Abstract
The MEK1,2 (MAPK/ERK kinase 1 and 2) pathway mediates the up-regulation of plasminogen activator inhibitor-1 (PAI-1) expression in vascular smooth muscle cells by a variety of hormones, including angiotensin II. Transfection of constitutively active MEKK-1, an upstream activator of the mitogen-activated protein (MAP) kinase pathways, was used to isolate an enhancer element located between -89 and -50 bp in PAI-1 promoter that was activated by MEKK-1 and selectively blocked by the MEK1,2 inhibitor PD98059. Mutational analysis revealed that the MEKK-1 response element (MRE) contained 2 cis-acting Sp1- and AP-1—like sequences, located between -75 to -70 and -63 to -52 bp, respectively. Overexpression of Sp1 enhanced MEKK-1—induced MRE promoter activity and a dominant-negative c-Fos blocked this Sp1 response. The combination of Sp1 and c-Jun or c-Fos was required to activate this MRE. Angiotensin II (Ang II) stimulation increased c-Fos, c-Jun, and Sp1 binding to the MRE by 100-, 4.9-, and 1.9-fold, respectively, and these responses were inhibited by PD98059 and AT1 receptor antagonist candesartan. Intravenous Ang II infusion in rats increased aortic c-Fos binding to the MRE. This MRE sequence mediated a 4-fold increase of MEK1,2-dependent PAI-1/luciferase mRNA expression by angiotensin II stimulation. This report identifies the MEK1,2 response element that mediates angiotensin II—stimulated PAI-1 promoter activation and shows that activation of this element requires Sp1 and AP-1 co-activation.
Introduction
Plasminogen activator inhibitor-1 (PAI-1) is the major regulator of both tissue and urokinase plasminogen activators.1,2 Inhibition of plasminogen activation by PAI-1 reduces the generation of plasmin and thereby impairs fibrinolysis. In addition, PAI-1 has also been suggested to reduce plasmin-mediated activation of matrix metalloproteases3 and compete with cellular αvβ3 integrin matrix interactions, which contribute to cellular migration.4 Overexpression of PAI-1 in transgenic mice or by adenovirus-mediated gene transfer increases thrombosis,5,6 and PAI-1 deficiency enhances fibrinolysis and neointimal remodeling.7,8 Increased levels of PAI-1 have been demonstrated in vascular lesions induced by balloon catheter injury and atherosclerosis.9-12 Within these vascular lesions, PAI-1 expression is prominently increased in neointimal smooth muscle cells.9,13 In previous animal studies using angiotensin-converting enzyme inhibition, angiotensin type 1 receptor antagonism and systemic angiotensin II (Ang II) infusion, our group and others have demonstrated that the Ang II/AT1 pathway plays an important role in regulating vascular PAI-1 gene expression in vivo.9,14-16
A host of agonists, including Ang II, transforming growth factor-β (TGF-β), very low density lipoprotein (VLDL), and phorbol ester, require the activity of MAPK/ERK kinase1 and 2 (MEK1,2) to induce PAI-1 mRNA expression.17-20 Although the MEK/ERK pathway plays a key role in controlling PAI-1 mRNA levels, the mechanism(s) that mediates the effects of MEK1,2 on PAI-1 transcription have not been identified. Previous reports have demonstrated that the MEK/ERK pathway can affect phosphorylation, nuclear transport, and transactivation of a number of transcription factors, including Elk-1, Sma- and Mad-related protein (SMAD), c-Jun, Sp1, and signal transducer and activator of transcription 3 (STAT3).21-25 In addition, Ras/MEK/ERK activation can lead to increased expression of intermediate-early response genes, such as c-fos, egr-1, and junB.26 Although the promoter of the PAI-1 gene contains an array of potential response elements for these transcription factors,27-29 the contributions of these transcriptional elements to the effects of MEK1,2 on the PAI-1 promoter activity have not yet been elucidated.
Given the importance of PAI-1 in vascular physiology and its possible contribution to vasculopathies, further understanding of how the MAP kinase pathways affect PAI-1 expression in vascular smooth muscle cells (VSMCs) is of considerable importance. In this study, we identified and characterized the MEK1,2 response element in the 5′ flanking region of PAI-1 gene in VSMCs. These studies identified 2 adjacent cis-acting AP-1—like and Sp1-like sequences that functionally cooperate in MEK-induced PAI-1 promoter activation. These results establish a mechanistic link between the MEK/ERK pathway and the activation of the PAI-1 promoter.
Materials and methods
Materials
Dominant-negative AP-1 (A-Fos), and CREB (CRE binding protein; A-CREB) expression vectors were provided by Dr Charles Vinson (National Cancer Institute, Bethesda, MD).30,31 pCMV-Sp1 was a gift from Dr Jonathan Horowitz (North Carolina State University, Raleigh, NC). pCMV—c-Jun was provided by Dr Tom Curran (St Jude Children's Research Hospital, Memphis, TN). pRSV—c-Fos was a gift from Dr Ronald Law (University of California at Los Angeles, CA). Plasmids for constructing adenovirus were gifts from Dr George King (Joslin Diabetes Center, Boston, MA). cDNA of constitutively active MEKK-1 was obtained from Stratagene (La Jolla, CA). pAP-1(PMA)-TA-Luc was purchased from Clontech Laboratories (Palo Alto, CA). PD98059, SP600125, SB203580, and Y27632 were purchased from Calbiochem (La Jolla, CA). Angiotensin II was purchased from Sigma-Aldrich (St Louis, MO). Candesartan CV 11974 was kindly provided by Dr Peter Morsing (Astra Hassle AB, Mölndal, Sweden).
Cell culture
Vascular smooth muscle cells were isolated from aortic explants from Sprague-Dawley rats, cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) as described previously.32
RNA isolation and Northern blot analysis
Cells were serum starved in medium containing 0.1% bovine serum albumin (BSA) overnight and then stimulated with 100 nM Ang II for 3 hours. Total RNA was isolated using TRI REAGENT (Molecular Research Center, Cincinnati, OH). RNA was separated by agarose gel electrophoresis and expressions of PAI-1; luciferase and acidic ribosomal phosphoprotein (36B4) mRNA levels were determined by Northern analysis, as described previously.32 Levels of mRNA expression were visualized and quantitated by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, CA).
Plasmid and adenovirus construction
Rat PAI-1 promoter -1800, -764 were isolated from rPAI-CAT(-1800) and rPAI-CAT(-764), kindly provided by Drs Thomas Gelehrter and Carolyn Bruzdzinski (University of Michigan, Ann Arbor),27 by SalI and BglII digestion, and then ligated into XhoI and BglII-digested pGL3-enhancer (Promega, Madison, WI). Obtained pGL3-en-PAI-1(-764) (PAI-1(-764)/Luc) was truncated by exonuclease III digestion (New England Biolab, Beverly, MA) to generate pGL3-en-PAI-1(-219) (PAI-1(-219)/Luc) and pGL3-en-PAI-1(-22) (PAI-1(-22)/Luc). To generate PAI-1(-88)/Luc, PAI-1(-65)/Luc, PAI-1(-65M)/Luc, and PAI-1(-50)/Luc, polymerase chain reactions using Taq polymerase (Promega), designated synthetic oligonucleotides (Invitrogen), and PAI-1(-764)/Luc as template were performed. Obtained fragments were digested by MluI and BglII and ligated into pGL3-enhancer digested by the same enzymes. Annealed synthetic oligonucleotide of MEKK-1 response element (MRE) was digested by MluI and SmaI and ligated into pGL3-promoter (Promega) digested by the same enzymes to generate pGL3-p-MRE (MRE/Luc) in the same orientation as wild-type PAI-1 promoter. MRE was repeatedly inserted into MRE/Luc in the same direction to generate (MRE)2/Luc and (MRE)4/Luc. A series of mutations was also constructed as for (MRE)2/Luc by using synthetic oligonucleotides containing trinucleotide substitutions of MRE (Invitrogen). All constructs were confirmed by sequencing (DNA Core Facility, Joslin Diabetes Center, Boston, MA). To generate adenoviruses, the MluI (5′)to BamHI (3′) fragments of PAI-1(-88)/Luc and PAI-1(-50)/Luc, and the NheI (5′) to BamHI (3′) fragments of PAI-1(-764)/Luc and the control vector, pGL3-enhancer, were inserted into pAdTrack and followed the instruction of AdEasy System.33
Reporter gene assay
VSMCs at about 70% confluence were starved in OPTI-MEM medium (Invitrogen) for overnight. Cells were then transfected with plasmids using Lipofectamine or Lipofectamine 2000 (Invitrogen) and harvested 24 hours after the start of transfection. Luciferase activities were assayed by using Dual Luciferase Reporter Assay System (Promega) and adjusted for transfection efficiency by normalizing firefly luciferase activities to the Renilla luciferase activities generated by cotransfected pRL-TK (Promega).
Nuclear protein extraction, pull-down assay, and coimmunoprecipitation
Confluent VSMCs were serum starved overnight and then stimulated with 100 nM Ang II. Aortas were collected from male Sprague-Dawley rats with 1-hour intrajugular vein infusion of saline or Ang II (100 ng/kg/min), as described previously.14 Nuclear protein extraction was performed as previously described34 for Sp1—AP-1 association assay, or using Nuclear Extract Kit (Active Motif, Carlsbad, CA) for pull-down assay. Double-stranded biotin-labeled MRE oligonucleotides (wild-type, 5′CAGAGTTAGAAGGTGGGTGGGGCTGGAACATGAGTTCATCTA3′; mutant, 5′CAGAGTTAGAAGGTGGGAGTCAGTGGAACAGTCAGTCAGTCA3′) (Invitrogen) bound to Streptavidin-agarose beads (Pierce, Rockford, IL) were incubated with 25 μg nuclear extract in gel shift binding buffer (Promega). The beads were then washed 3 times and boiled in sample buffer. Bound proteins were assayed by Western blot analysis with the use of anti—c-Fos, anti—c-Jun, and anti-Sp1 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). For coimmunoprecipitation, 200 μg nuclear extracts were immunoprecipitated with anti-Sp1 antibody-conjugated agarose (Santa Cruz Biotechnology) in 300 μL immunoprecipitation buffer as previously described except no NaCl was used.35 Sp1 and c-Jun content in immunoprecipitates were then quantitated by Western blot analysis.
Statistics
All statistical analyses were performed by one-way analysis of variance (ANOVA) using SigmaStat (SPSS, Chicago, IL). P values less than .05 were considered significantly different.
Results
Isolation of a MEK1,2 response element on the 5′ flanking region of PAI-1 gene
The roles of MEK1,2 and p38 MAP kinase in Ang II—induced PAI-1 expression in VSMCs were examined by using the MEK1,2 inhibitor PD98059 and the p38 inhibitor SB203580. Ang II stimulated an 11-fold increase in PAI-1 mRNA expression, and PD98059 reduced this Ang II response by 78% (P < .001). In contrast, pretreatment of cells with SB203580 did not inhibit Ang II—stimulated PAI-1 expression but caused a small increase in the effect of Ang II (P < .05) (data not shown). Neither PD98095 nor SB203580 had a significant effect on basal PAI-1 expression. These results show that the MEK1,2 pathway is required for the majority of Ang II—induced increase of PAI-1 mRNA expression.
MEK1,2 are the major MAP kinase kinases of the ERK-1, -2 pathway.36 MEK1,2 are activated by both the Ras/Raf-1 pathway and the MAP kinase kinase kinase-1 (MEKK-1) pathway.37,38 In addition to the MEK/ERK pathway, MEKK-1 also activates stress MAP kinase pathways by phosphorylating MKK4 and MKK7, which leads to the activation of the c-Jun N-terminal kinase (JNK) pathway39,40 and by phosphorylating MKK3 and MKK6, which leads to p38 MAP kinase activation.41 To examine the involvement of these MAP kinase pathways on PAI-1 promoter activity, the effects of upstream activation of these pathways using a constitutively active MAP kinase kinase kinase-1 (MEKK-1*) on PAI-1 promoter/luciferase reporter gene constructs were investigated.
A series of 5′ deletion of PAI-1 promoter/Luc constructs were generated to identify PAI-1 promoter sequence(s) that mediates PD98059-sensitive MEKK-1 response. MEKK-1* stimulated a 4- to 5-fold increase of promoter activity of PAI-1(-1800)/Luc, PAI-1(-764)/Luc, PAI-1(-219)/Luc, and PAI-1(-88)/Luc (P < .001) (Figure 1). Lower basal and MEKK-1—induced promoter activity was observed in PAI-1(-1800)/Luc compared with PAI-1(-764)/Luc, suggesting that the region between -1800 and -764 may contain a repressor element. Although PAI-1(-764)/Luc, PAI-1(-219)/Luc, and PAI-1(-88)/Luc displayed similar responses, further deletion of the 5′ PAI-1 promoter reduced its response to MEKK-1. The MEKK-1—stimulated promoter activities of PAI-1(-65)/Luc and PAI-1(-50)/Luc constructs were reduced by 70% and 90%, respectively, compared with the response from PAI-1(-88)/Luc. Further truncation of the PAI-1 promoter to -22 completely abolished the MEKK-1 response. These data indicated the major MEKK-1 response element encompasses a region between -89 and -50 of PAI-1 promoter. In addition, PD98059 caused a 93% decrease of both MEKK-1—stimulated PAI-1(-764) and PAI-1(-219) promoter activities (P < .001). Treatment of SB203580 decreased PAI-1/Luc activities of both constructs by about 56% (P < .001). These data suggest that the -219 bp at 5′ region from the transcription start site of the PAI-1 gene contains both MEK1,2 and p38 sensitive MEKK-1 response elements.
Effects of constitutively active MEKK-1 on PAI-1 promoter activity in VSMCs. A series of deletional PAI-1 promoter/Luciferase constructs, PAI-1(-1800)/Luc, PAI-1(-764)/Luc, PAI-1(-219)/Luc, PAI-1(-88)/Luc, PAI-1(-65)/Luc, PAI-1(-50)/Luc, and PAI-1(-22)/Luc, were cotransfected into VSMCs with pCDNA3.1 blank vector or with MEKK-1*. To examine if the activities of MEK1,2 or p38 were required in MEKK-1 stimulation, cells were treated with 50 μM PD98059 (PD) or 5 μM SB203580 (SB) 6 hours after the start of transfection. Renilla luciferase activities of cotransfected pRL-TK were used to normalize transfection efficiency. All transfections were performed in triplicate, and bar graph shows results from 3 or more separate experiments.
Effects of constitutively active MEKK-1 on PAI-1 promoter activity in VSMCs. A series of deletional PAI-1 promoter/Luciferase constructs, PAI-1(-1800)/Luc, PAI-1(-764)/Luc, PAI-1(-219)/Luc, PAI-1(-88)/Luc, PAI-1(-65)/Luc, PAI-1(-50)/Luc, and PAI-1(-22)/Luc, were cotransfected into VSMCs with pCDNA3.1 blank vector or with MEKK-1*. To examine if the activities of MEK1,2 or p38 were required in MEKK-1 stimulation, cells were treated with 50 μM PD98059 (PD) or 5 μM SB203580 (SB) 6 hours after the start of transfection. Renilla luciferase activities of cotransfected pRL-TK were used to normalize transfection efficiency. All transfections were performed in triplicate, and bar graph shows results from 3 or more separate experiments.
To further investigate the enhancer properties of the MEKK-1 response element (MRE), the MEKK-1 stimulation of this isolated sequence was examined. The MRE sequence was inserted upstream of the SV40 promoter in the pGL3 vector to create an MRE/Luc construct. In addition, multimers of the MRE were also generated to form (MRE)2/Luc and (MRE)4/Luc reporter constructs, containing 2 and 4 copies of this MRE, respectively. The presence of MRE, (MRE)2, and (MRE)4 did not affect basal activity of SV40 promoter. However, MEKK-1 activation stimulated the MRE response in a copy number—dependent manner, increasing luciferase activity from 2.5-fold for MRE/Luc to 5.7-fold for (MRE)2/Luc, and 24-fold for (MRE)4/Luc (Figure 2). Because MEKK-1 is an upstream activator of MEK1,2, JNK, and p38 pathways, we further investigated which pathway was responsible for the activation of this region. Figure 2 shows that, although MEK1,2 inhibitor PD98059 abolished MEKK-1—stimulated (MRE)2/Luc activity and reduced (MRE)4/Luc activity by 74%, neither p38 inhibitor SB203580 nor JNK inhibitor SP600125 affected MEKK-1—induced promoter activity of both constructs. These results indicated that the region between -89 and -50 bp of PAI-1 promoter contains the response element(s) to MEK1,2.
MRE responds to MEK1,2-dependent MEKK-1 stimulation in a copy number—dependent manner. Isolated MRE region was inserted into pGL3 promoter to generate MRE/Luc, (MRE)2/Luc, and (MRE)4/Luc. These constructs were transfected into VSMCs with pCDNA3.1 blank vector (hatched bars) or MEKK-1* (solid bars). Transfected cells were treated with 50 μM PD98059 (PD), 5 μM SB203580 (SB), or 20 μM SP600125 (SP) 6 hours after the start of transfection. Cells were lysed 1 day after the start of transfection, and firefly luciferase activities were normalized to Renilla luciferase activities of cotransfected pRL-TK. Results show normalized luciferase activity (mean ± SEM) from at least 3 separate experiments. Fold increases induced by MEKK-1* compared with blank vector, pCDNA3.1, are shown. **Statistical significance with P < .001.
MRE responds to MEK1,2-dependent MEKK-1 stimulation in a copy number—dependent manner. Isolated MRE region was inserted into pGL3 promoter to generate MRE/Luc, (MRE)2/Luc, and (MRE)4/Luc. These constructs were transfected into VSMCs with pCDNA3.1 blank vector (hatched bars) or MEKK-1* (solid bars). Transfected cells were treated with 50 μM PD98059 (PD), 5 μM SB203580 (SB), or 20 μM SP600125 (SP) 6 hours after the start of transfection. Cells were lysed 1 day after the start of transfection, and firefly luciferase activities were normalized to Renilla luciferase activities of cotransfected pRL-TK. Results show normalized luciferase activity (mean ± SEM) from at least 3 separate experiments. Fold increases induced by MEKK-1* compared with blank vector, pCDNA3.1, are shown. **Statistical significance with P < .001.
Both Sp1-like and AP-1—like sequences contribute to MEKK-1 stimulation of the MRE
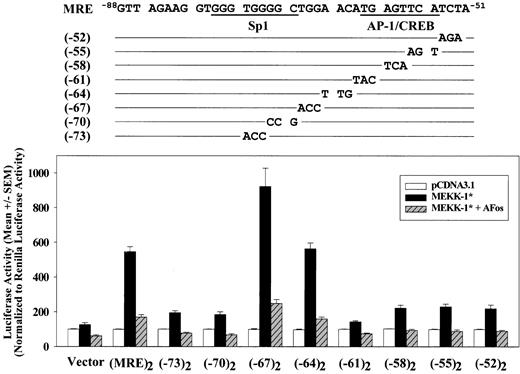
To further define the MRE region, a series of mutant constructs containing trinucleotide transversion substitutions in the region from -75 to -52 of both copies of MRE in (MRE)2/Luc were generated and their responses to MEKK-1 stimulation were studied. Compared with a 5.4-fold MEKK-1—induced increase of the promoter activity for wild-type (MRE)2/Luc, substitutions in the MRE sequence from -75 to -70 or -63 to -52 reduced MEKK-1 stimulation to 1- to 2-fold increase. In contrast, substitutions from -69 to -64 did not impair MRE activation by MEKK-1 (Figure 3). Because previous reports suggested that there is an AP-1—like site located in human PAI-1 promoter in a region analogous to the -62 to -55 region of the rat PAI-1 promoter,29,42,43 we also investigated the involvement of this AP-1—like site in MEKK-1 stimulation. To determine the role of the AP-1 in the native PAI-1 promoter, dinucleotide substitution (TGAGTTCA → TGAGTTTG) designed to disrupt this AP-1—like site in the MRE region within PAI-1(-65)/Luc construct was performed to generate PAI-1(-65M)/Luc. This substitution reduced MEKK-1—stimulated promoter activity to levels comparable to PAI-1(-50)/Luc (data not shown), suggesting that this AP-1—like element is required for the MEKK-1 activation of the sequence from -65 to -51 of PAI-1 promoter. To determine whether AP-1 binding protein was responsible for the MEKK-1 stimulation of this isolated MRE region, we examined the effects of a dominant-negative form of c-Fos, A-Fos, which binds and inactivates c-Jun,30 on MEKK-1 stimulation of these (MRE)2/Luc mutant constructs. A-Fos reduced MEKK-1—induced wild-type (MRE)2/Luc activation by 84% (Figure 3). In constructs with reduced response to MEKK-1 stimulation, coexpression of A-Fos completely blocked MEKK-1 stimulation. These results show that AP-1 is critical for MEKK-1 activation of this MRE. Moreover, mutations in an Sp1-like site (from -75 to -70) also impaired MRE activation. These results indicate that both Sp1-like and AP-1—like elements, separated by 7 base pairs, are required for MEKK-1—induced MRE activation.
Two separate regions in MRE contribute its response to MEKK-1 stimulation. A series of trinucleotide substitution were introduced into -75 to -52 region of both copies of MRE in (MRE)2/Luc, as shown in the upper panel. These mutant constructs were transfected into VSMCs with pCDNA3.1 blank vector or MEKK-1*. A dominant-negative AP-1, A-Fos, was also cotransfected with MEKK-1* to examine the contribution of AP-1. One day after the start of transfection, cells were harvested and luciferase activities were assayed. Luciferase activities were normalized to Renilla luciferase activities of cotransfected pRL-TK. Triplicate transfections were performed in each experiment, and the bar graph shows results from 3 or more separate experiments.
Two separate regions in MRE contribute its response to MEKK-1 stimulation. A series of trinucleotide substitution were introduced into -75 to -52 region of both copies of MRE in (MRE)2/Luc, as shown in the upper panel. These mutant constructs were transfected into VSMCs with pCDNA3.1 blank vector or MEKK-1*. A dominant-negative AP-1, A-Fos, was also cotransfected with MEKK-1* to examine the contribution of AP-1. One day after the start of transfection, cells were harvested and luciferase activities were assayed. Luciferase activities were normalized to Renilla luciferase activities of cotransfected pRL-TK. Triplicate transfections were performed in each experiment, and the bar graph shows results from 3 or more separate experiments.
The involvement of AP-1 and Sp1 binding proteins in MEKK-1—stimulated MRE activation was further examined by using the wild-type (MRE)2/Luc construct. Cotransfection of plasmids expressing Sp1, A-Fos, or control plasmid (pCMV) did not affect (MRE)2/Luc basal activity (Figure 4). However, cotransfection of these expression vectors with MEKK-1* revealed that the expression of Sp1 enhanced the MEKK-1 stimulation by 2.3-fold. Interestingly, the expression of dominant-negative AP-1, A-Fos, not only reduced 77% of MEKK-1 stimulation in the absence of Sp1 overexpression, but also completely inhibited this Sp1 enhancement of MEKK-1—induced MRE activation (Figure 4), suggesting that AP-1 is essential for Sp1 effects.
Dominant-negative AP-1 inhibits Sp1 enhancement of MEKK-1—stimulated MRE activation. (MRE)2/Luc was transfected into VSMCs with pCDNA3.1 blank vector or MEKK-1* in the presence of pCMV empty vector, Sp1, A-Fos, or Sp1 and A-Fos. Cells were lysed 1 day after the start of transfection and luciferase activities were assayed. Renilla luciferase activities of cotransfected pRL-TK were used to normalize transfection efficiency. All transfections were performed in triplicate, and the bar graph shows results from 3 separate experiments.
Dominant-negative AP-1 inhibits Sp1 enhancement of MEKK-1—stimulated MRE activation. (MRE)2/Luc was transfected into VSMCs with pCDNA3.1 blank vector or MEKK-1* in the presence of pCMV empty vector, Sp1, A-Fos, or Sp1 and A-Fos. Cells were lysed 1 day after the start of transfection and luciferase activities were assayed. Renilla luciferase activities of cotransfected pRL-TK were used to normalize transfection efficiency. All transfections were performed in triplicate, and the bar graph shows results from 3 separate experiments.
Sp1 and AP-1 cooperatively activate PAI-1 promoter
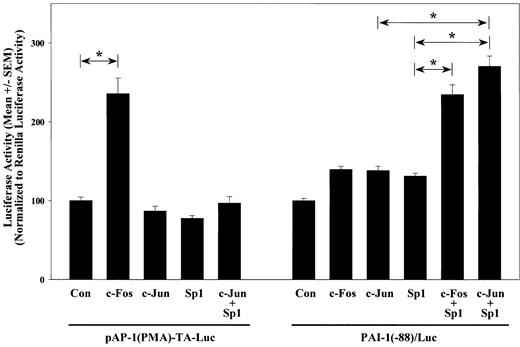
Because both the Sp1 site and the AP-1 site are required for full MRE activation and A-Fos—blocked Sp1 enhancement in MRE activation, the independent effects and potential interaction of AP-1 and Sp1 on the activation of PAI-1 promoter in VSMCs were investigated. Overexpressions of Sp1 and c-Jun or c-Fos were used to activate Sp1 and AP-1, respectively, and their effects on the activation of PAI-1(-88)/Luc were examined. For comparison, the effects of these transcription factors on pAP-1(PMA)-TA-Luc, which contains 6 tandem repeats of a putative AP-1 element, (TGACTAA)6, were also evaluated. Overexpression of c-Fos increased pAP-1(PMA)-TA-Luc activity by 2.4-fold (P < .05), whereas c-Jun and Sp1 had no effect. In contrast, overexpression of c-Fos, c-Jun, or Sp1 independently did not significantly increase PAI-1 promoter activity. However, coexpression of Sp1 with either c-Fos or c-Jun activated the PAI-1(-88)/Luc by 2.3- and 2.7-fold, respectively (P < .05) (Figure 5). On the contrary, coexpression of c-Jun and Sp1 had no effect on the AP-1 construct. These results show that this region of PAI-1 promoter does not function like a consensus AP-1. Instead, AP-1 transcription factors are required but not sufficient to induce PAI-1 promoter activity.
Sp1 and c-Jun cooperatively activate the PAI-1 promoter. VSMCs were transfected with PAI-1(-88)/Luc or pAP-1(PMA)-TA-Luc in the presence of pCDNA3.1 empty vector (Con), c-Fos, c-Jun, Sp1, or c-Jun and Sp1. Cells were lysed 1 day after the start of transfection, and luciferase activities were measured and normalized to Renilla luciferase activities of cotransfected pRL-TK. All transfections were performed in triplicate, and results from 3 or more separate experiments are shown in bar graph. *Statistical significance with P < .05 by ANOVA.
Sp1 and c-Jun cooperatively activate the PAI-1 promoter. VSMCs were transfected with PAI-1(-88)/Luc or pAP-1(PMA)-TA-Luc in the presence of pCDNA3.1 empty vector (Con), c-Fos, c-Jun, Sp1, or c-Jun and Sp1. Cells were lysed 1 day after the start of transfection, and luciferase activities were measured and normalized to Renilla luciferase activities of cotransfected pRL-TK. All transfections were performed in triplicate, and results from 3 or more separate experiments are shown in bar graph. *Statistical significance with P < .05 by ANOVA.
The cooperative effect of AP-1 and SP-1 on the PAI-1 promoter activation suggests that Sp1 and c-Jun/c-Fos might interact within the same transcriptional complex. Therefore, the physical association of Sp1 and c-Jun was investigated. Nuclear extracts from Ang II—stimulated VSMCs were immunoprecipitated with anti-Sp1 antibody. Western blotting revealed that Ang II stimulated a 3-fold increase of c-Jun—Sp1 association within 1 hour. The peak association was reached at an 11-fold increase 2 hours into the stimulation (P < .05), and reduced to 9-fold after 3 hours of induction (data not shown). These data show that Sp1 and c-Jun can associate in the absence of DNA.
The Sp1 and AP-1 cooperation also suggests that binding of both factors to the MRE is required to transactivate the PAI-1 promoter. Therefore, we investigated Sp1 and AP-1 transcription factor binding to the MRE region. Biotin-labeled double-stranded MRE oligonucleotide prebound to Streptavidin-agarose beads were used as bait to pull down binding protein from nuclear extracts of Ang II—stimulated VSMCs. Subsequent Western blotting revealed that Ang II—stimulated a 100-fold increase of c-Fos binding to the MRE in 1 hour (P < .001) (Figure 6A). Ang II stimulation of cells for 1 hour also induced the binding of c-Jun and Sp1 to MRE, 4.9-fold (P < .001) and 1.9-fold (P = .003), respectively (Figure 6B-C). In contrast, mutant MRE with both AP-1 and Sp1 binding sequences substituted (-63 to -52 and -75 to -70, respectively) did not bind c-Fos, c-Jun, or Sp1 (data not shown).
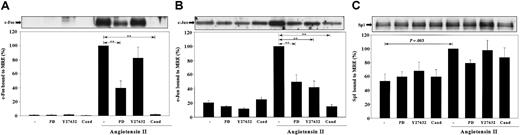
The effects of MEK1,2 and Rho-kinase inhibition, as well as AT1 receptor antagonism, on c-Fos, c-Jun, and Sp1 binding to the MRE. VSMCs were stimulated by Ang II for 1 hour with or without the pretreatment of 50 μM PD98059 (PD), 10 μM Y27632, or 1 μM candesartan (Cand). Nuclear proteins were extracted and incubated with biotinylated MRE prebound to Streptavidin-agarose beads. MRE-associated c-Fos (A), c-Jun (B), and Sp1 (C) was quantitated by Western blot analyses. Representative blots and bar graph quantitations from 3 or more separate experiments are shown. Statistical significance with P < .001 by ANOVA is marked as **.
The effects of MEK1,2 and Rho-kinase inhibition, as well as AT1 receptor antagonism, on c-Fos, c-Jun, and Sp1 binding to the MRE. VSMCs were stimulated by Ang II for 1 hour with or without the pretreatment of 50 μM PD98059 (PD), 10 μM Y27632, or 1 μM candesartan (Cand). Nuclear proteins were extracted and incubated with biotinylated MRE prebound to Streptavidin-agarose beads. MRE-associated c-Fos (A), c-Jun (B), and Sp1 (C) was quantitated by Western blot analyses. Representative blots and bar graph quantitations from 3 or more separate experiments are shown. Statistical significance with P < .001 by ANOVA is marked as **.
Because our group and others have shown that angiotensin AT1 antagonism by candesartan completely blocks Ang II—induced PAI-1 mRNA expression in rat VSMCs,14,17 the effect of candesartan on Ang II—stimulated AP-1 and Sp1 binding to the MRE was investigated. This study showed that candesartan completely blocked Ang II—induced c-Fos and c-Jun binding to the MRE (Figure 6A-B). In addition, Ang II stimulation in the presence of candesartan did not significantly increase Sp1 binding to the MRE compared with cells treated with candesartan alone (Figure 6C).
Because treatment of cells with the MEK1,2 inhibitor PD98059 reduced Ang II—stimulated PAI-1 mRNA expression by 78%, the roles of the MEK1,2 pathway in AP-1 and Sp1 binding to the MRE were investigated. Figure 6A-B shows that treatment of PD98059 reduced Ang II—induced c-Fos and c-Jun binding to the MRE by 60% (P < .001) and 50% (P < .001), respectively. In contrast, Ang II stimulation of PD98059-treated cells did not significantly increase Sp1 binding to the MRE compared with cells treated with PD98059 alone (Figure 6C). To further characterize the signaling pathways involved in effects of Ang II on the MRE, the role of the Rho-kinase pathway was investigated. The Rho-kinase activity has been previously shown to be required for Ang II—induced PAI-1 mRNA expression in rat VSMCs and in heart ventricle from Ang II—induced hypertensive rats.17,44 Figure 6B shows that treatment of cells with the Rho-kinase inhibitor Y27632 decreased c-Jun binding to the MRE by 58% (P < .001). However, Rho-kinase inhibition did not significantly reduce c-Fos or Sp1 binding to the MRE (Figure 6A-C).
Previously, it has been shown that intravenous Ang II infusion in rats increases aortic PAI-1 mRNA expression and that this response is mediated by way of the AT1 receptor.14,16 This experimental model was used to examine the in vivo effects of Ang II on AP-1 and Sp1 binding to the MRE. Nuclear proteins were extracted from aortas following intrajugular infusion of saline or Ang II (100 ng/kg/min) in Sprague-Dawley rats. c-Fos, c-Jun, and Sp1 bound to biotin-labeled double-stranded MRE oligonucleotide were isolated using Streptavidin and quantitated by Western blotting. Ang II infusion increased aortic c-Fos binding to the MRE by 17-fold (P = .017) compared with aortic nuclear extracts from saline-infused controls. Binding of aortic c-Jun and Sp1 to the MRE from Ang II—infused animals was increased by 1.7-fold and 1.1-fold, respectively, compared with saline-infused controls (data not shown).
Ang II—stimulated transcription activation of PAI-1 gene is mediated by MRE
To evaluate the functional importance of the MRE in PAI-1 mRNA expression, we studied the effects of deletional mutation of PAI-1/Luc reporter gene on its mRNA expression in response to Ang II stimulation in VSMCs. Adenoviral vectors containing PAI-1(-764)/Luc, PAI-1(-88)/Luc, PAI-1(-50)/Luc, and Luc without PAI-1 promoter sequence were constructed and used to increase the efficiency of gene transfer and provide an opportunity to directly measure PAI-1 transcriptional activity at the mRNA level by Northern blot analysis. VSMCs were infected with PAI-1/Luc-contained adenoviruses, serum-starved for 18 hours, and stimulated by Ang II in the presence or absence of PD98059. Northern blotting revealed that Ang II did not significantly activate PAI-1(-50)/Luc but increased luciferase mRNA expression of PAI-1(-88)/Luc and PAI-1(-764)/Luc by 4-fold (P < .001) (Figure 7). In comparison, Northern blotting for PAI-1 demonstrated that the responses of PAI-1/Luc constructs were comparable to those of the endogenous PAI-1 gene. Moreover, PD98059 pretreatment significantly reduced Ang II—stimulated luciferase mRNA of PAI-1(-88)/Luc and PAI-1(-764) by about 60% (P < .05 and P < .001, respectively). These results indicate that this MEK1,2 response element mediates PAI-1 response to Ang II stimulation.
MRE mediates Ang II—stimulated PAI-1 expression. VSMCs infected with adenoviruses containing PAI-1(-764)/Luc, PAI-1(-88)/Luc, PAI-1(-50)/Luc, as well as Luc without PAI-1 promoter (control), were starved and stimulated by Ang II for 3 hours with or without 50 μM PD98059 (PD) pretreatment. Northern blot analysis was performed by using a probe for luciferase and PAI-1, respectively. Representative blots of luciferase and PAI-1 mRNA expression and a bar graph quantitation of luciferase mRNA levels normalized to green fluorescence protein (GFP) mRNA of unstimulated cells from 3 separate experiments are shown. *P < .05 and **P < .001 by ANOVA.
MRE mediates Ang II—stimulated PAI-1 expression. VSMCs infected with adenoviruses containing PAI-1(-764)/Luc, PAI-1(-88)/Luc, PAI-1(-50)/Luc, as well as Luc without PAI-1 promoter (control), were starved and stimulated by Ang II for 3 hours with or without 50 μM PD98059 (PD) pretreatment. Northern blot analysis was performed by using a probe for luciferase and PAI-1, respectively. Representative blots of luciferase and PAI-1 mRNA expression and a bar graph quantitation of luciferase mRNA levels normalized to green fluorescence protein (GFP) mRNA of unstimulated cells from 3 separate experiments are shown. *P < .05 and **P < .001 by ANOVA.
Discussion
Although multiple lines of evidence suggest that the MEK/ERK pathway plays an important role in PAI-1 expression, the mechanism(s) and the PAI-1 promoter elements that mediate this response have not yet been defined. In this study, we isolate the MEK1,2 response element that is responsible for Ang II—stimulated PAI-1 promoter activation in VSMCs. The results showing that the MEK/ERK pathway mediates the up-regulation of PAI-1 expression by Ang II provide additional evidence for a critical role of this MAP kinase pathway in the regulation of PAI-1.17-19
By using a reporter gene approach, we identified an MEKK-1 response element (MRE) in the rat PAI-1 promoter, which is located from -88 to -51 bp relative to the transcription start site. A previous report has shown that activation of MEKK-1 can increase PAI-1 promoter activity in endothelial cells.45 Although a number of response elements have been identified in the region between -764 and -88,46-48 this region does not appear to play a critical role in the activation of the PAI-1 promoter by MEKK-1. Because the MEKK-1 activation of the MRE was completely blocked by MEK1,2 inhibition but was not affected by p38 or JNK inhibition, this sequence appears to selectively respond to stimulation from MEK1,2. This MRE responded to MEKK-1 both in the context of the wild-type PAI-1 promoter and when isolated and inserted upstream of an SV40 promoter. Moreover, the MRE responded to MEKK-1 in both a copy number—dependent manner and in the reverse orientation (data not shown). These results support its role as an MEK1,2 enhancer element.
Mutational analysis of this MRE revealed that there are 2 regions within this sequence that contribute to its response to MEKK-1. Results using a dinucleotide substitution of the AP-1—like site at -55 and -56 in PAI-1(-65)/Luc and a series of trinucleotide substitutions of the (MRE)2/Luc from -63 to -52 suggest that this AP-1—like sequence is required for the MEKK-1—induced PAI-1 expression. This sequence contains an AP-1—like palindrome of TGAGTTCA, but its 2 intervening bases deviate from the consensus AP-1 binding sequence (TGACTCA).49 Further evidence that an AP-1 element contributes to the MEKK-1 stimulation is that the AP-1 dominant-negative A-Fos inhibited MEKK-1—induced luciferase activities of (MRE)2/Luc (Figures 3, 4), PAI-1(-88)/Luc, and PAI-1 (-65)/Luc (data not shown). Although the region between -64 and -51 was originally described as a CRE (cyclic adenosine monophosphate [cAMP]—response element), due to its homology to a consensus CREB binding sequence (TGACGTCA),27 coexpression of a dominant-negative CREB (A-CREB)31 did not affect MEKK-1—stimulated PAI-1(-88)/Luc or PAI-1(-65)/Luc promoter activities (data not shown). The combination of results using substitution mutants and dominant-negative A-Fos demonstrates that this AP-1 site is critical for the activation of the PAI-1 promoter by MEKK-1 stimulation.
A second sequence in the MRE region, -78 to -70, also contributes to MEKK-1—induced MRE promoter activity. This sequence is GC rich, and the corresponding sequence on the human PAI-1 gene has been shown to bind Sp1.29,50-52 Comparison of the MEKK-1 induction of PAI-1(-88)/Luc and PAI-1(-65)/Luc indicated that an important MEKK-1 response component is located between -89 and -65. Trinucleotide substitution of the region from -75 to -70 impaired MEKK-1—stimulated activation of (MRE)2/Luc. A specific role of Sp1 in MEKK-1—induced MRE promoter activity is supported by results showing that coexpression of Sp1 enhances MEKK-1—stimulated MRE promoter activity. Previous reports also suggest that Sp1 binding to the sequence containing -78 to -69 of PAI-1 promoter contributes to PAI-1 expressions when stimulated by TGF-β, Ang II, glucose, glucosamine, and insulin.50,52-55
Roles for both the AP-1 and Sp1 elements in mediating Ang II—stimulated PAI-1 mRNA expression are consistent with previous reports using decoy oligodeoxynucleotides (ODNs) to compete for transcription factors binding. HVJ-liposome transfection of consensus AP-1 decoy ODN (TGAGTCA) into human vascular smooth muscle cells inhibits the combined effects of high glucose (22 mmol/L) and Ang II on endogenous PAI-1 mRNA expression and promoter activation of a -800-bp PAI-1/Luc reporter construct (p800/Luc) compared with cells maintained in low glucose (5.5 mmol/L).56 Because this AP-1 decoy ODN reduced PAI-1 expression was compared with high glucose treatment alone, it is likely that this AP-1 decoy ODN inhibited both the effect of Ang II and the effect induced by elevated extracellular glucose concentration. In this report, we identify an AP-1—like response element in the PAI-1 promoter within the MRE. This element binds c-Fos and c-Jun, and its activity is inhibited by dominant-negative c-Fos. However, unlike a consensus AP-1, activation of this AP-1—like element within the MRE requires both c-Fos or c-Jun and Sp1. Another report, also using a decoy ODN approach, has shown that transfection of a Sp1 (or Sp1-like) ODN inhibits Ang II—stimulated PAI-1 in mesangial cells.54 A critical role of Sp1 in PAI-1 expression is consistent with our findings that Sp1 binds to the MRE, mutation of the Sp1 sequence impairs MEKK-1—induced activation of the MRE, and overexpression of Sp1 enhances MEKK-1 activation of this element.
Our studies suggest that the Sp1 and AP-1 sites in the MRE cooperate in activating the PAI-1 promoter. This conclusion is supported by the findings that the enhancement of MEKK-1—induced (MRE)2 activity by coexpression of Sp1 was completely inhibited by A-Fos, a dominant-negative of AP-1 (Figure 4). Direct evidence for synergistic actions of AP-1 and Sp1 in the PAI-1 promoter is provided by results showing that overexpressions of c-Jun and Sp1 or c-Fos and Sp1 induced more luciferase activity from PAI-1(-88)/Luc than the additive effects of these transcription factors when provided individually. In contrast, this AP-1—Sp1 synergy was not observed with a specific AP-1 reporter construct (pAP-1(PMA)-TA-Luc). These results suggest that, although the AP-1 site in the MRE is critical for its activation, this response is enhanced by the availability of adjacent Sp1 site in VSMCs. This result is further supported by the nuclear protein pull-down studies, which shows that Ang II stimulation increases c-Fos, c-Jun, and Sp1 binding to the MRE. Although the mechanism by which Sp1 and AP-1 cooperate in increasing PAI-1 transcription has not yet been fully understood, the juxtaposition of Sp1 and AP-1 sites may suggest that direct interactions between these factors contribute to their synergistic effects. Consistently, Ang II stimulation caused an increase of Sp1—c-Jun association in VSMCs in the absence of DNA, providing a possible explanation for the role of A-Fos in inhibiting MEKK-1—induced activation of (MRE)2/Luc mutant constructs with AP-1 site substituted (from -63 to -52). Direct AP-1—Sp1 interactions may add the stability of transcription complexes or improve recruitment of transcriptional coactivator(s) to promoter elements, which results in enhanced promoter activity. Collectively, our data suggest that Sp1 facilitates c-Jun and c-Fos recruitment to the MRE to support the MEK1,2-specific activation of the MRE. The 100-fold increase of c-Fos binding to MRE, compared with 4.9-fold for c-Jun, suggests that c-Jun/c-Fos heterodimers may displace c-Jun homodimers or that c-Fos has additional partners in its heterodimeric structure. It is possible that additional transcription factors might associate with this MRE. Previous reports have shown that Ang II and MEKK-1 stimulation can activate Smad245,57 ; however, we were unable to detect Smad2 binding to the MRE (data not shown).
In this report, we show that treatment of cells with the MEK1,2 inhibitor PD98059 inhibited Ang II—stimulated activation of PAI-1 promoter containing the MRE and Ang II—induced c-Fos and c-Jun binding to the MRE. In addition, PD98059 inhibited constitutively active MEKK-1—induced MRE activity in the context of the endogenous PAI-1 5′ flanking promoter and as an isolated enhancer element inserted upstream of an SV40 promoter. These results support the conclusions that the MRE is an MEK1,2 response element and that this element mediates PAI-1 promoter activation by Ang II.
Previous reports demonstrated that Rho-kinase inhibition reduced Ang II—stimulated PAI-1 expression without affecting the phosphorylation of ERK.17,44 Our studies show that Rho-kinase inhibition reduced Ang II—stimulated binding of c-Jun, but not c-Fos or Sp1, to the MRE. Because Ang II—stimulated c-Jun binding to the MRE was decreased by either MEK1,2 or Rho-kinase inhibition, it appears that both of these pathways contribute to AP-1 activation within the MRE. In contrast, PD98059 inhibited Ang II—stimulated c-Fos binding to the MRE, whereas Y27632 did not significantly affect this response. Because both of these compounds inhibit PAI-1 expression, these results suggest that increased c-Fos alone is not sufficient to activate the MRE. This finding is consistent with the result that overexpression of c-Fos by itself does not activate the PAI-1(-88)/Luc construct. Moreover, neither the MEK 1,2 inhibitor nor the Rho-kinase inhibitor completely blocked c-Fos or c-Jun binding to the MRE. These results suggest that both MEK1,2-dependent and -independent pathways contribute to Ang II—stimulated effects on the MRE in the PAI-1 promoter.
The physiologic relevance of the effect of Ang II on the MRE was examined by using nuclear extracts isolated from rat aortas. One-hour Ang II intravenous infusion in rats induced aortic c-Fos binding to the MRE by 17-fold, which was comparable with the robust c-Fos response to Ang II from cultured VSMCs. In addition, aortic c-Jun and Sp1 binding to the MRE was observed in rats subjected to either saline or Ang II infusion. These in vivo studies show that rat aortic c-Fos, c-Jun, and Sp1 bind to the MRE. Among these factors, Ang II induced a significant increase in c-Fos/MRE association by using nuclear extracts from aorta and cultured VSMCs. These results suggest that the c-Fos response on the MRE plays an important role in activating the PAI-1 promoter both in vivo and in vitro.
In this report, we showed that the AT1 receptor antagonist candesartan blocked Ang II—stimulated c-Jun, c-Fos, and Sp1 binding to MRE. These results are consistent with reports from our group and others showing that AT1 receptor antagonism inhibits PAI-1 expression in VSMCs, endothelial cells, and vascular tissues.14-17,32,58 Although there is considerable evidence for a role of the AT1 receptor in affecting vascular PAI-1 expression, the role of the Ang II/AT1 pathway in regulating circulating PAI-1 antigen remains controversial. Although clinical studies have shown that both angiotensin-converting enzyme (ACE) inhibitors and AT1 receptor blockers can lower plasma PAI-1 levels,59-61 some comparative studies have indicated that ACE inhibitors are more effective.59,62 The differential responses that have been observed regarding the relative efficacy of ACE inhibitor and AT1 antagonist in affecting plasma PAI-1 could be related to the etiology for the increased PAI-1 levels in a particular group of subjects and/or the specific antagonist used. In regard to the latter, our group has shown that losartan partially blocks Ang II—induced PAI-1 in VSMCs, whereas candesartan completely blocks this response.14,32 Although AT1 antagonism decreases aortic and cardiac PAI-1 expression in rats,14-16 the effects of AT1 blockage on PAI-1 expression in human vascular tissues in vivo remain to be determined. Moreover, the relative contribution of PAI-1 derived from vascular tissues to the total pool of plasma PAI-1 is unknown.
By using adenovirus for gene transfer, we demonstrate that the 38—base pair MRE region of PAI-1 promoter is accountable for its response to Ang II stimulation. In addition, the involvement of MEK1,2 pathway in the activation of the MRE region is also confirmed by PD98095 inhibition. Previously, the sequences of PAI-1 promoter which functionally responds to the stimulation of TGF-β, Ang II, glucose, glucosamine, and fatty acid, were reported.50,52-54,63 However, the sequence that mediates the response of PAI-1 to Ang II stimulation in VSMCs was not reported, partially because of low transfection efficiency of primary cell culture by cationic lipid reagents such as Lipofectamine, about 5% maximum based on estimate from transfected GFP-expressing cells (data not shown), and could only generate less than 50% response to Ang II stimulation during transient transfection.17 By using adenoviral vectors we were able to infect more than 80% of cells with PAI-1/Luc reporter constructs and show that the effect of Ang II on the MRE region is comparable to its effect on endogenous PAI-1 mRNA.
In summary, this report demonstrates that MEKK-1/MEK1,2 pathway can activate PAI-1 promoter activity in VSMCs. This response is mediated by the cooperative effects of neighboring Sp1 and AP-1 binding sequences in the 5′ flanking region of the PAI-1 gene. Because MEK1,2 inhibition blocks the induction of PAI-1 mRNA by a number of vasoactive hormones and factors, this pathway may have important physiologic significance for the control of vascular PAI-1 expression.
Prepublished online as Blood First Edition Paper, December 4, 2003; DOI 10.1182/blood-2003-05-1737.
Supported in part by National Institutes of Health (grants DK 48358 [E.P.F.] and DK 36836 [Joslin's Diabetes and Endocrinology Research Center Grant]) and the Juvenile Diabetes Foundation International. H-C.C. was a recipient of a Mary K. Iacocca Fellowship.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Thomas Gelehrter and Carolyn Bruzdzinski for rat PAI-1 promoter DNA, Dr Charles Vinson for A-Fos and A-CREB expression plasmids, Dr Jonathan Horowitz for Sp1 expression plasmid, Dr Ronald Law for c-Fos expression plasmid, Dr Tom Curran for c-Jun expression plasmid, and Dr George King for adenovirus generating plasmids. We thank Allen C. Clermont and Dr Junichi Takahashi for assistance with intrajugular infusion.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal