have MWS,18 but the clinical features of this patient were recently reported to be more consistent with CINCA syndrome.38 This example highlights the limitations of such an approach if an overlap exists in phenotype classification. However, this study clearly shows that none of the mutations identified to date in patients with the most severe expression of disease (CINCA syndrome with chronic meningitis and arthropathy) were observed in patients with the mildest phenotype (FCU). Although this analysis deals with a limited number of patients, these data indicate a relative phenotype/genotype correlation, suggesting that distinct mutations differently affect cryopyrin function and expression.
Mapping CIAS1 mutations on a 3D model of the NACHT nucleotide-binding domain
In the absence of experimental data, a 3D model of the structure of the CIAS1 NACHT domain can be used to further evaluate the molecular impact of mutations and to investigate the function of CIAS1. Walker A (P-loop) and Walker B (Mg2+-binding site) motifs clearly identify NACHT domains as NTPases.3 However, apart from these 2 signatures, NACHT domain sequences are fairly different from those of typical NBDs, for which experimental 3D structures have been identified. NACHT domains are larger, with a predicted α fold (most α-helices; data not shown) in the C-terminal region. We used a battery of sequence analysis/structure prediction methods, including the sensitive hydrophobic cluster analysis,24,25 to generate a relevant model for part of the CIAS1 NACHT domain. These methods made it possible to detect significant relationships at low levels of sequence identity (less than 20%), supported by relevant statistical scores, and to align the CIAS1 sequence accurately with sequences of known 3D structures constituting templates for homology modeling.
The CIAS1 NBD fold is predicted to consist of a typical 5-stranded β-sheet surrounded by α-helices, as observed in AAA+ ATPases, the structures of which have been used as templates for modeling (Figure 2). Walker A, Walker B, and a “sensor” motif are found at the end of the parallel strands, forming the nucleotide-binding site, which in AAA+ ATPases also involves residues from a second α-helical domain following the NBD. NACHT domains may have a similar structure, but the lack of accurate alignment for the α-helical domain precluded its modeling on AAA+ ATPase templates.
Strikingly, most of the CIAS1 mutations in the NBD (Figure 2A, arrows) are located on 1 side of this domain (Figure 2B) along the nucleotide-binding cleft or in prolongation of the cleft. They are found in loops next to the parallel β-strands (loop after β-strand S2: R260, L264; loop after β-strand S3, near the Walker B motif: D303, L305, Q306, F309; loop after β-strand S4: T348, corresponding to the “sensing” residue of the sensor motif for the detection of nucleotide binding and hydrolysis, close to the ATP γ-phosphate in AAA+ ATPases19,40 ). Mutations are also found in α-helix H5 (A352, L353, E354, H358) and in the N-terminus of helix H1B (V198). Most of these regions are also involved in oligomeric interactions in AAA+ ATPases. Superimposition of our 3D model of CIAS1 NBD on the NBD subunit of the AAA+ nsf ring-forming hexamer (data not shown) suggests that CIAS1 may be involved in similar oligomeric organization. Thus, mutations within the NBD located outside the structure core are most likely to affect nucleotide binding and hydrolysis or to disturb conformational changes affecting the quaternary structure.30
Other CIAS1 mutations are located in the C-terminal part of the NACHT domain, after the NBD (residues A374, T405, T436, A439, F523, G569, Y570, F573, E627, L632, and M662). Although no accurate alignment or model can be built for this region, the predicted secondary structures are consistent with a mainly α-helical domain following the NBD, which should run from residue 374 to residue 451.
Discussion
FCU, MWS, and CINCA syndrome, 3 conditions associated with CIAS1 mutations, are inherited as dominant traits. Almost 50 independent mutations, including those described in this study, have now been characterized. All these mutations are missense mutations affecting exon 3 of CIAS1, causing a wide spectrum of disease expression. These findings strongly suggest that the mutated protein exerts a dominant-negative or a gain-of-function effect over the wild-type product and that the null mutation of one allele would probably have no effect or would lead to a different phenotypic expression because of haploinsufficiency. Although we cannot rule out an effect of unknown modifier genes in phenotypic expression, specific CIAS1 mutations seem likely to affect disease expression, as shown by some degree of genotype/phenotype correlation observed within this spectrum of phenotypic expression. This correlation is particularly clear if we consider the extreme groups defined by the magnitude of phenotypic expression (ie, FCU and the severe CINCA syndrome). Patients from different groups do not share mutations whereas, within each group, several unrelated patients carry the same mutation, occurring as an independent event in each case. In contrast, a few patients from contiguous groups, such as FCU/MWS or MWS and milder forms of CINCA syndrome, may share mutations. In such patients, the moderate expression of symptoms, as for chronic meningitis, might have been missed, or patients might not yet have developed the features used to discriminate between the various groups. Fine analysis of a larger number of patients with each condition is required to confirm and strengthen these observations. If confirmed, these data may be of utmost importance for prognostic assessment and for adjustment of treatment for patients with CIAS1-associated diseases. Our analysis also confirms the previously suggested genetic heterogeneity of these disorders,13,20 because mutations in CIAS1 were identified in only 60% of the patients analyzed.
To localize mutated residues at the 3D level and to investigate further the function of CIAS1, we constructed a model of the cryopyrin NACHT domain by homology modeling based on known structures of NBD domains. The NACHT domain of cryopyrin can be accurately aligned with nucleotide-binding domains from members of the AAA+ class of proteins, which generally have an α-helical domain following the NBD. The proposed model for the NACHT domain of cryopyrin is highly consistent with the NB-ARC domain model of CED4 protein proposed by Jaroszewski et al.41 These 2 nucleotide-binding related domains share specific features, including a sensor 1 region within motif 4 and a highly conserved proline in motif 5 that distinguishes these proteins from the rest of the ATPases.42 These 2 families of domains differ, however, in their C-terminal regions, which cannot be aligned. Because the amino acid sequences of CIAS1 differ considerably from those of the templates used for modeling and despite the adapted methodology and cautions applied to this analysis, we cannot totally exclude the possibility that in, some places, the alignment, and thus the structure prediction, remains imperfect. However, because most mutations in the NBD are located in conserved regions close to invariant regular secondary structures, the model may be considered reliable. Experimental structure determination would be necessary to refine the alignment in some regions, to obtain atomic details, and to determine the fold(s) adopted by region following NBD, in which several other mutations are located. Although members of the AAA+ ATPases have different cellular activities,43,44 they all function as regulatory subunits of macromolecular protein complexes.43,45 Their function depends on the binding and hydrolysis of a nucleotide, resulting in conformational changes that promote assembly, disassembly, or functional operation of another part of the protein complex. In addition, many of these proteins form hexamers, in a process demonstrated to be ATP dependent in the case of nsf-D2.35 On the basis of the similarity of the NACHT cryopyrin sequence to the sequence of these proteins, it is tempting to speculate that nucleotide binding to cryopyrin induces conformational change in this protein or promotes its oligomerization. Although the similarity to AAA+ ATPases suggests that the CIAS1 NACHT domain forms a hexamer structure, we cannot exclude the possibility that CIAS1 oligomers, if indeed they exist, display different stochiometries or arrangements.
Twelve of the 23 different substitutions identified affect residues of the NACHT nucleotide-binding domain. Remarkably, all are clustered on one side of the protein, near the nucleotide-binding cleft, within a region possibly involved in oligomeric interactions, based on the known oligomeric structures of AAA+ ATPases. Based on sequence similarity, the mutations appear to affect residues directly involved in “sensing” of the nucleotide-binding state, in predicted subunit interactions or residues that are located close to the Walker A and B motifs. None of these mutations target highly conserved positions intimately involved in the binding of the metal ion or of the nucleotide. Although we cannot rule out the possibility that nucleotide binding is impaired, this observation may suggest that defective hydrolysis and conformational change and oligomerization of the protein, in particular, may be the main mechanism by which the mutated protein exerts its dominant effect. Because half the cryopyrin monomers expressed in patients' cells are translated from the wild-type allele, the mutated monomer should exert a transdominant effect over the normal protein function. This may occur through the formation of oligomers containing mixtures of active and inactive monomers that fail to support protein activity. Further attempts to correlate the location of the mutations with disease severity were uninformative. This may be explained by the fact that residues clustered in the same loop, such as L305 and Q306, predicted to be either buried (L305) or exposed at the surface (Q306), differentially affect the stability of the loop and its ability to interact with potential partners. In addition, the nature of the substitution at a given position may determine the extent to which it impairs protein function or oligomerization. Several substitutions beyond A374N have been identified in the CIAS1 sequence, in a region for which no accurate structural information is available. These mutations are also associated with a relative genotype/phenotype correlation. Most of the mutations in this region (such as F523L or Y570C) leads to the most severe expression of the disease. Based on sequence similarities with AAA ATPases, this region may regulate the oligomerization process.
One of the main functions of AAA+ proteins is to form and to regulate transient macromolecular complexes. The pyrin domain and LRR repeats of cryopyrin are expected to mediate intermolecular interactions. The pyrin domain can interact with the ASC adaptor, which in turn recruits effector protein through its CARD domain to generate a heterocomplex.2 The CARD domain ASC has been shown to bind to that of procaspase-1, inducing the processing and activation of caspase-1 and the activation of NF-κB. Furthermore, the LRRs of cryopyrin may contain a ligand-binding domain that may constitute a molecular on/off switch, as reported for NOD proteins. By analogy to the NALP1/ASC/caspase-1 and caspase-5 protein complexes, which assemble to form the inflammasome,9 NACHT domain oligomerization of cryopyrin may be essential for the formation of a macromolecular heterocomplex, bringing into proximity several effectors and inducing their activation. However, such a mechanism remains to be demonstrated. Given the phenotypic expression of cryopyrin-associated disorders and the in vitro studies performed with this protein, the function of this complex is likely to be connected with critical processes such as apoptosis regulation, NF-κB activation, caspase-1 activation, and cytokine secretion, all elements of inflammatory responses.
Prepublished online as Blood First Edition Paper, November 20, 2003; DOI 10.1182/blood-2003-07-2531.
Supported by grants from l'Institut National de la Santé et de la Recherche Médicale (INSERM) and by a fellowship from the Fondation pour la Recherche Médicale (B.N.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the patients and their families for their cooperation. We also thank Cécile Dumont, Stéphanie Certain, and Nathalie Lambert for excellent technical assistance and Jean-Paul Mornon for helpful discussion and critical reading of this manuscript.

![Figure 2. Sequence alignment and mutation mapping of CIAS1 NBD. (A) Alignment of the CIAS1 sequence with 4 NBD sequences of known 3D structures corresponding to proteins of the AAA+ ATPases superfamily. p97: d1 AAA domain of membrane fusion ATPase p97 (PDB identifier, 1e32[chain A])36 nsf: D2 hexamerization domain of N-ethylmaleimide-sensitive factor (PDB 1d2n [chain A])34,35; Cdc6p (PDB 1fnn [chain A])33; and RuvB (PDB 1hqc [chain A]).40 Alignment was generated by threading and PSI-BLAST procedures and was carefully refined and extended using the sensitive hydrophobic cluster analysis (HCA) (“Patients and methods”). Vertical arrows indicate the positions of the CIAS1 mutations. Amino acids of nsf involved in intersubunit contacts35 are boxed in red. Identical amino acids are shaded in black, whereas similar amino acids are shaded in gray with white and black letters for hydrophobic (or amino acids that can substitute for them) and nonhydrophobic amino acids, respectively. Green circles indicate the positions most frequently occupied by hydrophobic amino acids in the NACHT family of domains. These residues mainly correspond to amino acids that, in the aligned NBDs, are buried within the considered structures and that can serve as anchors for the alignment procedure. Moreover, positions of the observed regular secondary structures (underlined and labeled under the sequence (S indicates β-strand; H, helix) in most cases match those of the predicted secondary structures of CIAS1 with respect to its sequence (E indicates extended β-strand; H, helix; C, coil). No accurate structural alignment could be obtained for helix H5 (indicated in brackets). However, the N-termini of the corresponding sequences could be aligned, highlighting the conservation of 2 hydrophobic amino acids. The main original features of the CIAS1 NBD fold with respect to the NBD core structures shown in (A) are (1) the presence of another helix, H3C, after helix H3B (nsf labeling; in this respect, the predicted structure of CIAS1 is thought to be similar to that of Cdc6p,33 in which a longer helix H3C is also present between helix H3B and strand S3 [yellow]) and (2) a large loop linking strand S3 to helix H4. This loop is 7 amino acids longer than the corresponding loop in nsf,34,35 but it also contains an isoleucine-glycine-proline (IGP) sequence, which in the nsf structure forms a tight turn involved in hexamer interactions.34,35 (B) Mapping of mutations on the 3D model of CIAS1 NBD. Two orthogonal views are shown in ribbon representation. The model was constructed based of the alignment shown in panel A. Strands and helices are labeled and colored as in panel A. Positions of mutations are shown and labeled, as are positions of the Walker A T231 (blue on helix H2) and Walker B D300 (orange in strand S3) motifs and as are positions of ATP and of the magnesium ion, as in the nsf structure.35 Note that the conformation of the loop linking strand S3 to helix H4B is hypothetical. Cα-trace of the α-helical domain following NBD in AAA+ ATPases (nsf D235) is shown on right to illustrate its position with respect to the NDB. According to HCA (data not shown), T405 can be tentatively located in the C-terminal end of an extended structure, after 2 α-helices, lying near the ATP-binding site (gray circle).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/7/10.1182_blood-2003-07-2531/6/m_zh80070458930002.jpeg?Expires=1763677413&Signature=2Z0LrxqWbUemjtINa-7UNKWBaQXkNUe1Cg5-TqhcRV5ooE1hlt6qJgkdZegEIim8yCQxNIKrI~-g7b9AGxkeDjs1EBpMQ1-mYnEzp6Uaz8sMzs1Sv3pOkXofy~MN97lhty8Jztf3b0CMgGp0~GXDhWKQWppO1mcxH0LfVKgXQaNxuThY7Ad20RhJGMFohwtt~DkfWf6B2-MU42X4rpm-qu0QQseFK0WAm3lehgIOjpZmbj~6Zy1x3aVJAJ0etY204zHjz-t0ygUL8RAUITp2DjP~QHhnq800zGxpVO5y-jF9PGiG3N-um9rn8jWvJIp0EKMOPQ32PKUaZD-aTo37gg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal