Abstract
Using loss of heterozygosity (LOH) and X-chromosome inactivation, we compared peripheral blood (PB) plasma with bone marrow (BM) cells in detecting genomic abnormalities in patients with acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). We detected LOH in the PB plasma of all 45 patients who had cytogenetically documented chromosomal abnormalities (5q-, 7-, +8, 17-, or 20-). BM cells from the same patients showed LOH in 89% of patients with MDS and 70% of patients with AML. Posttherapy samples from 16 of these patients demonstrated complete concordance between LOH and cytogenetics in detecting residual disease in 15 samples. Of the 16 samples, 4 showed LOH in plasma with normal BM morphology. Using X-chromosome inactivation, clonality was detectable in 19 (73%) of 26 BM samples, whereas all PB plasma samples showed clonality. These data support the conclusion that PB plasma is enriched by tumor-specific DNA and can replace BM cells for studying genomic abnormalities. (Blood. 2004;103:2799-2801)
Introduction
Bone marrow (BM) cells are currently considered the most reliable source for tumor DNA in patients with acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). Obtaining BM cells can be hindered by the presence of marrow fibrosis or by the patchiness of the disease. Sorting blast cells is frequently used to enrich samples for leukemia-specific DNA. However, this is particularly difficult in MDS since the neoplastic cells can differentiate and differentiated leukemic cells should not be excluded.1
We hypothesized that peripheral blood (PB) plasma or serum may contain most of the cellular components of leukemic cells, including DNA. The presence of tumor-specific DNA in PB plasma has already been extensively studied in solid tumors.2-5 Rare reports suggested that plasma or serum DNA can be used for the study of molecular abnormalities in hematologic malignancies.6,7 Dasi et al8 reported that plasma is adequate to study telomerase reverse transcriptase RNA levels in patients with colorectal cancer and lymphoma. We have previously reported that PB plasma is better than bone marrow cells for the detection of the internal tandem repeat of the FLT-3 gene in AML and MDS.9 Here, we demonstrate that PB plasma is enriched by tumor-specific DNA. Because chromosomal abnormalities such as 5q-,7-, +8, 17-, and 20- are the most common abnormalities seen in patients with AML and patients with MDS,9-15 we selected a number of microsatellite markers in the most commonly deleted regions of these chromosomes and analyzed loss of heterozygosity (LOH). We compared LOH in PB plasma with that in BM cells from paired samples obtained from patients with cytogenetically confirmed abnormality in these chromosomes. We also used a clonality assay based on X-chromosome activation using the human androgen receptor locus (HUMARA)16,17 in female patients to compare PB plasma with BM cells for detecting clonality in patients with confirmed AML or MDS.
Study design
Normal PB samples were obtained from 30 healthy volunteers under the age of 30 years to reduce the likelihood of malignant disease in this control group. All samples were collected under protocols approved by the institutional review board of the University of Texas MD Anderson Cancer Center. CD3+ cells were separated from PB samples using magnetic beads from Miltenyi Biotec and AutoMACS (Auburn, CA).
Sixteen microsatellite markers from 5q-, 7q-, +8, 17p-, and 20q- chromosomal regions were selected for analysis on the basis of their high incidence of being abnormal. The markers were D5S644, D5S433, D5S2027, D5S471, D7S630, D7S657, D7S661, D8S264, D8S505, D17S849, D17S831, D17S921, D20S119, D20S110, D20S961, and 1PLC1-1. As a control and to quantify the amount of DNA in the plasma, the Ras oncogene DNA was amplified using fam-labeled primers.18 Quantitative analysis of LOH and Ras amplification products was performed using ABI Prism 310 and 3100 genetic analyzers (Applied Biosystems, Foster City, CA). LOH and X-chromosome clonality were calculated using the formula (AR = (L1/L2) / (N1/N2), where L1 and L2 are the peaks for leukemic samples/plasma (ie, non CD3+) and N1 and N2 are the peaks for nonleukemic control samples (CD3+). LOH was defined as a ratio of less than 0.5 or more than 1.5.
Results and discussion
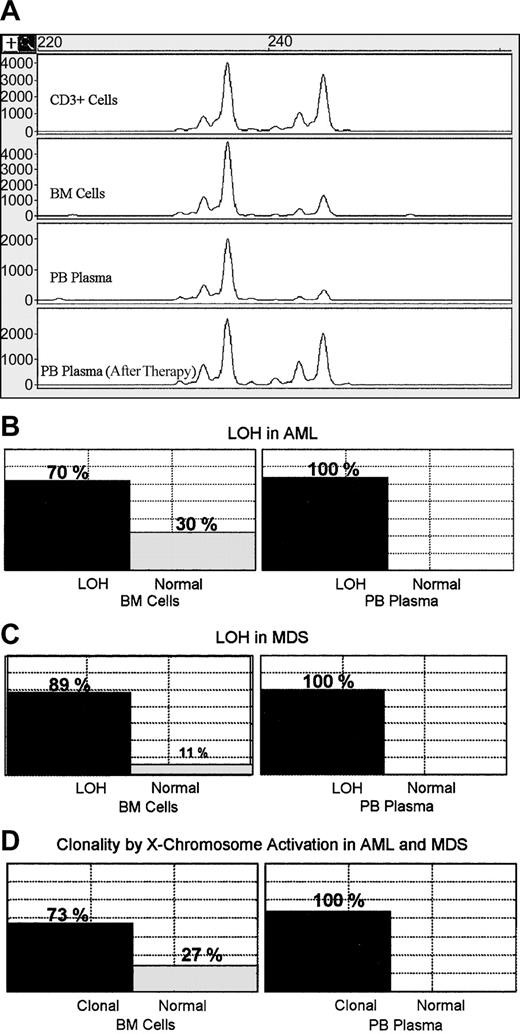
Samples from 45 patients with cytogenetically confirmed abnormalities were analyzed. Some of the patients had abnormalities in more than one chromosome. LOH was detected in 88.9% (24 of 27) and 70.3% (26 of 37) of the chromosomal abnormalities using BM cell samples in patients with MDS and patients with AML, respectively (Figure 1). In contrast, we found LOH in 100% of the abnormalities when PB plasma samples were used in both MDS and AML (25 of 25 and 27 of 27, respectively; Figure 1). In almost all samples LOH was easily detectable in the plasma samples as compared with BM cells. The changes in the ratios between the 2 alleles in PB plasma samples were significantly higher than those detected in BM cells (P < .001, Wilcoxon matched pairs test). From 16 of these patients, paired posttherapy follow-up PB and BM samples were collected and analyzed by cytogenetic, LOH, and morphology (Table 1). Of the 16 samples, 10 showed karyotypic evidence of residual disease, and 6 showed neither karyotypic nor LOH evidence of residual disease. Except for one case, there was concordance between BM karyotype and PB plasma LOH. In 4 samples, there was evidence of residual disease by karyotype and LOH, but not by morphology. In one case there was cytogenetic evidence of residual disease but not by plasma LOH or morphology.
Detection of LOH and X-chromosome inactivation in bone marrow cells and peripheral blood plasma. (A) Representative example showing LOH in bone marrow cells and PB plasma before therapy and disappearance of the LOH when the patient was in remission. (B) Box plots showing positive and negative cases for LOH as detected in 37 BM cell samples from patients with AML. All tested corresponding PB plasma samples (27 samples) showed LOH. (C) Positive and negative cases for LOH detected in 27 samples of BM cells from patients with MDS, while all tested corresponding PB plasma samples (25 samples) showed LOH. (D) Monoclonality and polyclonality were detected by X-chromosome inactivation in BM cell samples from 26 patients with AML or MDS, whereas all tested plasma samples (22 samples) were monoclonal.
Detection of LOH and X-chromosome inactivation in bone marrow cells and peripheral blood plasma. (A) Representative example showing LOH in bone marrow cells and PB plasma before therapy and disappearance of the LOH when the patient was in remission. (B) Box plots showing positive and negative cases for LOH as detected in 37 BM cell samples from patients with AML. All tested corresponding PB plasma samples (27 samples) showed LOH. (C) Positive and negative cases for LOH detected in 27 samples of BM cells from patients with MDS, while all tested corresponding PB plasma samples (25 samples) showed LOH. (D) Monoclonality and polyclonality were detected by X-chromosome inactivation in BM cell samples from 26 patients with AML or MDS, whereas all tested plasma samples (22 samples) were monoclonal.
Follow-up samples with cytogenetic and PB plasma LOH analysis
Patient . | Diagnosis* . | % BM blasts . | % positive metaphases† . | Plasma LOH . |
|---|---|---|---|---|
| 1 | RAEB-T | 54 | 73.3 | L |
| 2 | RAEB-T | 26 | 100 | L |
| 3 | RAEB | 12 | 100 | L |
| 4 | RAEB | 0 | 5 | L |
| 5 | RAEB | 3 | 10 | N |
| 6 | RA | 1 | 28.6 | L |
| 7 | RAEB | 2 | 100 | L |
| 8 | RAEB | 70 | 100 | L |
| 9 | RAEB | 0 | 0 | N |
| 10 | RAEB-T | 5 | 100 | L |
| 11 | AML | 2 | 0 | N |
| 12 | AML | 29 | 10 | L |
| 13 | AML | 5 | 0 | N |
| 14 | AML | 0 | 0 | N |
| 15 | AML | 7 | 0 | N |
| 16 | AML | 1 | 0 | N |
Patient . | Diagnosis* . | % BM blasts . | % positive metaphases† . | Plasma LOH . |
|---|---|---|---|---|
| 1 | RAEB-T | 54 | 73.3 | L |
| 2 | RAEB-T | 26 | 100 | L |
| 3 | RAEB | 12 | 100 | L |
| 4 | RAEB | 0 | 5 | L |
| 5 | RAEB | 3 | 10 | N |
| 6 | RA | 1 | 28.6 | L |
| 7 | RAEB | 2 | 100 | L |
| 8 | RAEB | 70 | 100 | L |
| 9 | RAEB | 0 | 0 | N |
| 10 | RAEB-T | 5 | 100 | L |
| 11 | AML | 2 | 0 | N |
| 12 | AML | 29 | 10 | L |
| 13 | AML | 5 | 0 | N |
| 14 | AML | 0 | 0 | N |
| 15 | AML | 7 | 0 | N |
| 16 | AML | 1 | 0 | N |
BM indicates bone marrow; LOH, loss of heterozygosity; RAEB-T, refractory anemia with excess of blasts transformed; RAEB, refractory anemia with excess of blasts; RA, refractory anemia; AML, acute myeloid leukemia; L, loss of heterozygosity; N, normal pattern.
Diagnosis is based on the bone marrow findings at the time of presentation.
The percentage of abnormal metaphases is based on analyzing 20 metaphases.
There was significant variation among patients in the levels of DNA in plasma as determined using the Ras DNA amplification products. Equal amounts of plasma were used for extracting DNA and equal amounts of DNA solution were used for this assay in a fashion similar to that used for the LOH. The levels of plasma DNA did not correlate with the detection of LOH. Control PB samples yielded no abnormalities at any of the markers assayed.
Using HUMARA and X-chromosome inactivation, we analyzed clonality in PB plasma and BM cells from 26 female patients with AML12 or MDS.14 Only 19 (73%) of the 26 BM samples showed clonality (12 of 14 MDS and 7 of 12 AML), whereas all analyzable PB plasma samples showed clonality. The T cells (CD3+) were used as normal controls. However, we observed that PB plasma from some patients may show no methylation in both alleles, despite the adequate amplification of control microsatellite markers. This suggests that methylation may not be stable in plasma. This phenomenon was not observed in PB plasma from the 15 healthy controls.
These data suggest that PB plasma is a reliable source of DNA to test for genomic abnormalities in patients with AML or MDS. Unlike BM aspiration samples, the dilution effect of residual normal cells does not appear to be an issue when PB plasma is used. We do not know the reason for this enrichment of PB plasma by leukemia-specific DNA. However, we speculate that since the leukemic cells have a higher turnover rate than normal cells, they do not process their contents properly, pouring most of their contents into circulation. In contrast, normal cells undergo orderly apoptosis and disintegration, allowing the reticuloendothelial system to absorb most of their products. Plasma is more convenient to obtain and is not influenced by fibrosis or patchiness of the disease. The use of plasma as a source of tumor DNA is particularly important in patients with MDS, since the leukemic cells are capable of differentiating, and at many stages of this disease it is difficult to determine whether the disease is responding to therapy when only BM morphology is used. Evaluating the changes of cytogenetic abnormalities using PB plasma is a potential tool that can be used to monitor these patients.
Prepublished online as Blood First Edition Paper, October 23, 2003; DOI 10.1182/blood-2003-06-1840.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal