Abstract
We analyzed mutations of 7 vitamin K—dependent protein and cytochrome P450 2C9 genes in 45 patients and investigated whether any contribute to the large interpatient variability in the warfarin dose-effect relationship. Total clearance and daily dose, INR and INR/Cp, were used as pharmacokinetic and pharmacodynamic indexes, respectively. Patients were grouped by genotype based on a single polymorphism and combinations of polymorphisms. Among the 30 sequence variants identified, CYP2C9*3, 165Thr → Met of the factor II gene, -402G → A, (37-bp repeat)n, and -746T → C of the factor VII gene, and (CAA repeat)n of the γ-glutamyl carboxylase gene were selected as candidate polymorphisms. As the analysis of single polymorphisms implied, the highest INR/Cp mean values and the lowest warfarin maintenance doses were observed in patients homozygous for the 165Met, -402G, (37-bp repeat)6 and -746T alleles. Multiple regression analysis revealed that warfarin sensitivity was independently associated with -402G → A, (CAA repeat)n, CYP2C9*3, and 165Thr → Met, which accounted for 50% of variance. These results suggest that part of the considerable interpatient variation is attributable to genetic variation, and the combined genotyping of CYP2C9 and certain vitamin K—dependent protein genes is useful for predicting anticoagulant responses.
Introduction
Warfarin is the most widely prescribed anticoagulant drug for the prevention and treatment of arterial and venous thromboembolic disorders.1-3 Because of large interpatient variability in the dose-anticoagulant effect relationship and a narrow therapeutic index,4 careful dosage adjustment based on INR (prothrombin time expressed as international normalized ratio) is essential. Warfarin is available as a racemic mixture of 2 enantiomers, (S)- and (R)-warfarin. In contrast to (R)-warfarin, which is metabolized by multiple cytochrome P450s (CYPs), including CYP1A2 and CYP3A4, (S)-warfarin is predominantly metabolized to 7-hydroxywarfarin by polymorphic CYP2C9.5,6 Since the potency of (S)-warfarin is much higher than that of (R)-warfarin, about 3- to 5-fold, any change in the activity of CYP2C9 is likely to have a significant influence on the anticoagulant response.4,7 Previous in vitro and in vivo findings revealed that certain variants in the CYP2C9 gene are associated with large interindividual differences in the pharmacokinetic and pharmacodynamic outcomes of warfarin therapy.8-11 Three major alleles have been found to date in humans: Arg144/Ile359, Cys144/Ile359, and Arg144/Leu359, which have been designated CYP2C9*1 (wild-type or reference allele), CYP2C9*2, and CYP2C9*3, respectively, by the Nomenclature Committee.7 Patients having at least one CYP2C9*3 allele require a lower dose of warfarin on average than homozygotes for the CYP2C9*1 allele due to an impaired metabolic capacity.4,7 However, the CYP2C9*3 allele provides important genetic information for the use of warfarin: the frequency of the allele is extremely low (∼2%) in Japanese subjects, and large interpatient variability in the dose-anticoagulant effect relationship remains even when the pharmacogenetic concept of CYP2C9 is introduced into the clinical setting. These findings suggest that additional factor(s) contribute to the relationship.
Warfarin functions as a vitamin K antagonist, decreasing the activities of the vitamin K—dependent procoagulant components.12 These proteins include factor II (prothrombin), VII, IX, and X as well as the coagulation inhibitors protein C and protein S. In addition, γ-glutamyl carboxylase (GGC) is a key enzyme in the production of γ-carboxyglutamate, which has a profound influence on the normal function of vitamin K—dependent proteins.12,13 Thus, it is hypothesized that functional polymorphisms of the genes encoding these components are associated with the large variability in the warfarin dose-effect relationship.
Despite the identification of variants of vitamin K—dependent protein genes that cause bleeding disorders, such as hereditary deficiencies of proteins,14-17 no study has examined the contributions of variants to the warfarin dose-effect relationship. The initial aim of this study was the identification of polymorphisms of 7 genes of interest (those for factors II, VII, IX, and X; proteins C and S; and GGC). The second and major aim of this study was to evaluate the impact of variants in 8 genes (including CYP2C9) on the pharmacokinetic and pharmacodynamic outcomes of warfarin therapy.
Patients, materials, and methods
The investigation was approved by the Review Board of Tottori University Hospital, and all subjects gave informed consent to participate in this study.
Patients
Forty-five unrelated patients (27 males and 18 females; mean age, 63.1 years; age range, 35 to 82 years) treated at the Department of Second Surgery, Tottori University Hospital, participated. They received warfarin therapy for the prevention or treatment of thromboemboric diseases (valve replacements, atrial fibrillation, and ischemic heart failure) with fixed maintenance doses for at least 2 months before the study. The average warfarin dose and INR were 3.8 mg/d (range, 1 to 8.5 mg/d) and 2.32 (range, 1.52 to 3.84), respectively. Biochemical and hematologic tests performed before the study revealed no evidence of hepatic or renal impairments. Because the patients were elderly, some of them also were taking other drugs; however, none of the patients were taking any medications that might have interfered with the pharmacokinetics or pharmacodynamics of warfarin.18,19
Study protocol and clinical data collection
A single nonfasting blood sample (approximately 10 mL) was obtained from each patient, just before the morning dose of warfarin. Each blood sample was divided into 3 portions: (1) for measuring concentrations of warfarin enantiomers in plasma, (2) for determining INR, and (3) for extracting DNA for genotyping.
Data collection consisted of a review of inpatient and outpatient medical records. We used the electronic medical database available in the hospital to obtain precise information on the INR value, the warfarin daily dose, type of prescription drugs, and bleeding events. We collected these data prospectively for each patient for at least 7 months from the day the sample was collected.
Assay of (S)- and (R)-warfarin plasma concentrations
(S)- and (R)-warfarin concentrations in serum were measured by high performance liquid chromatography (HPLC).20 Internal standard [diclofenac, 100 μL of a 10 μg/mL solution in a 10/90 (vol/vol) mixture of isopropanol-hexan solution], 1 mL of HCL (2 N), and 5 mL of ether were added to a 0.5-mL plasma sample. The mixture was shaken for 10 minutes and then centrifuged at 1500g for 10 minutes. A 4-mL aliquot of the organic layer was transferred to a glass tube and evaporated to dryness under a stream of nitrogen. The residue was reconstituted with 200 μL of 10% (vol/vol) isopropanol in hexane, and 40 μL of the solution was then injected into the HPLC system. The sensitivity of the assay of both compounds was 20 ng/mL. The coefficient of the intra- and interassay variation was within 10%. Recovery of both compounds ranged from 95% to 102%. A Shimadzu LC-10A system (Shimadzu, Kyoto, Japan) equipped with an LC-10AS pump and an ultraviolet detector (SPD-10A) was used. A Chiralcel OD analytical column (10 μm, 250Å∼4.6 mm ID, Daicel, Tokyo, Japan) coupled with a Chiralcel OD guard column (50 × 4.6 mm ID) was used with a mobile phase that consisted of a 18/0.5/81.5 (vol/vol) mixture of isopropanol, acetic acid, and hexane delivered at a flow rate of 1.2 mL/min. The column temperature was maintained at 25°C.
Identification of variants in vitamin K—dependent proteins, proteins C and S, and GGC genes
Genomic deoxyribonucleic acid (DNA) was isolated from blood samples using the Toyobo blood kit on a Toyobo HMX-2000 robot (Toyobo, Osaka, Japan). The primer design was based on the sequence of the 5′-flanking region and/or the intron/exon junction of the 7 genes of interest. Almost all of the primers were designed to divide all 14 exons of the factor II gene, the promoter region, and 8 exons of the factor VII gene, 8 exons of the factor IX gene, 8 exons of the factor X gene, 15 exons and intron 6 of the GGC gene, 15 exons of the protein S gene, and the promoter region and 9 exons of the protein C gene into fragments of approximately 350 bp, for the screening of mutations by subsequent single-strand conformation polymorphism (SSCP) analysis. Fragments over 350 bp in length were digested with appropriate restriction enzymes prior to SSCP analysis.
SSCP analysis was performed using the GenePhor system (Amersham Pharmacia Biotech AB, Uppsala, Sweden) as recommended by the manufacturer. The polymerase chain reaction (PCR) product (3-6 μL) was mixed with 5 μL of 20 mM EDTA (ethylenediaminetetraacetic acid), 95% formamide, and 0.05% bromophenol blue, and this mixture was heated at 95°C for 5 minutes and then quick-chilled in an ice-water bath. The resulting single-stranded DNA (5.5 μL) was then loaded on a 12.5% polyacrylamide gel (GeneGel excel 12.5/24 kit; Amersham Pharmacia Biotech AB). Electrophoresis was carried out at 450V of constant power at 15°C for 2 to 4 hours, depending on the fragment size. After electrophoresis, gels were stained using an automated gel stainer with PlusOne (Amersham Pharmacia Biotech AB).
DNA sequence
PCR products were sequenced either directly or after subcloning on an ABI 3100 automatic sequencer (Applied Biosystems, Foster City, CA) using a Big-Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems). If the direct sequencing was incomplete, each amplified PCR product was subcloned into the vector pGEM (Promega, Madison, WI) and introduced into competent JM109 cells (Promega). Prior to the sequencing, reaction mixtures were purified with a DyeEx Spin kit (QUIAGEN GmbH, Hilden, Germany). The sequencing primers were those used in the PCR amplifications.
Genotyping
Genotyping of CYP2C9 was performed with the Sequence Detection System (ABI PRISM 7000; Applied Biosystems) using a commercially available Taqman probe and primer set (Applied Biosystems).
Pharmacokinetic analysis
Apparent oral clearance (CL, mL/kg/min) of the respective warfarin enantiomers was used as an index for pharmacokinetic evaluation and was calculated as follows: dose/2 (mg/kg)/[Plasma (S)— or (R)—warfarin concentration (Cp, μg/mL) × 1000/1440], in which dose/2 is the daily dose of respective warfarin enantiomers. We also calculated the INR response per warfarin plasma concentration, termed the warfarin sensitivity index (INR/Cp).
Statistical analysis
All data are presented as the mean ± standard deviation (SD). The statistical differences between 2 groups were evaluated using Student t test and between more than 2 groups using ANOVA (with Tukey-Kramer multiple comparison test), as appropriate. A χ2 test was used to compare the allele frequency of each variant with that expected for a population in Hardy-Weinberg equilibrium. Stepwise multiple regression analysis was used to determine the relative effects of candidate gene polymorphisms on warfarin sensitivity. Each mutation was treated as a categorical value (eg, homozygote for the reference allele = 0; heterozygote for the mutant allele = 1; and homozygote for the mutant allele = 2). A P value less than .05 was taken to be the minimum level of statistical significance.
Results
Variants of vitamin K—dependent protein genes
Before the functional characterization, variants in 7 genes of interest were identified by SSCP analysis. Altogether, over 90 PCRs were performed for each patient. The results are summarized in Table 1. Of the 29 sequence variants identified, 26 were single nucleotide polymorphisms (SNPs) and 3 were insertions: the decanucleotide insertion [(cctatatcct)0→1] at position -323 (promoter region) and (37-base pair repeat)6 → 7 in intron 7 of the factor VII gene, and (CAA repeat)10 → 11 or 13 in intron 6 of the GGC gene. Allele frequencies of these insertions were as follows: -323(cctatatcct)0/1 (0.94/0.06), (37-bp repeat)6/7 (0.80/0.20), and (CAA repeat)10/11/13 (0.62/0.34/0.03). The frequency of the (37-bp repeat) 6 → 7 of the factor VII gene deviated significantly from the expected Hardy-Weinberg distribution (P < .05). Interestingly, 3 mutations in the promoter region of the factor VII gene (-401G → T, -323(cctatatcct)0/1, and -122T → C) occurred simultaneously, indicating a haplotype; -401G-323(cctatatcct)0-122T (haplotype A1) and -401T-323(cctatatcct)1-122C (haplotype A2). Of the 26 SNPs, 15 were located in noncoding regions, 8 were synonymous mutations, and 3 were nonsynonymous mutations. Three nonsynonymous mutations, a cytosine-to-thymine substitution at nucleotide 494 (494C → T) in the factor II gene, 1238G → A in the factor VII gene, and 1378G → A in the GGC gene, resulted in an amino acid change from threonine to methionine at codon 165 (165Thr → Met), 353Arg → Gln, and 460Val → Ile, with an allele frequency of 0.62, 0.04, and 0.02, respectively. By the screening of databases, 460Val → Ile was identified as a novel variant.
Genetic variants of the 7 vitamin K-dependent proteins in our study population
. | . | . | Nucleotide sequence . | . | . | Genotype . | . | . | Allele frequency . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene . | Location . | Position* . | Reference (Re) . | Mutant (Mu) . | Amino acid substitution . | Re/Re . | Re/Mu . | Mu/Mu . | Re . | Mu . | ||||
| Factor II | ||||||||||||||
| (Prothrombin) | intron 4 | +36 | acatGgagg | acatAgagg | — | 36 | 9 | 0 | 0.90 | 0.10 | ||||
| intron 5 | +90 | tcctCttcc | tcctGttcc | — | 3 | 19 | 23 | 0.28 | 0.72 | |||||
| exon 6 | 494 | accaCggga | accaTggga | Thr165Met | 8 | 18 | 19 | 0.38 | 0.62 | |||||
| intron 7 | –69 | ggaaCtggg | ggaaTtggg | — | 19 | 18 | 8 | 0.62 | 0.38 | |||||
| exon 12 | 1602 | ggccGgtct | ggccAgtct | Pro534Pro | 36 | 9 | 0 | 0.90 | 0.10 | |||||
| intron 13 | +88 | gcagAggaa | gcagGggaa | — | 1 | 8 | 36 | 0.11 | 0.89 | |||||
| Factor VII | promoter | –402 | atacGgtct | atacAgtct | — | 14 | 22 | 9 | 0.56 | 0.44 | ||||
| promoter | –401 | tacgGtctt | tacgTtctt | — | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| promoter | –323 | (cctatatcct)0 | (cctatatcct)1 | — | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| promoter | –122 | ggtgTtcag | ggtgCtcag | — | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| intron 4 | –58 | ggggGctgg | ggggActgg | — | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| exon 5 | 525 | gccaCgagg | gccaTgagg | His115His | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| exon 5 | 549 | cagaCgggg | cagaTgggg | Asp123Asp | 44 | 1 | 0 | 0.99 | 0.01 | |||||
| intron 7 | –788 | (37-bp repeat)6 | (37-bp repeat)7 | — | 36 | 0 | 9 | 0.80 | 0.20 | |||||
| intron 7 | –746 | ctctTccct | ctctCccct | — | 16 | 20 | 9 | 0.58 | 0.42 | |||||
| intron 7 | –206 | ctccGctgt | ctccActgt | — | 41 | 1 | 3 | 0.92 | 0.08 | |||||
| intron 7 | –20 | ctgaGgggg | ctgaAgggg | — | 41 | 4 | 0 | 0.96 | 0.04 | |||||
| exon 8 | 1238 | taccGgggc | taccAgggc | Arg353Gln | 41 | 4 | 0 | 0.96 | 0.04 | |||||
| Factor IX | intron 8 | +1401 | ctttTtgtg | ctttCtgtg | — | 40 | 3 | 2 | 0.92 | 0.08 | ||||
| Factor X | exon 7 | 792 | gaacCattc | gaacTattc | Thr264Thr | 3 | 21 | 21 | 0.30 | 0.70 | ||||
| Protein C | exon 9 | 891 | ccgaCaatg | ccgaTaatg | Asp297Asp | 43 | 2 | 0 | 0.98 | 0.02 | ||||
| Protein S | intron 12 | +3 | gtaaTagat | gtaaCagat | — | 32 | 12 | 1 | 0.84 | 0.16 | ||||
| exon 15 | 2001 | gtccAtcag | gtccGtcag | Pro667Pro | 7 | 20 | 18 | 0.38 | 0.62 | |||||
| GGC | intron 2 | –67 | gtgcAgtga | gtgcTgtga | — | 21 | 23 | 1 | 0.72 | 0.28 | ||||
| intron 2 | –33 | ccccGcaca | ccccAcaca | — | 31 | 14 | 0 | 0.84 | 0.16 | |||||
| intron 6 | –152 | (CAA)10 | (CAA)11 | — | 15 | 24 | 3 | 0.62 | 0.34 | |||||
| –152 | (CAA)13 | — | 3 | 0 | 0.03 | |||||||||
| exon 9 | 1218 | cccgTtccc | cccgCtccc | Arg406Arg | 4 | 21 | 20 | 0.32 | 0.68 | |||||
| exon 9 | 1242 | tcacCtacc | tcacTtacc | Thr414Thr | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| exon 10 | 1378 | taatGtcac | taatAtcac | Val460Ile | 43 | 2 | 0 | 0.98 | 0.02 | |||||
. | . | . | Nucleotide sequence . | . | . | Genotype . | . | . | Allele frequency . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene . | Location . | Position* . | Reference (Re) . | Mutant (Mu) . | Amino acid substitution . | Re/Re . | Re/Mu . | Mu/Mu . | Re . | Mu . | ||||
| Factor II | ||||||||||||||
| (Prothrombin) | intron 4 | +36 | acatGgagg | acatAgagg | — | 36 | 9 | 0 | 0.90 | 0.10 | ||||
| intron 5 | +90 | tcctCttcc | tcctGttcc | — | 3 | 19 | 23 | 0.28 | 0.72 | |||||
| exon 6 | 494 | accaCggga | accaTggga | Thr165Met | 8 | 18 | 19 | 0.38 | 0.62 | |||||
| intron 7 | –69 | ggaaCtggg | ggaaTtggg | — | 19 | 18 | 8 | 0.62 | 0.38 | |||||
| exon 12 | 1602 | ggccGgtct | ggccAgtct | Pro534Pro | 36 | 9 | 0 | 0.90 | 0.10 | |||||
| intron 13 | +88 | gcagAggaa | gcagGggaa | — | 1 | 8 | 36 | 0.11 | 0.89 | |||||
| Factor VII | promoter | –402 | atacGgtct | atacAgtct | — | 14 | 22 | 9 | 0.56 | 0.44 | ||||
| promoter | –401 | tacgGtctt | tacgTtctt | — | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| promoter | –323 | (cctatatcct)0 | (cctatatcct)1 | — | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| promoter | –122 | ggtgTtcag | ggtgCtcag | — | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| intron 4 | –58 | ggggGctgg | ggggActgg | — | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| exon 5 | 525 | gccaCgagg | gccaTgagg | His115His | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| exon 5 | 549 | cagaCgggg | cagaTgggg | Asp123Asp | 44 | 1 | 0 | 0.99 | 0.01 | |||||
| intron 7 | –788 | (37-bp repeat)6 | (37-bp repeat)7 | — | 36 | 0 | 9 | 0.80 | 0.20 | |||||
| intron 7 | –746 | ctctTccct | ctctCccct | — | 16 | 20 | 9 | 0.58 | 0.42 | |||||
| intron 7 | –206 | ctccGctgt | ctccActgt | — | 41 | 1 | 3 | 0.92 | 0.08 | |||||
| intron 7 | –20 | ctgaGgggg | ctgaAgggg | — | 41 | 4 | 0 | 0.96 | 0.04 | |||||
| exon 8 | 1238 | taccGgggc | taccAgggc | Arg353Gln | 41 | 4 | 0 | 0.96 | 0.04 | |||||
| Factor IX | intron 8 | +1401 | ctttTtgtg | ctttCtgtg | — | 40 | 3 | 2 | 0.92 | 0.08 | ||||
| Factor X | exon 7 | 792 | gaacCattc | gaacTattc | Thr264Thr | 3 | 21 | 21 | 0.30 | 0.70 | ||||
| Protein C | exon 9 | 891 | ccgaCaatg | ccgaTaatg | Asp297Asp | 43 | 2 | 0 | 0.98 | 0.02 | ||||
| Protein S | intron 12 | +3 | gtaaTagat | gtaaCagat | — | 32 | 12 | 1 | 0.84 | 0.16 | ||||
| exon 15 | 2001 | gtccAtcag | gtccGtcag | Pro667Pro | 7 | 20 | 18 | 0.38 | 0.62 | |||||
| GGC | intron 2 | –67 | gtgcAgtga | gtgcTgtga | — | 21 | 23 | 1 | 0.72 | 0.28 | ||||
| intron 2 | –33 | ccccGcaca | ccccAcaca | — | 31 | 14 | 0 | 0.84 | 0.16 | |||||
| intron 6 | –152 | (CAA)10 | (CAA)11 | — | 15 | 24 | 3 | 0.62 | 0.34 | |||||
| –152 | (CAA)13 | — | 3 | 0 | 0.03 | |||||||||
| exon 9 | 1218 | cccgTtccc | cccgCtccc | Arg406Arg | 4 | 21 | 20 | 0.32 | 0.68 | |||||
| exon 9 | 1242 | tcacCtacc | tcacTtacc | Thr414Thr | 40 | 5 | 0 | 0.94 | 0.06 | |||||
| exon 10 | 1378 | taatGtcac | taatAtcac | Val460Ile | 43 | 2 | 0 | 0.98 | 0.02 | |||||
The reference allele for each gene had the following GenBank accession number; M17262 for factor II; J02933 for factor VII; K02402 for factor IX; AH002727 for factor X; M11228 for protein C; NT_022625 and M57853 for protein S; U65896 for GGC. — indicates synonymous mutation.
Position is relative to the ATG start site except for the factor VII gene. In the factor VII gene, residue +1 is the first amino acid of the mature protein33
Effect of genetic polymorphisms on the pharmacokinetic and pharmacodynamic outcomes of warfarin therapy
CYP2C9 gene. Among the 45 patients, 2 were heterozygous carriers for the CYP2C9*3 allele. Allele frequencies for CYP2C9*1 and CYP2C9*3 were 0.978 and 0.022, respectively, and were consistent with those of previous studies.4,21 As shown in Figure 1A, although the time course of change in mean INR values did not differ between the 2 genotype groups, the mean warfarin daily dose was 40% lower in the CYP2C9*3 group than in the reference CYP2C9*1 group throughout the observation period. When pharmacokinetic backgrounds were compared between the 2 groups, mean plasma (S)-warfarin clearance was found to be 33% lower in the CYP2C9*3 group. On the other hand, the mean INR/Cp value was 43% higher in the CYP2C9*3 group (Figure 1B).
Polymorphisms of the CYP2C9 gene and in vivo pharmacodynamic and pharmacokinetic outcomes of warfarin. (A) Time course of changes in warfarin daily dose and INR values during the observation period (7 months) in patients with and without the CYP2C9*3 mutant allele. (B) Comparisons of warfarin clearance and INR/Cp between the 2 CYP2C9 genotypic groups. Bars represent the mean.
Polymorphisms of the CYP2C9 gene and in vivo pharmacodynamic and pharmacokinetic outcomes of warfarin. (A) Time course of changes in warfarin daily dose and INR values during the observation period (7 months) in patients with and without the CYP2C9*3 mutant allele. (B) Comparisons of warfarin clearance and INR/Cp between the 2 CYP2C9 genotypic groups. Bars represent the mean.
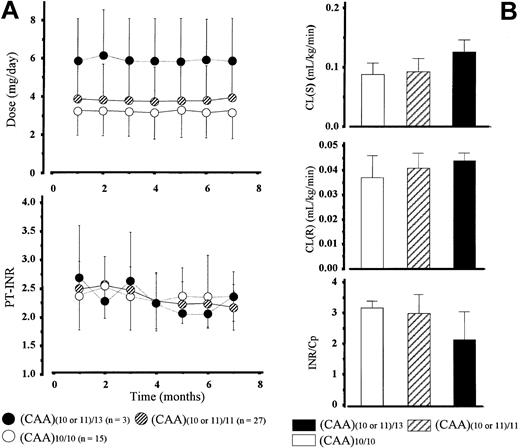
GGC gene. We divided patients into 3 groups with regard to the number of microsatellites in intron 6: (CAA)10/10 (n = 15), (CAA)(10 or 11)/11 (n = 27), and (CAA)(10 or 11)/13 (n = 3). The mean daily dose of warfarin increased together with the number of microsatellites at comparable mean INR values in the 3 genotype groups (Figure 2A). However, the increase did not reflect an elevation in plasma warfarin clearance, because these values did not differ significantly among the 3 groups. The mean INR/Cp value was almost 32% lower in patients with a (CAA)13 allele than those homozygous for (CAA)10, a reference allele, and patients heterozygous for the (CAA)11 allele having an intermediate value (Figure 2B).
Polymorphisms of the γ-glutamylcarboxylase gene (CAA repeats in intron 6) and in vivo pharmacodynamic and pharmacokinetic outcomes of warfarin. (A) Time course of changes in warfarin daily dose and INR values during the observation period (7 months) in patients with various patterns of the microsatellite (CAA repeat) in the GGC gene. (B) Comparisons of warfarin clearance and INR/Cp among the 3 GGC genotypic groups. Bars represent the mean ± SD.
Polymorphisms of the γ-glutamylcarboxylase gene (CAA repeats in intron 6) and in vivo pharmacodynamic and pharmacokinetic outcomes of warfarin. (A) Time course of changes in warfarin daily dose and INR values during the observation period (7 months) in patients with various patterns of the microsatellite (CAA repeat) in the GGC gene. (B) Comparisons of warfarin clearance and INR/Cp among the 3 GGC genotypic groups. Bars represent the mean ± SD.
Other genes. The contributions of the mutations in other vitamin K—dependent protein genes to mean INR/Cp values are shown in Table 2. In the factor II gene, homozygotes for the nonsynonymous mutant allele (494T, tentatively designated the M2 allele, M2/M2 genotype) had significantly higher mean INR/Cp values than patients having a reference allele (494C, M1 allele) with homozygosity (M1/M1 genotype). In contrast, homozygous carriers of the -402A allele (B2 allele, B2/B2 genotype) of the factor VII gene had significantly lower mean INR/Cp values than those of the reference allele (-402G, B1 allele, B1/B1 genotype), and heterozygotes (B1/B2 genotype) had values intermediate between those of the 2 homozygous groups. Similarly, homozygous carriers for the (37 bp repeat)7 (b/b genotype) or -746C allele (N2/N2 genotype) in the factor VII gene had lower mean INR/Cp values than those for the (37 bp repeat)6 (a/a genotype) or -746T allele (N1/N1 genotype). Taking these findings into consideration, all of the mutations in the vitamin K—dependent protein genes except 494C → T in factor II were associated with a lower INR/Cp value, suggesting reduced warfarin sensitivity. In contrast to the INR/Cp values, there were no significant differences in the warfarin daily doses for each genotype.
Mean INR/Cp values and warfarin daily dose in various genotypes determined from single and multiple gene polymorphisms
Gene . | Allele . | Genotype . | n . | INR/Cp . | Warfarin dose, mg (range) . |
|---|---|---|---|---|---|
| Factor II | 494C (M1) | M1/M1 | 8 | 2.62 ± 0.54 | 3.3 ± 1.1 (1.5-5.0) |
| 494T (M2) | M1/M2 | 18 | 3.00 ± 0.39 | 3.9 ± 1.8 (1.5-7.5) | |
| M2/M2 | 19 | 3.26 ± 0.18* | 3.8 ± 2.1 (1.0-8.5) | ||
| Factor VII | –402G (B1) | B1/B1 | 14 | 3.42 ± 0.82 | 3.5 ± 2.0 (1.5-8.0) |
| –402A (B2) | B1/B2 | 22 | 3.09 ± 0.43† | 3.9 ± 1.9 (1.0-8.5) | |
| B2/B2 | 9 | 2.39 ± 0.18* | 3.8 ± 1.3 (1.5-6.0) | ||
| (37-bp repeat)6 (a) | a/a | 36 | 3.14 ± 0.55 | 3.8 ± 1.9 (1.0-8.5) | |
| (37-bp repeat)7 (b) | b/b | 9 | 2.58 ± 0.21 | 3.5 ± 1.3 (1.5-5.0) | |
| –746T (N1) | N1/N1 | 16 | 3.34 ± 0.66 | 3.6 ± 1.9 (1.5-8.0) | |
| –746C (N2) | N1/N2 | 20 | 3.00 ± 0.51 | 4.0 ± 2.0 (1.0-8.5) | |
| N2/N2 | 9 | 2.58 ± 0.21* | 4.0 ± 1.3 (1.5-5.0) | ||
| Combination | I | (M1/M1)(B2/B2)(b/b)(N2/N2) | 5 | 2.14 ± 0.31 | 3.6 ± 1.4 (1.5-5.0) |
| II | (M1/M2)(B1/B1)(a/a)(N1/N1) | 7 | 3.56 ± 0.41 | 3.0 ± 1.3 (1.5-4.5) | |
| III | (M1/M2)(B1/B2)(a/a)(N1/N2) | 3 | 2.63 ± 0.72 | 5.3 ± 2.6 (2.5-7.5) | |
| IV | (M2/M2)(B1/B1)(a/a)(N1/N1) | 3 | 4.48 ± 0.83 | 2.3 ± 1.4 (1.5-4.0) | |
| V | (M2/M2)(B1/B2)(a/a)(N1/N2) | 9 | 2.93 ± 0.41 | 4.3 ± 1.9 (2.5-8.5) |
Gene . | Allele . | Genotype . | n . | INR/Cp . | Warfarin dose, mg (range) . |
|---|---|---|---|---|---|
| Factor II | 494C (M1) | M1/M1 | 8 | 2.62 ± 0.54 | 3.3 ± 1.1 (1.5-5.0) |
| 494T (M2) | M1/M2 | 18 | 3.00 ± 0.39 | 3.9 ± 1.8 (1.5-7.5) | |
| M2/M2 | 19 | 3.26 ± 0.18* | 3.8 ± 2.1 (1.0-8.5) | ||
| Factor VII | –402G (B1) | B1/B1 | 14 | 3.42 ± 0.82 | 3.5 ± 2.0 (1.5-8.0) |
| –402A (B2) | B1/B2 | 22 | 3.09 ± 0.43† | 3.9 ± 1.9 (1.0-8.5) | |
| B2/B2 | 9 | 2.39 ± 0.18* | 3.8 ± 1.3 (1.5-6.0) | ||
| (37-bp repeat)6 (a) | a/a | 36 | 3.14 ± 0.55 | 3.8 ± 1.9 (1.0-8.5) | |
| (37-bp repeat)7 (b) | b/b | 9 | 2.58 ± 0.21 | 3.5 ± 1.3 (1.5-5.0) | |
| –746T (N1) | N1/N1 | 16 | 3.34 ± 0.66 | 3.6 ± 1.9 (1.5-8.0) | |
| –746C (N2) | N1/N2 | 20 | 3.00 ± 0.51 | 4.0 ± 2.0 (1.0-8.5) | |
| N2/N2 | 9 | 2.58 ± 0.21* | 4.0 ± 1.3 (1.5-5.0) | ||
| Combination | I | (M1/M1)(B2/B2)(b/b)(N2/N2) | 5 | 2.14 ± 0.31 | 3.6 ± 1.4 (1.5-5.0) |
| II | (M1/M2)(B1/B1)(a/a)(N1/N1) | 7 | 3.56 ± 0.41 | 3.0 ± 1.3 (1.5-4.5) | |
| III | (M1/M2)(B1/B2)(a/a)(N1/N2) | 3 | 2.63 ± 0.72 | 5.3 ± 2.6 (2.5-7.5) | |
| IV | (M2/M2)(B1/B1)(a/a)(N1/N1) | 3 | 4.48 ± 0.83 | 2.3 ± 1.4 (1.5-4.0) | |
| V | (M2/M2)(B1/B2)(a/a)(N1/N2) | 9 | 2.93 ± 0.41 | 4.3 ± 1.9 (2.5-8.5) |
Other genotypes of patients in groups I through V were fixed as follows: CYP2C9*1/*1, haplotype A1 in the factor VII gene, and (CAA repeat)10 or 11 in the GGC gene.
P < .05 versus homozygotes for the reference allele
P < .05 versus homozygotes for the mutant allele
Further analysis was performed with these 4 mutations combined. Combining the 4 polymorphisms allowed around 60% of the population to be grouped in 5 genotypes (I through V in Table 2), each found in at least 3 subjects. As the analysis of single polymorphisms implies, the highest INR/Cp mean values were observed in patients homozygous for the M2, B1, and N1 alleles (genotype IV), whereas the lowest values were associated with the combined genotypes that include homozygous M1, B2, and N2 alleles (genotype I). Patients with genotype IV had mean INR/Cp values (4.48 ± 0.83) 2 times as high as patients with genotype I (2.14 ± 0.31). Furthermore, patients with genotype IV had the lowest mean warfarin daily dose (2.3 ± 1.4) among the 5 genotypic groups: the mean warfarin dose was 1.3 to 2.3 times lower in patients with genotype IV than patients in other groups.
In order to assess the contributions of all selected candidate gene polymorphisms to warfarin sensitivity, a stepwise multiple regression analysis was carried out. In the analysis, the only variables that were significantly and independently associated with warfarin sensitivity were -402G → A in the factor VII gene (partial r2 = 0.35), (CAA repeat)n in the GGC gene (partial r2 = 0.09), CYP2C9*3 (partial r2 = 0.04), and 494C → T in the factor II gene (partial r2 = 0.01), together accounting for approximately 50% of variance (multiple r = 0.70). The other 2 mutations in the factor VII gene, (37-bp repeat)n and -746T → C, had poor explanatory capacities compared with the -402G → A mutation (data not shown), suggesting that -402G → A was the main predictor for warfarin sensitivity among the 3 mutations.
Discussion
In the present study, we analyzed mutations in the genes of 7 vitamin K—dependent coagulation proteins (factor II, VII, IX, and X; protein C and S; and GGC) in 45 patients, and then investigated whether these mutations contribute to the large interpatient variability in the dose-anticoagulant effect relationship. In addition to CYP2C9*3, a variant associated with a decrease in hepatic metabolism and an increase in sensitivity to warfarin, 494C → T (165thr → Met) of the factor II gene, -402G → A, (37-bp repeat)n, and -746T → C of the factor VII gene, and (CAA repeat)n of the GGC gene were selected as candidate polymorphisms.
Several studies have shown that certain mutations in the factor VII gene contribute to the variability in factor VII plasma activity; most of the data available are on the -402G → A, 353Arg → Gln, and (37-bp repeat)n polymorphisms.22-25 Recent study revealed that the -401T allele was responsible for decreased transcriptional activity, whereas the -402A allele was associated with an increased rate of transcription thus leading to higher factor VII levels.22 Because of high linkage disequilibrium among certain mutations, the net influence of (37-bp repeat)n on factor VII levels might be partially masked. In order to estimate the independent contribution of the (37-bp repeat)n polymorphism, Pinotti et al established eukaryotic cells generating various numbers of (37-bp repeat) and demonstrated that higher numbers of repeats were associated with higher mRNA expression levels.25 Since warfarin is a competitive antagonist of vitamin K in the processes leading to the synthesis of factor VII (maintained in the inactive form), and since high levels of factor VII may be related to a hypercoagulable state, less sensitivity to warfarin therapy would be expected in patients with mutations leading to high factor VII levels. In contrast to these 2 polymorphisms, the 353Arg → Gln variant in the catalytic domain was reported to be associated with an approximately 25% reduction in factor VII coagulant activity in heterozygous carriers and a 50% reduction in homozygous carriers.26 However, no contribution of this variant to warfarin sensitivity was observed in the present study. The lack of any apparent effect may be explained by the fact that all our patients were heterozygotes.
Among various mutations identified in the present study, only the (37-bp repeat)6 → 7 insertion polymorphism in the factor VII gene was not in Hardy-Weinberg equilibrium because of a deficit of heterozygotes. The sequence of this portion was confirmed for all patients by at least 2 independent PCR amplifications to ensure that the polymorphisms identified were not PCR-based artifacts. These results were in contrast to recent findings by Shimokata et al, who reported that approximately 42% of 380 unrelated Japanese individuals were heterozygous carriers for the (37-bp repeat)7 allele.27 In the present study, there were no clinical differences (eg, disease state and biochemical tests) between the 2 homozygous groups (data not shown). Thus, the significance of this deviation was not clear, and further genetic analysis will be necessary to confirm this finding; however, since most mutations in the present study were in Hardy-Weinberg equilibrium, the findings obtained should be valid for the general population.
In the GGC gene, one novel nonsynonymous variant and 3 patterns of (CAA) repeats were observed. To date, at least 2 nonsynonymous mutations, 394Arg → Leu and 501Trp → Ser, leading to a combined congenital deficiency of all vitamin K—dependent coagulation factors, have been reported.15,16,28 These mutations were responsible for weak procoagulant activity, and oral vitamin K1 administration was effective in resolving the clinical symptoms, indicating that carboxylase activity plays a critical role in the warfarin dose-effect relationship. As shown in Figure 2A, the mean daily dose of warfarin increased as the number of microsatellites increased even when mean INR values were comparable among the 3 genotype groups. Patients with 13 repeats [(CAA)13] had the lowest mean INR/Cp values, suggesting the possibility of reduced sensitivity to warfarin in these patients. Theoretically, the large dose of warfarin necessary to reach the target INR value in these patients may, at least partially, be explained by increased GGC activity. However, there is no data to support this hypothesis, and more in vivo and in vitro experiments are required to elucidate the role of the microsatellites in carboxylation capability.
As shown in Table 2, patients with different genotypes in conjunction with 4 polymorphisms from the 2 genes showed up to 2-fold differences in mean INR/Cp values, thus part of the considerable interpatient variation in the dose-effect relationship was attributable to genetic variation, which may be of assistance in the interpretation of INR/Cp levels on an individual basis. It is worth noting that patients with genotype IV had the highest mean INR/Cp values among the 5 groups, suggesting high sensitivity to warfarin. Moreover, an intergenotypic difference was observed in the maintenance dose of warfarin; the mean dose was 1.3 to 2.3 times lower in patients with genotype IV than those in other genotype groups. These results were also supportive of a contribution of individual genetic status to the relationship.
Among various candidate gene polymorphisms, it is of interest which mutation(s) contributed most to, and to what extent, the overall variation in the INR/Cp values. In order to address these issues, we carried out a stepwise multiple regression analysis. Four functional polymorphisms (ie, CYP2C9*3, 494C → T, -402G → A, and [CAA repeat]n) together accounted for 50% of overall between-patient variability in warfarin sensitivity. In contrast to the extensive contribution of -402G → A and (CAA repeat)n, CYP2C9*3 and 494C → T explained only approximately 5% of the variance. However, although the CYP2C9*3 allele is extremely rare in Japanese populations (ie, 2%), it accounted for 4% of the variance in warfarin sensitivity. Since the frequency of the *3 allele is known to be higher in Caucasian subjects than in Japanese subjects,4 the *3 allele is expected to be the major factor responsible for the between-patient variability in warfarin sensitivity in Caucasian patients. Prothrombin (factor II) is composed of 3 structural regions as follows: fragment 1, which contains the γ-carboxyglutamic acid (Gla) domain (residues 1 to 40) and kringle 1 domain (residues 41 to 155); and fragment 2, which contains mainly the kringle 2 domain (residues 156 to 271); and a serine protease precursor domain (residues 272 to 597).29,30 Previous studies showed that the kringle 2 domain plays an important role in the binding of prothrombin to factor Xa and factor Va.31,32 A cytosine to thymine transition at nucleotide 494 in the factor II gene, resulting in the substitution at codon 165 of an uncharged polar side chain amino acid “threonine” with a nonpolar side chain amino acid “methionine,” is located in the kringle 2 domain of prothrombin.
The limitations of our study need to be addressed. First, we calculated the warfarin sensitivity index (INR/Cp) based on the total plasma concentration (bound plus unbound). Both warfarin enantiomers are extensively bound to plasma protein (ie, 99%), and no enantioselective differences in the binding have been observed.4 Since only the unbound fraction can reach and/or be equilibrated with that at the site of action (eg, γ-glutamyl carboxylase), one must be careful that the interindividual variability in INR/Cp is dependent on the status of the plasma protein binding of the drug. Most studies with regard to phenotypic and genotypic correlations measured various vitamin K—dependent protein plasma levels. Unfortunately, we did not measure plasma antigenic levels and so were unable to assess the changes in protein levels due to genetic polymorphisms. An extended large population study that combines the genotyping of the 4 candidate polymorphisms with corresponding phenotype indexes should be conducted to confirm the present findings.
Prepublished online as Blood First Edition Paper, December 4, 2003; DOI 10.1182/blood-2003-09-3043.
Supported by a grant from the Ministry of Education, Science, Sports, and Culture of Japan.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal