Abstract
Non-germinal center small B-cell lymphomas represent a heterogeneous group of non-Hodgkin lymphomas, the most frequent histologic subtypes being small lymphocytic lymphoma (SLL), splenic marginal zone B-cell lymphoma (MZL), and mantle cell lymphoma (MCL). In order to identify genomic signatures specific for each disease, we analyzed 128 primary tumors using high-density microarrays. Several clusters of genes significantly discriminated the 3 histologic subtypes. Genes associated with cell adhesion, angiogenesis, and inhibition of apoptosis were up-regulated in SLL. Genes associated with intracellular signaling via the AKT1 pathway were up-regulated in splenic MZL. Genes associated with cell cycle control and multidrug resistance were up-regulated in MCL. Using 44 genes selected within the gene clusters discriminant for the 3 lymphoma subtypes, we generated a class prediction score that allowed us to classify the 3 entities in 96% of the cases, including borderline cases. Whereas specific transcriptional profiles easily distinguished all MZL samples, SLL samples, and most of the MCL samples into separate groups, few MCL cases exhibited MZL-type transcriptional profiles. This study demonstrates that SLL, splenic MZL, and MCL possess specific transcriptional profiles that may be relevant to the pathogenesis and the diagnosis of these histologic subtypes. (Blood. 2004;103:2727-2737)
Introduction
Gene-expression profiling has been used extensively for diffuse large B-cell lymphoma analysis during the past 3 years,1-3 contributing to a dramatic increase of knowledge for B-cell lymphoid tumorogenesis.4 Lymphoma oncogenesis involves disruption of normal B-cell differentiation at different stages. The non-germinal center small B-cell lymphomas5 represent a heterogeneous group of B-cell lymphoid neoplasms related to different cells of origin, the most frequent subtypes being small lymphocytic lymphoma (SLL) which is molecularly and clinically related to chronic lymphocytic leukemia (CLL), marginal zone B-cell lymphoma (MZL), and mantle cell lymphoma (MCL), which is associated with t(11;14) (q13;q32) resulting in cyclin D1 overexpression.
These 3 lymphoma entities have a very different clinical outcome but may be difficult to distinguish either histologically or clinically. Moreover, in spite of multifaceted phenotypic characterization, there are some “borderline” cases without definitive classification. The recent studies using gene-expression profiling have established that lymphomas and leukemias that are difficult to distinguish morphologically can nevertheless be distinguished molecularly.1-3,6-8
The goal of our study was to characterize the molecular pathways specifically involved in SLL, splenic MZL, and MCL using gene-expression profiling. The results showed that different biologic pathways were altered in these malignancies, each disease being characterized by a specific genomic signature. According to the expression of 44 genes selected within the gene clusters discriminant for the 3 lymphoma subtypes, we generated a class prediction score in order to classify all the cases involved in the study, including the few borderline cases. Moreover, MZL and SLL samples, and most of the MCL samples, could be distinguished according to their transcriptional profiles as distinct entities. However, a few MCL cases exhibited MZL-type transcriptional profiles.
Patients, materials, and methods
Selection of patients
Fresh-frozen tumor biopsies and clinical data were obtained retrospectively from 121 untreated patients of 3 different institutions after complete morphologic analysis, including cytologic, immunologic, cytogenetic (conventional cytogenetic and fluorescent in situ hybridization [FISH]), and/or molecular analysis, to assess the diagnosis of typical SLL, typical MZL, or MCL. All patients had signed informed consent for biopsy analysis.
Morphologic characteristics of the patients are shown in Table 1. Only 2 patients with SLL had more than 10 × 109/L circulating peripheral blood lymphoma cells9 (13 × 109/L and 14 × 109/L, respectively). All patients with MZL had a splenic MZL except 2 who had nodal MZL.10 Two patients with splenic MZL with the expected immunophenotype and morphology presented a t(11;14) translocation with cyclin D1 overexpression and unmutated IgVH gene status. All MCL samples were controlled for cyclin D1 overexpression by competitive reverse transcriptase-polymerase chain reaction (RT-PCR).11 Three of the patients with MCL lacked CD5 expression but were typical MCL when considering histology, cytology, and cytogenetic analysis. This group of patients corresponded to the preliminary group or learning set.
Phenotypic characteristics of the 122 patients whose biopsy has been hybridized on microarrays
Patients Preliminary group Validation group . | Total N = 122 93 29 . | B-CLL/SLL n = 31 . | . | Splenic MZL*n = 37 . | . | MCL n = 56 . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Preliminary group, n = 19 (%) . | Validation group, n = 10 (%) . | Preliminary group, n = 29 (%) . | Validation group, n = 8 (%) . | Preliminary group, n = 45 (%) . | Validation group, n = 11 (%) . | |||
| Samples | ||||||||||
| Lymph node | 77 (63) | 19 (100) | 9 (90) | 2 (6)† | 0 | 38 (85) | 9 (82) | |||
| Spleen | 33 (27) | 0 | 1 (10) | 19 (66) | 8 (100) | 5 (11) | 0 | |||
| Blood | 9 (7) | 0 | 0 | 8 (28) | 0 | 1 (2) | 0 | |||
| Others: tonsil | 3 (3) | 0 | 0 | 0 | 0 | 1 (2) | 2 (18) | |||
| Immunophenotype | 122 (100) | |||||||||
| CD19/CD20+ | 19 (100) | 9 (100) | 29 (100) | 8 (100) | 38 (100) | 9 (100) | ||||
| CD5+ | 19 (100) | 9 (100) | 5 (17) | 0 | 35 (92) | 9 (100) | ||||
| CD23+ | 19 (100) | 9 (100) | 0 | 0 | 0 | 0 | ||||
| CD43+ | 19 (100) | 9 (100) | 0 | 0 | 38 (100) | 9 (100) | ||||
| CD10+ | 0 | 0 | 0 | 0 | — | — | ||||
| Cytogenetic features | 96 (79) | 19 | 2 | 29 | 2 | 34 | 2 | |||
| del 13q | 1 (5) | 0 | 1 (3) | 0 | 0 | 0 | ||||
| +12 | 9 (47) | 0 | 0 | 0 | 0 | 0 | ||||
| del 11q | 4 (21) | 0 | 0 | 0 | 1 (3) | 0 | ||||
| +3 or +3q | 0 | 0 | 8 (27) | 0 | 2 (6) | 0 | ||||
| del 7q | 0 | 0 | 5 (17) | 0 | 0 | 0 | ||||
| t(11;14) | 0 | — | 2 (11) | 0/2 | 34/34 (100) | 2/2 (100) | ||||
| Molecular features | ||||||||||
| Cyclin D1 +++ | 58 (46) | — | — | 2 (11) | 0 | 45 (100) | 9 (100) | |||
| Ig mutation status | ||||||||||
| Mutated (<98) | 36 (29) | 5 (26) | 1 (10) | 19 (66) | 4 (50) | 7 (15) | 0 | |||
| Unmutated (≥ 98%) | 63 (52) | 12 (63) | 8 (80) | 8* (27) | 4 (50) | 22 (49) | 9 (82) | |||
| Not done | 23 (19) | 2 (11) | 1 (10) | 2 (7) | 0 | 16 (36) | 2 (8) | |||
| Circulating peripheral | ||||||||||
| blood lymphoma | ||||||||||
| cells > 10 × 109/L | 2/19 (10) | 0/8 | 13 (45) | 0/6 | 1/45 | 0/7 | ||||
Patients Preliminary group Validation group . | Total N = 122 93 29 . | B-CLL/SLL n = 31 . | . | Splenic MZL*n = 37 . | . | MCL n = 56 . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Preliminary group, n = 19 (%) . | Validation group, n = 10 (%) . | Preliminary group, n = 29 (%) . | Validation group, n = 8 (%) . | Preliminary group, n = 45 (%) . | Validation group, n = 11 (%) . | |||
| Samples | ||||||||||
| Lymph node | 77 (63) | 19 (100) | 9 (90) | 2 (6)† | 0 | 38 (85) | 9 (82) | |||
| Spleen | 33 (27) | 0 | 1 (10) | 19 (66) | 8 (100) | 5 (11) | 0 | |||
| Blood | 9 (7) | 0 | 0 | 8 (28) | 0 | 1 (2) | 0 | |||
| Others: tonsil | 3 (3) | 0 | 0 | 0 | 0 | 1 (2) | 2 (18) | |||
| Immunophenotype | 122 (100) | |||||||||
| CD19/CD20+ | 19 (100) | 9 (100) | 29 (100) | 8 (100) | 38 (100) | 9 (100) | ||||
| CD5+ | 19 (100) | 9 (100) | 5 (17) | 0 | 35 (92) | 9 (100) | ||||
| CD23+ | 19 (100) | 9 (100) | 0 | 0 | 0 | 0 | ||||
| CD43+ | 19 (100) | 9 (100) | 0 | 0 | 38 (100) | 9 (100) | ||||
| CD10+ | 0 | 0 | 0 | 0 | — | — | ||||
| Cytogenetic features | 96 (79) | 19 | 2 | 29 | 2 | 34 | 2 | |||
| del 13q | 1 (5) | 0 | 1 (3) | 0 | 0 | 0 | ||||
| +12 | 9 (47) | 0 | 0 | 0 | 0 | 0 | ||||
| del 11q | 4 (21) | 0 | 0 | 0 | 1 (3) | 0 | ||||
| +3 or +3q | 0 | 0 | 8 (27) | 0 | 2 (6) | 0 | ||||
| del 7q | 0 | 0 | 5 (17) | 0 | 0 | 0 | ||||
| t(11;14) | 0 | — | 2 (11) | 0/2 | 34/34 (100) | 2/2 (100) | ||||
| Molecular features | ||||||||||
| Cyclin D1 +++ | 58 (46) | — | — | 2 (11) | 0 | 45 (100) | 9 (100) | |||
| Ig mutation status | ||||||||||
| Mutated (<98) | 36 (29) | 5 (26) | 1 (10) | 19 (66) | 4 (50) | 7 (15) | 0 | |||
| Unmutated (≥ 98%) | 63 (52) | 12 (63) | 8 (80) | 8* (27) | 4 (50) | 22 (49) | 9 (82) | |||
| Not done | 23 (19) | 2 (11) | 1 (10) | 2 (7) | 0 | 16 (36) | 2 (8) | |||
| Circulating peripheral | ||||||||||
| blood lymphoma | ||||||||||
| cells > 10 × 109/L | 2/19 (10) | 0/8 | 13 (45) | 0/6 | 1/45 | 0/7 | ||||
Blood samples were used for microarray hybridization when frozen material (spleen biopsy [n = 8] for splenic MZL or node biopsy [n = 1] for MCL) was not available for RNA extraction. All blood samples contained more than 50% lymphoma cells.—indicates not done.
bcl-6 immunostaining was negative.
Two patients presented a nodal MZL and not splenic MZL at diagnosis.
Before sample processing, morphologic review of all the cases was performed (F.B., P.F., P.G., E.C.-B.) and 6 cases were classified as borderline (Table 2). Among them, 4 MZL cases were secondarily classified as borderline SLL-MZL because of a very diffuse pattern of splenic infiltration and CD5+ expression for 3 of those cases, and 2 MCL cases were secondarily classified as borderline MCL-MZL because of lack of MCL morphologic characteristics. These 6 borderline cases were then analyzed separately. The presence of somatic mutations of IgVH genes was determined in all available samples.12
Clinical and morphologic characteristics of the borderline cases
. | Case 1 SLL/MZL . | Case 2 SLL/MZL . | Case 3 SLL/MZL . | Case 4 SLL/MZL . | Case 5 MZL/MCL . | Case 6 MZL/MCL . |
|---|---|---|---|---|---|---|
| Clinical features | ||||||
| Sites of involvement | Spleen | Spleen | Spleen | Spleen | Nodes | Nodes |
| Bone marrow | Bone marrow | Bone marrow | Bone marrow | spleen | spleen | |
| Blood | Blood | Bone marrow | Bone marrow | |||
| Lymphocytosis | 3 × 109/L | 12 × 109/L | 8 × 109/L | 80 × 109/L | 4 × 109/L | 3 × 109/L |
| LDH | Elevated | Elevated | Normal | Elevated | Normal | Elevated |
| B2 microglobulin, g/L | 3.1 | 6 | 1.9 | 3.2 | 4.3 | 3.1 |
| Monoclonal component | M Kappa-29.3 g/L | G Lamda-4 g/L | 0 | 0 | 0 | 0 |
| Follow-up, years | 2.26 | 148 days | 3.55 | 1.94 | 121 days | 2.83 |
| Status at last follow-up | Alive | Dead (Neuro) | Alive | Dead | Dead | Alive |
| Histology | Diffuse infiltration of red pulp by small lympho-plasmacytic cells | Diffuse infiltration of small plasmacytoid cells | Micronodular infiltration of small cells | Diffuse infiltration of red pulp by small round cells; nodular pattern in splenic hilar LN | Micronodular and diffuse infiltration of round cells and centrocytic cells | Diffuse infiltration of small round cells, numerous mitoses |
| Lymphocyte cytology | Lympho-plasmacytic cells, MZL type | Atypical CLL cells (clumped chromatin, plasmacytoid differentiation, smudge cells) | Clumped chromatin, no smudge cells | Clumped chromatin, some smudge cells | Heterogeneous in size (small to large), basophilic cytoplasm | Small cells with regular nucleus |
| Immunophenotype of B cells | CD5+ CD23− | CD5+ CD23− CD43+ wk | CD5+ CD23+ wk | CD5+ CD23− CD43− | CD5+ CD23− CD43− | CD5+ CD23− CD43+ |
| CD43+ wk | Slg+++, MC7+, CD22++ | |||||
| Slg+++ | ||||||
| FMC7+, CD22++ | ||||||
| Cytogenetics | +12 | Complex without recurrent abnormalities | +12 | Complex with +3q | Complex with t(11;14) | No t(11;14) but very rare mitoses |
| del13q | +18 | t(1;12) | ||||
| IgVH mutational status | Mutated | Mutated | Mutated | Mutated | Mutated | Not mutated |
| Cyclin D1 | Not applicable | Not applicable | Not applicable | Not applicable | ++ | ++ |
| Site of biopsy | Spleen | Spleen | Spleen | Spleen | Spleen | Node |
. | Case 1 SLL/MZL . | Case 2 SLL/MZL . | Case 3 SLL/MZL . | Case 4 SLL/MZL . | Case 5 MZL/MCL . | Case 6 MZL/MCL . |
|---|---|---|---|---|---|---|
| Clinical features | ||||||
| Sites of involvement | Spleen | Spleen | Spleen | Spleen | Nodes | Nodes |
| Bone marrow | Bone marrow | Bone marrow | Bone marrow | spleen | spleen | |
| Blood | Blood | Bone marrow | Bone marrow | |||
| Lymphocytosis | 3 × 109/L | 12 × 109/L | 8 × 109/L | 80 × 109/L | 4 × 109/L | 3 × 109/L |
| LDH | Elevated | Elevated | Normal | Elevated | Normal | Elevated |
| B2 microglobulin, g/L | 3.1 | 6 | 1.9 | 3.2 | 4.3 | 3.1 |
| Monoclonal component | M Kappa-29.3 g/L | G Lamda-4 g/L | 0 | 0 | 0 | 0 |
| Follow-up, years | 2.26 | 148 days | 3.55 | 1.94 | 121 days | 2.83 |
| Status at last follow-up | Alive | Dead (Neuro) | Alive | Dead | Dead | Alive |
| Histology | Diffuse infiltration of red pulp by small lympho-plasmacytic cells | Diffuse infiltration of small plasmacytoid cells | Micronodular infiltration of small cells | Diffuse infiltration of red pulp by small round cells; nodular pattern in splenic hilar LN | Micronodular and diffuse infiltration of round cells and centrocytic cells | Diffuse infiltration of small round cells, numerous mitoses |
| Lymphocyte cytology | Lympho-plasmacytic cells, MZL type | Atypical CLL cells (clumped chromatin, plasmacytoid differentiation, smudge cells) | Clumped chromatin, no smudge cells | Clumped chromatin, some smudge cells | Heterogeneous in size (small to large), basophilic cytoplasm | Small cells with regular nucleus |
| Immunophenotype of B cells | CD5+ CD23− | CD5+ CD23− CD43+ wk | CD5+ CD23+ wk | CD5+ CD23− CD43− | CD5+ CD23− CD43− | CD5+ CD23− CD43+ |
| CD43+ wk | Slg+++, MC7+, CD22++ | |||||
| Slg+++ | ||||||
| FMC7+, CD22++ | ||||||
| Cytogenetics | +12 | Complex without recurrent abnormalities | +12 | Complex with +3q | Complex with t(11;14) | No t(11;14) but very rare mitoses |
| del13q | +18 | t(1;12) | ||||
| IgVH mutational status | Mutated | Mutated | Mutated | Mutated | Mutated | Not mutated |
| Cyclin D1 | Not applicable | Not applicable | Not applicable | Not applicable | ++ | ++ |
| Site of biopsy | Spleen | Spleen | Spleen | Spleen | Spleen | Node |
+, ++, and +++ correspond to the expression level of the immunostaining.
LN indicates lymph node; LDH, lactodehydrogenase.
Isolation of total RNA
Tumor material was snap frozen in liquid nitrogen. Frozen sections were stained with hematoxylin and eosin and only samples that had more than 50% tumor cells were selected. Ten 50-μm sections were used for the isolation of RNA. Total RNA was isolated using a guanidium isothiocyanate-based method (Trizol reagent; Gibco BRL, Grand Island, NY). Total RNA obtained from 2 cell lines of mantle cell type (GRANTA and NCEB) was also extracted. Total RNA was treated with DNAse to minimize genomic DNA contamination (DNA free; Ambion, Cambridgeshire, United Kingdom), the RNA was then dissolved in RNase-free water to a final concentration of 0.5μg/μL. Quality and quantity controls were assessed by optical density (OD) 260/280 ratios and by electrophoresis (Agilent 2100 Bioanalyzer; Agilent, Massy, France). Twenty-two samples from the learning set were not accepted for further experimentation, 3 because of insufficient quantity and 19 because of insufficient quality. A total of 101 samples were available for microarray hybridization (99 patient samples [93 typical and 6 borderline cases] and 2 cell lines).
cDNA microarrays
The microarrays included 6692 clones of complementary DNA, 2500 of which were related with carcinogenesis, and 427 controls.13 The cDNA clones were obtained through a selection of IMAGE clones using expressed sequence tag (EST) databases, and were amplified as previously described. PCR products were spotted onto nylon membranes using a 64 plain pins Microgrid II printer (Apogent Discoveries, Hudson, NY). The amounts of target cDNA accessible to probe hybridization were controlled in each spot using the hybridization of an oligonucleotide common to each cDNA13 performed previously to the sample hybridization.
Labeling and hybridization to the microarray
cDNA synthesis, radioactive-labeling, hybridization, and washing of the cDNA microarray membranes were performed according to procedures available on the web at http://tagc.univ-mrs.fr/pub. Briefly, 2.0μg total RNA was used as a template for cDNA synthesis in the presence of 33[P] dATP (Amersham Pharmacia Biotech, Bucks, United Kingdom) to produce labeled cDNA. The labeled targets were purified using spin columns (Sephadex G50; Roche Diagnostics, Meylan, France). Hybridizations were carried out during 48 hours at 68°C in a rotation hybridization oven using 30 × 106 cpm of the probe in a final volume of 300 μL buffer. The membranes were then washed at 68°C twice during 3 hours, exposed for 24 and 48 hours on a radioimager, and scanned (Fuji Bas 5000; Raytest, Paris, France). Signal intensities on scanned images were quantified (ArrayGauge software v1.3; Fuji, Paris, France). The values were corrected for background level, normalized for the amount of spotted cDNA and the variability of experimental conditions, and log-transformed.14 From the initial 6692 clones, 2533 were significantly expressed in a majority of the samples.
Validation strategy
To investigate the diagnostic value of the gene-expression profile, an additional 29 untreated patients were independently selected for a validation group on the same criteria as those described for the preliminary group. Morphologic characteristics are detailed in Table 1. Samples were processed independently from the preliminary group using the same experimental protocol and the assignment to lymphoma subtypes was allocated in a blinded fashion.
Statistical analysis
Before analysis, the reproducibility of the experiments was verified by comparing 2 hybridizations of samples prepared from the same RNA on 2 different arrays. Twenty-one samples were then hybridized twice. The reproducibility of the experiments between the preliminary group and the validation group was also verified with 2 samples hybridized in both groups. Regarding the analysis, these 2 samples were included in the preliminary group and not in the validation group. In every case, the results showed good reproducibility with correlation coefficients of 0.98 (data not shown).
Average linkage, unsupervised, hierarchical clustering was applied to preliminary group samples to investigate relationships between samples and relationships between genes. We used the Cluster program and displayed results with the Treeview program (M. Eisen, University of California, Berkeley; http://www.microarrays.org/software).15
Genes discriminating particular lymphomas subtypes (SLL, splenic MZL, MCL) were searched in the preliminary samples using a signal-to-noise calculation with a discriminating score (DS) equal to (μ1-μ2)/(σ1+σ2),16,17 where μ1 and σ1 represent, respectively, mean and standard deviation of the expression levels of the genes in the group of the lymphoma subtype 1, and μ2 and σ2 represent, respectively, mean and standard deviation of the expression levels of the genes in the other samples. One hundred random permutations of the samples were used to calculate the significant level of risk at 1 of 10 000, resulting in less than one false-positive gene. A given gene was considered to characterize a particular subtype when it was overexpressed in this subtype in comparison to the 2 other subtypes. A very similar set of genes could also be detected by a t statistic with Bonferroni correction.18
A multiple class predictor was built using the method described by Sorlie et al:19 (1) clusters of genes discriminating tumor subtypes were selected, (2) a mean profile was determined for each tumor subtype, and (3) correlations between individual samples and tumor subtypes were calculated. A high correlation between one sample and one tumor-subtype mean profile reflects the similarity between that sample and the tumor subtype. Selected clusters were those containing more than 80% of genes distinguishing a particular lymphoma subtype and associated with a correlation superior to 0.66. This correlation threshold allowed the separation between spleen and MZL signatures, which were strongly associated with the tissue of origin of the MZL samples (mainly spleen). Subtype-specific mean profiles were determined using the mean expression values observed in all samples of this subtype for each selected gene. Pearson correlations between the expression of the selected genes in each sample and the 3 mean profiles were calculated (RSLL, RMCL, RMZL). Correlations of more than 0.4 were considered as significant with a 5% risk. This threshold was calculated by generating bootstrapped profiles, where expression of each gene was randomly chosen from all the observed values for that gene in samples of any subtype. Samples exhibiting no significant correlation with any subtype profile were considered as unpredictable. None of the samples exhibited a significant correlation with 2 subtype profiles.
Protein validation (immunohistochemistry)
The potential extension of microarray-based diagnosis prediction was further explored using immunohistochemistry. We chose one molecule for each lymphoma subtype because of the very high discriminating score for each specific disease and because of the commercial availability of antibodies: L-selectin for SLL (1/20; Dako SA, Trappes, France), pAKT1 protein for splenic MZL (1/100; Upstate Cell Signaling, Euromedex, Strasbourg, France), and glutathione S-transferase (GSTpi) (1/200; Tebu International, Le Perray en Yveline, France) for MCL. We used formalin-fixed, paraffin-embedded sections for glutathione S-transferase pi and pAKT1 analysis, and frozen sections for L-selectin analysis. The formalin-fixed, paraffin-embedded sections were subjected to antigen retrieval by boiling in a microwave for 30 minutes in 0.01 M sodium citrate buffer (pH 6.0) and then exposed to 3% hydrogen peroxide diluted in water for 20 minutes at room temperature. Immunochemistry analysis was realized using a Ventana system according to the manufacturer's instructions, except for pAKT1 incubated at 4°C overnight and for which a streptavidin-biotin system (Dako Duet kit) was used. Fifteen samples (5 for each entity) were analyzed.
Results
Categorization of gene-expression signatures
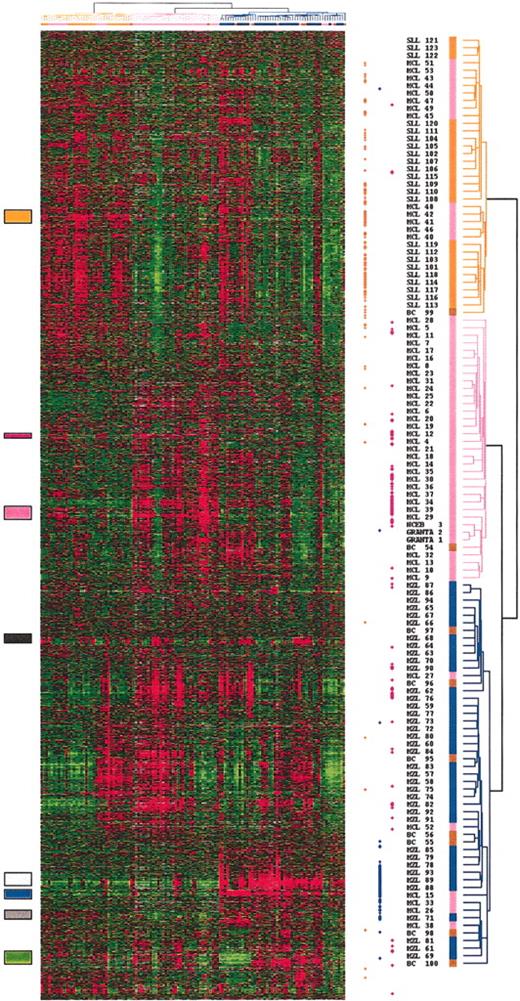
Total RNA of each normal and malignant sample was isolated and used to generate a radio-labeled cDNA, which was hybridized to microarrays containing 6692 human cDNA clones. Radioactive dot intensities of scanned images were quantified. The overall expression patterns were first analyzed using hierarchical clustering in an unsupervised fashion to group the 122 analyses of the 101 preliminary group samples according to the gene-expression profiles of the 2533 expressed genes. Figure 1 presents the variation in gene-expression patterns in the samples. Examination of the data on the vertical axis highlighted clusters of correlated genes defining gene-expression signatures corresponding either to the cell type or the tissue type in which its component was expressed (eg, the “T-cell” signature, the “SPLEEN” signature) or to the biologic process in which its component genes are known to function (eg, the “EARLY RESPONSE” signature; Figure 2). The “SPLEEN” cluster-grouped genes expressed only in spleen samples, whatever the disease subtype, and included genes related to red cells (hemoglobin zeta ζ, EPB4.1-like2, KLF1, for example). A “STROMAL” cluster associating genes such as COL1A1, COL1A2, COL3A1, MMP2, MMP9, TIMP3, and VCAM1 was expressed in nodal and splenic samples as compared with the cell lines and the blood samples. Cyclin D1 gene expression correlated with that of other genes including MACS, ETV4, JUP (junction plakoglobin), EHD1 to constitute an independent and small gene cluster. Three lymphoma subtype-specific clusters could be detected: the SLL cluster (interleukin 4 [IL4] receptor, L-selectin, RAS-GAP1, titin), the MZL cluster (AKT1, AGER, S100A9, ZFP36), and the MCL cluster (GST pi, DNMT1, PCNA, CDK4).
Hierarchical clustering of gene-expression data. Measurements of gene expression from 122 microarray analyses of the 101 nonfollicular small B-cell lymphoma samples of the preliminary group, and the 2 mantle cell lines (NCEB; GRANTA) are depicted. Each column represents the genomic profile of the 2533 expressed genes for one tumor, and each row represents the relative level of expression of one cDNA clone. Red indicates a high level of expression of messenger RNA of the given gene in the tumor, as compared with the median value of this gene, and green indicates a low level of expression, according to the color scale. Gray indicates missing data. Correlated genes are grouped across the samples into clusters localized by the colored boxes. The dendrogram, reported also at the top of the hierarchical clustering, lists the samples studied and provides a measure of the relatedness of gene expression in each sample. SLL cases are colored in orange, MZL cases in blue, MCL cases in pink, and cases reclassified as borderline cases in brown. The dots at the right side of the hierarchical clustering represent the genes that significantly discriminate the 3 entities. The orange dots represent the genes that discriminated the SLL entity, the blue dots the MZL entity, and the pink dots the MCL entity.
Hierarchical clustering of gene-expression data. Measurements of gene expression from 122 microarray analyses of the 101 nonfollicular small B-cell lymphoma samples of the preliminary group, and the 2 mantle cell lines (NCEB; GRANTA) are depicted. Each column represents the genomic profile of the 2533 expressed genes for one tumor, and each row represents the relative level of expression of one cDNA clone. Red indicates a high level of expression of messenger RNA of the given gene in the tumor, as compared with the median value of this gene, and green indicates a low level of expression, according to the color scale. Gray indicates missing data. Correlated genes are grouped across the samples into clusters localized by the colored boxes. The dendrogram, reported also at the top of the hierarchical clustering, lists the samples studied and provides a measure of the relatedness of gene expression in each sample. SLL cases are colored in orange, MZL cases in blue, MCL cases in pink, and cases reclassified as borderline cases in brown. The dots at the right side of the hierarchical clustering represent the genes that significantly discriminate the 3 entities. The orange dots represent the genes that discriminated the SLL entity, the blue dots the MZL entity, and the pink dots the MCL entity.
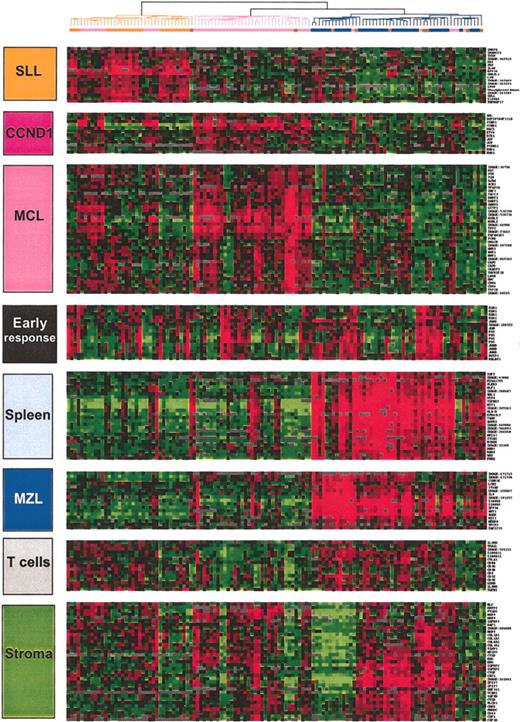
Characteristic gene clusters. Some of the correlated genes grouped into clusters and noted at the left side of the hierarchical clustering figures are reported. They correspond either to a specific type of contaminating cell (T cells) or a tissue type (stroma, spleen). The gene expression of 3 clusters were more specifically related to the 3 lymphoma entities: SLL, MZL, and MCL. SLL indicates small lymphocytic lymphoma; MZL, marginal zone lymphoma; MCL, mantle cell lymphoma; BC, borderline cases; CCND1, cyclin D1. Gene names are available on the Blood website as supplemental data (Table S1); see the Data Set link at the top of the online article.
Characteristic gene clusters. Some of the correlated genes grouped into clusters and noted at the left side of the hierarchical clustering figures are reported. They correspond either to a specific type of contaminating cell (T cells) or a tissue type (stroma, spleen). The gene expression of 3 clusters were more specifically related to the 3 lymphoma entities: SLL, MZL, and MCL. SLL indicates small lymphocytic lymphoma; MZL, marginal zone lymphoma; MCL, mantle cell lymphoma; BC, borderline cases; CCND1, cyclin D1. Gene names are available on the Blood website as supplemental data (Table S1); see the Data Set link at the top of the online article.
Although no information on the identity of the samples was used in the clustering, the algorithm could segregate, with few exceptions, the SLL, splenic MZL, and MCL subtypes. Clearly, it appeared that several of the previously identified gene clusters were differentially expressed between the SLL, the splenic MZL, and the MCL categories. SLL samples overexpressed the SLL gene cluster. Splenic MZL samples overexpressed genes grouped in a cluster including AKT1, S100A proteins, and AGER, and genes grouped in the cluster related to the splenic tissue. The MCL cluster genes were overexpressed in the MCL samples, as well as in the MCL cell lines. MCL samples also overexpressed genes related to the stroma.
Gene-expression-based diagnostic predictor
We used the preliminary collection of samples belonging to known classes to create a “diagnostic predictor” based on the gene-expression profiles. Using a discriminating score16 and a permutation test to assess each gene's individual ability to distinguish the lymphoma subtype,20 we found that 415 genes were significantly (P < .0001, 0.75 false-positive gene) correlated with the SLL, splenic MZL, or MCL subtype (Table 3). We ranked the informative genes into 3 groups, the higher score indicating the greater ability to differentiate one sample between the 3 profiles. The list of genes is available on the web at http://tagc.univ-mrs.fr/pub.
Distribution of the subtype-discriminating genes within specific clusters
Gene-expression variable . | No. of microarray features in signature n = 2532 . | No. of microarray features significantly associated (P < .00001) with diagnosis of SLL n = 178 . | No. of microarray features significantly associated (P < .00001) with diagnosis of MZL n = 83 . | No. of microarray features significantly associated (P < .00001) with diagnosis of MCL n = 154 . | Representative genes . | GenBank accession no. . |
|---|---|---|---|---|---|---|
| SLL cluster | 735 | 163 | 1 | 12 | L-selectin | AJ246000 |
| IL4-R | C75499 | |||||
| TNF-R17 | ||||||
| Cyclin D1 cluster | 13 | 0 | 0 | 13 | CCND1 | NM 053056 |
| MCL cluster | 349 | 1 | 1 | 75 | PCNA | AF508260 |
| CDK4 | NM_007671 | |||||
| MYBL2 | ||||||
| Early response cluster | 20 | 0 | 0 | 0 | Fos | V01512 |
| Spleen cluster | 54 | 0 | 36 | 0 | HBZ | J00182 |
| KLF1 | NM_006563 | |||||
| EPB41L2 | M_001431 | |||||
| MZL cluster | 17 | 0 | 11 | 0 | AKT1 | AF283829 |
| AGER | NM_173982 | |||||
| MXI1 | BC012907 | |||||
| T-cells cluster | 15 | 0 | 4 | 0 | CD3A | — |
| CD3G | X06026 | |||||
| Stroma cluster | 38 | 0 | 3 | 5 | SPARC | J03040 |
| MMP9 | J05079 | |||||
| Others | 1291 | 14 | 27 | 49 | — |
Gene-expression variable . | No. of microarray features in signature n = 2532 . | No. of microarray features significantly associated (P < .00001) with diagnosis of SLL n = 178 . | No. of microarray features significantly associated (P < .00001) with diagnosis of MZL n = 83 . | No. of microarray features significantly associated (P < .00001) with diagnosis of MCL n = 154 . | Representative genes . | GenBank accession no. . |
|---|---|---|---|---|---|---|
| SLL cluster | 735 | 163 | 1 | 12 | L-selectin | AJ246000 |
| IL4-R | C75499 | |||||
| TNF-R17 | ||||||
| Cyclin D1 cluster | 13 | 0 | 0 | 13 | CCND1 | NM 053056 |
| MCL cluster | 349 | 1 | 1 | 75 | PCNA | AF508260 |
| CDK4 | NM_007671 | |||||
| MYBL2 | ||||||
| Early response cluster | 20 | 0 | 0 | 0 | Fos | V01512 |
| Spleen cluster | 54 | 0 | 36 | 0 | HBZ | J00182 |
| KLF1 | NM_006563 | |||||
| EPB41L2 | M_001431 | |||||
| MZL cluster | 17 | 0 | 11 | 0 | AKT1 | AF283829 |
| AGER | NM_173982 | |||||
| MXI1 | BC012907 | |||||
| T-cells cluster | 15 | 0 | 4 | 0 | CD3A | — |
| CD3G | X06026 | |||||
| Stroma cluster | 38 | 0 | 3 | 5 | SPARC | J03040 |
| MMP9 | J05079 | |||||
| Others | 1291 | 14 | 27 | 49 | — |
—indicates no unique accession number.
SLL discriminating genes grouped 178 genes related to cell adhesion (P- and L-selectin, integrin α5, LAMR1, COL4A4, COL18A1, CCR2, MIF), to angiogenesis (angiopoietin-like 3, FIGF), and to cell fate (notch 4, IL4R, TNFR17). Several of these genes (BCL2, survivin, and TNFR10) were related to inhibition of apoptosis, a major feature in CLL/SLL. Several genes related to the cell cycle such as cyclin H and cyclin D2 and D3, RB1, ELF1, and TGFB1&2 were also overexpressed.
MCL discriminating genes regrouped 154 genes. Most of these genes could be related to 3 functions: cell proliferation, gene transcription, and drug resistance. Cell proliferation genes represented the most important group of genes including PCNA, CDK4, cyclins F, DNMT1, CDC46, MYBL2, and topoisomerase A2. Genes related to transcription activity were TFAP2A, TCF3, SRF, NFE2L3, SMARCA4. Several genes related to drug resistance were overexpressed in MCL: GST pi and 2 MDR/ATP-binding cassette (ABC) membrane proteins (ABCG2 and ABCC5). All MCL samples overexpressed cyclin D1 cluster independently from the other specific MCL genes.
Splenic MZL discriminating genes regrouped 83 genes from 2 clusters. One of the 2 clusters was specifically associated with the MZL signature, whatever the site of the biopsy (spleen, node, or blood). These genes were involved in intracellular signaling (AKT1, AGER) or transcription (TFCP2, MXI1). Several S100A proteins (S100 A8 and A9 [caligranulin a and b]) positively discriminated the MZL samples. The second splenic MZL signature corresponded to the “SPLEEN” cluster and was related to genes expressed by cells others than lymphoma B-cells such as red cells (eg, hemoglobin zeta) and T cells, considered as contaminant cells interfering with the MZL signature. Genes belonging to this cluster were excluded in the final diagnostic predictor. This led to a total of 47 MZL discriminating genes, and these genes shared weaker discriminating scores than SLL or MCL discriminating genes. For example, the top 100 discriminating scores contained only 6 genes discriminating MZL samples.
In order to create a multiclass predictor, we used a method derived from Sorlie et al.19 This method first selects genes from subtype-specific clusters to generate the predictor. Second, the inclusion of a gene in a given class is determined by the correlation between individual samples and subtype mean profiles. It allowed a balanced selection of genes discriminating between lymphoma subtypes, and provided a sample prediction in more than 2 classes. Using this strategy, a minimal set of 44 genes from the SLL (16 genes), MCL (15 genes), MZL (9 genes), and CCND1 (4 genes) clusters were selected (Figure 3B). This set of genes included most of the best discriminating genes for the 3 lymphoma subtypes, and selection of larger sets of genes—up to 115 genes—gave very similar results (data not shown).
Partition for SLL, MZL, MCL, and borderline samples using the 44-genes predictor. (A) Preliminary group samples were correlated to SLL, MZL, and MCL mean profiles. The horizontal axis represents correlations with the MCL mean profile (RMCL) and the vertical axis represents correlations with the SLL mean profile (RSLL). The SLL samples are represented in orange, the splenic MZL samples in blue, and the MCL samples in pink. Cases reclassified as borderline cases are represented in brown. Three of the 6 cases were hybridized twice. Each axis represents the positive value of one subtype. (B) Gene-expression data of the 44 selected genes used for the diagnostic predictor. All samples, the preliminary group, and the validation group samples were clustered according to the expression data of the 44 genes of the diagnostic predictor. Validation group samples were blind analyzed and are marked with a black dot. Note the small branch on the left side of the dendogram containing 4 unpredictable samples. See Table S2 for gene names.
Partition for SLL, MZL, MCL, and borderline samples using the 44-genes predictor. (A) Preliminary group samples were correlated to SLL, MZL, and MCL mean profiles. The horizontal axis represents correlations with the MCL mean profile (RMCL) and the vertical axis represents correlations with the SLL mean profile (RSLL). The SLL samples are represented in orange, the splenic MZL samples in blue, and the MCL samples in pink. Cases reclassified as borderline cases are represented in brown. Three of the 6 cases were hybridized twice. Each axis represents the positive value of one subtype. (B) Gene-expression data of the 44 selected genes used for the diagnostic predictor. All samples, the preliminary group, and the validation group samples were clustered according to the expression data of the 44 genes of the diagnostic predictor. Validation group samples were blind analyzed and are marked with a black dot. Note the small branch on the left side of the dendogram containing 4 unpredictable samples. See Table S2 for gene names.
Sample classification using the gene-expression-based diagnostic predictor
Correlations of individual samples with MCL and SLL mean profiles are shown in Figure 3A. The 19 SLL samples and the 27 splenic MZL samples formed homogeneous groups which were well separated from each other. Most of the MCL samples grouped together, but 3 of them shared MZL profiles and 2 samples could not be classified (see the 2 pink squares in the middle of the figure). All the borderline cases could be easily partitioned.
The validation group was blind analyzed. All the 10 SLL cases, 5 of the 8 MZL cases, and 8 of the 11 MCL cases were correctly classified in their respective entity. Two MCL samples exhibited an MZL gene-expression profile that ranked them with a predominant MZL score. Survival of these 5 misclassified MCL cases (3 in the preliminary group and 2 in the validation group) was not significantly different from that of the other patients with MCL although the median survival was more than 4.4 years (range, 2.3-5.6 years) compared with 2.6 years (range, 0.17-11.0 years) in the other MCL cases. Three MZL samples and one MCL sample in the validation group exhibited a profile that did not allow classification (Figure 3B). In these cases, hybridization signals were low, with gene-expression values that were not discriminant enough to determine a class prediction score.
The 6 borderline cases were analyzed separately. The 4 patients with borderline SLL-MZL clustered clearly within the MZL group and not in the SLL group. Among the 2 patients with borderline MCL-MZL, one clustered within the MCL group and the other within the MZL group. These data further demonstrated the interest for our genomic diagnostic-predictor for classifying borderline cases.
Patients with typical morphology but with presence of atypical cytogenetic or immunologic characteristics all fell into the appropriate category. Samples of the 2 patients with MZL with t(11;14) translocation, unmutated IgVH status, and cyclin D1 overexpression expressed the MZL hallmark genes such as AKT1, AGER, S100A proteins, and CCDN1 cluster genes but not the MCL hallmark genes such as proliferation genes or drug resistance genes signatures. In the 2-dimensional representation, these 2 patients were located inside the MZL group. The 5 splenic MZL cases expressing the CD5 marker and the 3 MCL cases lacking the CD5 marker expressed the expected signatures corresponding to their subtype and were in their correct group. The 2 nodal MZL cases were located inside the MZL group with high expression of the MZL gene signature, independently from the type of tissue biopsy. The 8 MCL cases with mutated IgVH presented a typical MCL profile. All MZL, SLL, and MCL samples presenting either mutated or unmutated IgVH status were grouped according to their histologic subtype.
Immunohistochemistry results
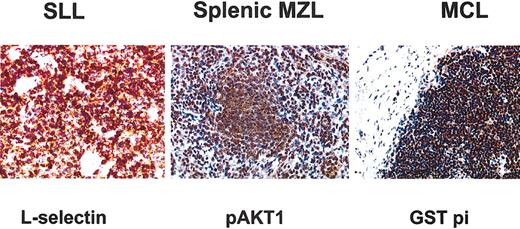
We confirmed at the protein level that L-selectin was closely associated to SLL, AKT1 to MZL, and GST pi to MCL (Figure 4). L-selectin immunostaining showed a strong signal in all 5 SLL cases whereas no expression was detected in MZL and MCL cases. The 4 MZL/SLL borderline cases were all negative for L-selectin. GST pi immunostaining displayed a very strong signal in MCL samples whereas no expression was detected in MZL and detection in SLL was very weak as expected. Among the 2 MZL cases with t(11;14) one exhibited no GST pi expression whereas the other displayed very weak expression. The immnunohistochemistry results of the 6 borderline cases were consistent with their genomic profile (data not shown). AKT1 immunostaining displayed a strong signal in MZL samples.
Immunohistochemical staining for the 3 antibodies chosen for their specificity from the microarray data. GST pi immunostaining showed a strong signal in all cyclin D1-positive MCL samples. L-selectin protein expression was specifically expressed in SLL samples. pAKT1 immunostaining showed a strong signal in MZL samples. Original magnification, × 250.
Immunohistochemical staining for the 3 antibodies chosen for their specificity from the microarray data. GST pi immunostaining showed a strong signal in all cyclin D1-positive MCL samples. L-selectin protein expression was specifically expressed in SLL samples. pAKT1 immunostaining showed a strong signal in MZL samples. Original magnification, × 250.
Discussion
SLL, splenic MZL, and MCL lymphomas display specific genomic signatures
The recent development of DNA microarrays provides an opportunity to take the genome-wide approach to extend current biologic insights into diseases. Using this approach, we were able to assign specific gene-expression signatures to non-germinal center small B-cell lymphomas, SLL, splenic MZL, and MCL. The results reflected the involvement of different biologic pathways.
The major oncogenic dysregulation that we observed in SLL in comparison to the 2 other lymphoma subtypes was the altered level of expression of genes whose function is related to induce resistance to apoptosis: bcl-2, already known as the major CLL dysregulation,21 but also overexpression of other molecules such as survivin and TNFR10 known to have an antiapoptotic function. Bax expression was deregulated, which is consistent with what has been reported, bax expression being related to bcl2 expression for the regulation of B-CLL cell survival.22 In addition to genes preventing cell death, an overexpression of several genes encoding cell cycle proteins such as cyclin D2, D3, and cyclin H was found in these low-rate proliferating cells. The IL4 receptor pathway was overexpressed in all SLL samples as has been reported in CLL analysis.8 IL4 receptor is the ligand for IL4 that promotes B-cell growth and survival via transcription nuclear factor (NF)-kappaB.23 IL4 receptor stimulation is also known to induce CD23 expression24 which is a marker of the SLL/CLL cells.
Comparison of gene-expression profiles between B-CLL reported in several studies6,8,25 and SLL in our study showed that patients with B-CLL and patients with SLL shared a common gene signature, including L-selectin and P-selectin, titin, IL4 receptor, CCR, adenylate kinase, diacylglycerol kinase, cyclin D2, and bcl-2 overexpression. This suggests a common normal cell precursor, suggested by others to be related to memory B cells.6,8
Splenic marginal zone B-cell lymphomas were specifically characterized by the overexpression of a gene cluster containing AKT1, which is localized in a hot spot region for chromosomal abnormalities in lymphomas: 14q32.3. AKT1 is a serine threonine kinase and belongs to a major cell survival pathway, the PI 3-kinase/AKT pathway that regulates survival of many cells by inhibiting the action of certain proapoptotic proteins (BAD, FKHR, caspase 9).26 In the same gene cluster, the analysis showed S100A proteins and AGER grouped together with the same overexpression among the MZL samples. S100 proteins belong to a group of EF-hand calcium-binding proteins and are polypeptides present at sites of inflammation. Expression of S100A11 has been reported to be elevated in colorectal cancer compared with normal colorectal mucosa27 , and in breast cancer-derived metastatic axillar lymph nodes compared with normal lymph nodes or breast fibroadenomas.28 It has been demonstrated that AKT1 is involved in the regulation of S100A protein expression.29 AGER (advanced glycosylation end product-specific receptor) protein, or RAGE, is a multiligand member of the immunoglobulin superfamily,30 and particularly a central receptor for the S100/calgranulin superfamily.31 Engagement of RAGE by a ligand activates key cell signaling pathways, such as p21(ras), MAP kinases, NF-kappaB, and cdc42/rac.32,33 Expression of several other genes related to transcription were significantly correlated in this AKT1 cluster, such as ZFP36 which codes for a zinc finger protein related with TNFα induced-inflammatory process, MXI1, a max-interacting protein, and TFCP2, a transcription factor for CP2 interacting with the alpha-globin gene promotor.34
Comparison of the MCL-specific signature to SLL and MZL signatures showed high levels of expression of cell cycle progression genes that function in the G1 phase, such as cyclin D1 and CDK4, as well as in the G2/M phase, such as cyclin F, or in both, such as PCNA, responsible for controlling the transition from G1 phase to the S phase and from the G2 phase to the M phase. This deregulation of several genes that are involved at different points of the cell cycle is in accordance with studies reporting that overexpressed cyclin D1 in MCL cooperates with other regulatory cell cycle genes.7,35,36 Moreover, when considering MCL analysis, clustering resulted in the separation of 2 signatures: the cyclin D1 signature and a signature including all the other MCL genes. This suggests that, if cyclin D1 overexpression is important and considered as characteristic of MCL, cyclin D1 overexpression is not sufficient alone to provide an MCL phenotype and that the control of cyclin D1 expression is independent from that of other MCL-specific genes. This result is in accordance with the existence of cyclin-D1-negative MCL recently reported.7 In this case, entry into S phase is suggested to depend on the overexpression of other cyclins such as cyclin D2 or cyclin D3.7 We observed in every case that gene-expression-increased cell cycle activity was correlated to an overexpression of genes encoding for protein machinery (actin beta and gamma genes; TUBB2, beta 2-tubulin gene; DDBN1 or Debrin1, an actin-binding protein involved in the regulation of the growth process; BMP4 or bone morphogenetic protein 4, SMARCA 4 an actin-dependent regulator of chromatin) and, in parallel, genes encoding for cell proliferation (FYN, ABL, PTK, and kit encoding for tyrosine kinase proteins). Interestingly, several genes encoding for membrane transporters and implicated in drug resistance were found to be overexpressed: 2 GST pi and 2 MDR/ATP-binding cassette (ABC) membrane proteins (ABCG2 and ABCC5). The gene GST pi is located on chromosome 11,37 very close to Bcl1, and has been suggested38 to be rearranged within the t(11;14) translocation characterizing MCLs.39 This overexpression of several genes related to multidrug resistance is consistent with the known MCL clinical evolution which is marked by treatment failure leading to fatal recurrence with a median survival close to 3 years.40
Genomic profiling and clinical implications for SLL, splenic MZL, and MCL
Our study allowed us to distinguish with great precision the SLL, splenic MZL, and MCL entities with 44 genes grouped into 4 gene-expression signatures. We were able to classify each patient by using a score that allowed us to perform a genomic prediction of the diagnosis.
Several samples in our series were either borderline cases, which occur in routine practice,10,41 or cases exhibiting atypical immunophenotypic or cytogenetic characteristics. The analysis of the 6 borderline cases displayed specific MZL or MCL genomic profiles without any ambiguity that were consistent with the clinical outcome of the patients. Histologic immunostaining using the 3 selected antibodies (GST pi, L-selectin analysis) showed consistent results with the respective genomic score obtained in these borderline cases.
The translocation t(11;14)(q13;q32) has been described in about 15% of patients with splenic MZL.42-44 This feature is considered to be characteristic of MCL although it can also be seen in plasma-cell leukemia, multiple myeloma, hairy cell leukemias, and B-prolymphocytic leukemia,45 and is missing in MCL.7,46 We have demonstrated that the cyclin D1 signature was a feature independent from the MCL signature. Interestingly, and confirming these data, the 2 splenic MZL samples exhibiting a t(11,14)(q13;q32) translocation shared a common genomic profile with that of the typical MZL samples. None of the genes specific for the MCL signature except cyclin D1 were expressed in these 2 MZL samples. Furthermore, expression values of GST pi transcript whose gene is located near the cyclin D1 locus were not as elevated as GST pi expression values in the MCL samples (data not shown), as confirmed by the lack or weak expression of GST pi protein in these samples. This supports the hypothesis of different bcl-1 breakpoints in MCL and in MZL.44,47 The existence of a t(11;14) translocation in MZL does not seem to induce a more aggressive clinical course (Troussard et al44 and C. T., personal data, February 2003) supporting the hypothesis that occurrence of t(11,14) in MZL should not suffice to alter diagnostic classification or to consider these t(11;14)-positive patients with MZL as a distinct lymphoma group.
The development of a specific microarray with 44 genes could be proposed as a supplementary tool in routine practice. Our results showed that 4 samples (3%) were not classified using the genomic predictor because of poor quality of RNA. In clinical practice, obtaining high-quality RNA for all samples is difficult. Investigations at the protein level could be an easier way. The use of supplementary markers, such as the 3 selected in our study—after confirmation of the presented preliminary results—in addition to the classical immunostaining panel for B-cell lymphomas diagnosis might be considered in pathology laboratories, in order to improve the diagnosis of B-cell lymphomas.
In conclusion, molecular analysis of malignancies using gene-expression analysis provide substantial information: non-germinal center small B-cell malignancies result from an accumulation of oncogenic alterations that alter the homeostatic control of B-cell proliferation, apoptosis, and differentiation via different biologic pathways which are specific for each disease. These differences can have direct clinical implications such as helping to classify the borderline cases. It may also help to better understand these diseases and provide specific potential targets for therapy, as it has been demonstrated for diffuse large B-cell lymphomas.48
Prepublished online as Blood First Edition Paper, November 20, 2003; DOI 10.1182/blood-2003-06-2160.
Supported by the Ligue Contre le Cancer: Départements du Rhône, de la Saône et Loire, de la Drôme et de l'Ardèche. V.N. was supported by the Association pour la Recherche sur le Cancer and the Paoli-Calmettes Institut.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr M. A. Shipp and Dr R. D. Gascoyne for helpful discussions, and Dr C. Dumontet for critical review of the manuscript. We thank Nadege Granger, Claudine Barrios, Karine Castellano, and Frédérique Rebouillat for their excellent technical help.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal