Abstract
Acquired mutations in megakaryocyte transcription factor GATA1 have recently been reported in Down syndrome (DS), transient myeloproliferative disorder (TMD), and acute megakaryoblastic leukemia (AMKL). To provide novel insight into GATA1 mutations in DS, genomic DNA was assayed from 12 AMKL and 4 TMD cases (including neonatal, prediagnosis samples in 4 of 16), neonatal blood spots from 21 DS children without clinically evident TMD or AMKL, and 62 non-DS cord blood samples, using techniques not previously employed with such samples. GATA1 mutations were present in all TMD and AMKL cases and at birth in 3 of 4 children without known clinical TMD, who later developed AMKL. They were present at birth in 2 of 21 DS neonates, who have not yet, but could still, develop AMKL (now 26 and 31 months). GATA1 mutations were not detected in 62 non-DS cord blood samples. In 4 AMKL patients multiple independent GATA1 mutations were observed. These data show GATA1 mutations occur in utero in most DS TMD and AMKL, that they may occur without clinical signs of disease, and that multiple separate GATA1 mutant clones can occur in an individual. The findings have implications for pathogenesis of DS TMD and AMKL and highlight parallels between DS AMKL and other childhood leukemias.
Introduction
Children with Down syndrome (DS) are uniquely predisposed to clonal disorders affecting the megakaryocyte lineage; specifically, transient myeloproliferative disorder (TMD), also known as transient leukemia (TL) and acute megakaryoblastic leukemia (AMKL) (reviewed in Lange,1 Taub and Ravindranath,2 Zipursky,3 Gamis and Hilden,4 and Zipursky et al5 ). TMD is reported to occur in 10%-20% of DS newborns.6,7 It has a variable clinical presentation ranging from infants who have clinical abnormalities to infants who are well but have circulating blasts detected as an incidental finding. As blood counts and smears are not routinely performed on all DS newborns, it is possible that the reported incidence of TMD has been underestimated either because a blood count is not performed or because subtle abnormalities may be overlooked. In most cases TMD resolves spontaneously. But in up to 30% of cases AMKL develops, either by overt progression or after apparent remission for a number of months or years. Alternatively, and rarely, TMD itself is the direct cause of death as a result of massive hepatic fibrosis secondary to megakaryoblast infiltration. Given this characteristic clinical phenotype, it has been suggested that TMD is a disorder of fetal hematopoiesis.4
DS children have a 500-fold increased risk of developing AMKL compared to the general pediatric population.5,8,9 Several features set DS-associated AMKL apart from non-DS AMKL. First, DS AMKL nearly always presents during a narrow temporal window before 4 years of age.10 Second, in some, but not all cases of DS AMKL, there is documented evidence of previous TMD, and the megakaryoblasts in AMKL and TMD are morphologically, immunophenotypically, and ultrastructurally similar.11-14 Third, DS AMKL has an unusually good prognosis, and finally, DS AMKL has characteristic karyotypic abnormalities.1,15 Taken together, these observations suggest both a biologic relationship between TMD and AMKL and a distinct pathogenetic basis for these conditions.2,3
GATA1 is a phosphorylated, double zinc finger, DNA-binding transcription factor encoded on the X-chromosome. Recently, acquired somatic mutations in one copy of GATA1 were demonstrated both in TMD and in DS AMKL, while such mutations were absent in non-DS AMKL and in DS acute lymphoid leukemia.16-20 All reported mutations occur in the 5′ end of the gene, mainly in the first coding exon, exon 2. Mutations either introduce a stop codon before codon 84 or disrupt splicing of exon 2 to the rest of GATA1 mRNA and thus remove the first translational start codon. In either case, the predicted GATA1 protein would be translated from an alternative translational start codon, codon 84, and GATA1 protein would be truncated at the N-terminus. From a mechanistic point of view, involvement of GATA1 in DS AMKL is compatible with recent proposed models of leukemogenesis, where mutations in transcription factors block differentiation.21 Thus, in DS TMD and AMKL, mutation of GATA1 would help determine the lineage phenotype of the leukemia.21,22
Involvement of GATA1 in DS AMKL and TMD is consistent with its role in hematopoiesis as a key regulator of megakaryocyte, erythroid, eosinophil, and mast cell differentiation (reviewed in Shivdasani and Orkin,23 Orkin,24 and Cantor and Orkin25 ). In mice, germ line ablation of megakaryocyte-specific GATA1 expression results in perturbed megakaryocyte differentiation with increased numbers of arrested megakaryoblasts in bone marrow and spleen and abnormal platelet maturation.26,27 In humans, germ line missense mutations in the highly conserved bifunctional N-terminal zinc finger of GATA1 have established its central role in regulating megakaryoblast numbers and ensuring normal megakaryocyte and platelet differentiation.28-32
These studies, together with the clinical and genetic data in DS AMKL and TMD, strongly implicate GATA1 in the pathogenesis of abnormal megakaryopoiesis in DS. In addition, the presence of GATA1 mutations in TMD suggests that these mutations arise in utero. However, a number of questions remain about the exact role of GATA1 in the clinical expression and natural history of DS-associated AMKL and TMD. In particular, are GATA1 mutations present at birth in all cases of AMKL, including those without a clear clinical history of antecedent TMD? More generally, are GATA1 mutations detectable at birth in both unselected DS neonates and healthy non-DS newborns?
In this study we set out to specifically address the questions mentioned above by analyzing samples from DS individuals with and without clinically overt TMD and AMKL, including, where available, sequential samples from birth. A number of approaches were employed to detect minor populations of cells containing GATA1 mutations to identify the earliest stages of the disease process associated with GATA1 abnormality. Results from this analysis provide novel insight into the likely sequence of molecular and cellular events that underlie DS TMD and AMKL. More generally, the results may have implications for management of DS infants and children.
Patients, materials, and methods
Subjects studied
Twelve DS patients with AMKL and 4 with TMD were studied (Tables 1, 2). One of the TMD patients subsequently developed AMKL (L14). For 8 patients' sequential samples, including neonatal blood spots, diagnostic and remission materials were obtained. Diagnosis of AMKL and TMD was based on morphology and immunophenotype. Some patients with AMKL were identified as they had participated in the United Kingdom MRC AML X and XII trials. In these cases the diagnosis was confirmed by independent review. Neonatal blood spots were obtained from 21 randomly selected DS children who were under 5 years of age at the time of the study. Clinical records on all children were examined. This study was approved by local (Oxford LREC) and national United Kingdom ethics (Thames Valley MREC) committees. In all cases informed consent was obtained from the parents.
GATA-1 mutations in AKML patients with single samples
Pt . | Sex . | Diagnosis . | Age at diagnosis (mo) . | DNA source . | WBC count, × 109/L (% blast) . | Karyotype . | Sequence in genomic DNA*† . | Cloned sequence from genomic DNA (no. mutant clones/no. total clones)† . | Radiolabeled GATA1 exon 2 PCR products . | Cloned sequence of bands cut from polyacrylamide gels (no. mutant clones/no. total clones)† . | Consequence of mutation† . | Clinical status/time since diagnosis . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L02 | M | AMKL | 30 | Diagnosis BM | 9.3 (25) | 47, XY, +21(c)[10] | 267 ins 10 | Wild type (0/5) | Wild-type band and faint insertion band | 267 ins 10 (4/12) | 267 ins 10 - Stop codon before Met 84 | CCR/27 mo |
| 49, XY, +8, +21, +21(c)[2] | ||||||||||||
| 48, XY, der(1)add (p32)[2] | ||||||||||||
| L03 | M | AMKL | 32 | Diagnosis BM | 41.3 (60) | del(3)(q21q26),i(7)(q10), del(16) (q22q24), +21c[7]/47, 21c[8] | 317 del 13 | ND | Wild-type band and prominent deletion band | 317 del 13 (2/2) | 317 del 13 - Exon 2 splice mutant | CCR/15 mo |
| L07 | M | AMKL | 39 | Diagnosis BM | 6.2 (51) | 48,XY,der(4)t(1;4) (q25q35),del(6) (q15q21),+11, +21c[10] | 324 del 9 | ND | Wild-type band and deletion band 1; faint deletion band 2 | 324 del 9 (2/3); 269 del 23 (3/6) | 324 del 9 - exon 2 splice mutant; 269 del 23 - stop codon before Met 84 | Died of sepsis during treatment |
| 47,XY,+21c[2] | ||||||||||||
| L08 | F | AMKL | 13 | Diagnosis BM | 23.1 (58) | 47,XX,t(4;11;13),+21c[9] | 299 ins 1 | Wild-type (6/8); 299 ins 1 (2/8) | Single wild-type band | ND | 299 ins 1 - stop codon before Met 84 | CCR/124 mo |
| L10 | M | AMKL | 17 | Diagnosis BM | 14.1 (40) | 48,XY,+21,+8[11] | 317 del 13 | ND | Wild-type band and deletion band | 317 del 13 (5/9) | 317 del 13 - exon 2 splice mutant | CCR/55 mo |
| L11 | M | AMKL | 21 | Diagnosis BM | 63.7 (65) | 47,XY,+21(c)[8] | 299 ins 1 | Wild-type (1/4); 299 ins 1 (3/4) | Wild-type band | ND | 299 ins 1 - stop codon before Met 84 | Died of sepsis during treatment |
| T03 | F | TMD | Birth | PB | 200 (10) | 47,XY,+21(c)[10] | 161C>T | ND | Wild-type band | ND | 161 C>T - stop codon before Met 84 | Died at day +4; liver failure secondary to fibrosis |
Pt . | Sex . | Diagnosis . | Age at diagnosis (mo) . | DNA source . | WBC count, × 109/L (% blast) . | Karyotype . | Sequence in genomic DNA*† . | Cloned sequence from genomic DNA (no. mutant clones/no. total clones)† . | Radiolabeled GATA1 exon 2 PCR products . | Cloned sequence of bands cut from polyacrylamide gels (no. mutant clones/no. total clones)† . | Consequence of mutation† . | Clinical status/time since diagnosis . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L02 | M | AMKL | 30 | Diagnosis BM | 9.3 (25) | 47, XY, +21(c)[10] | 267 ins 10 | Wild type (0/5) | Wild-type band and faint insertion band | 267 ins 10 (4/12) | 267 ins 10 - Stop codon before Met 84 | CCR/27 mo |
| 49, XY, +8, +21, +21(c)[2] | ||||||||||||
| 48, XY, der(1)add (p32)[2] | ||||||||||||
| L03 | M | AMKL | 32 | Diagnosis BM | 41.3 (60) | del(3)(q21q26),i(7)(q10), del(16) (q22q24), +21c[7]/47, 21c[8] | 317 del 13 | ND | Wild-type band and prominent deletion band | 317 del 13 (2/2) | 317 del 13 - Exon 2 splice mutant | CCR/15 mo |
| L07 | M | AMKL | 39 | Diagnosis BM | 6.2 (51) | 48,XY,der(4)t(1;4) (q25q35),del(6) (q15q21),+11, +21c[10] | 324 del 9 | ND | Wild-type band and deletion band 1; faint deletion band 2 | 324 del 9 (2/3); 269 del 23 (3/6) | 324 del 9 - exon 2 splice mutant; 269 del 23 - stop codon before Met 84 | Died of sepsis during treatment |
| 47,XY,+21c[2] | ||||||||||||
| L08 | F | AMKL | 13 | Diagnosis BM | 23.1 (58) | 47,XX,t(4;11;13),+21c[9] | 299 ins 1 | Wild-type (6/8); 299 ins 1 (2/8) | Single wild-type band | ND | 299 ins 1 - stop codon before Met 84 | CCR/124 mo |
| L10 | M | AMKL | 17 | Diagnosis BM | 14.1 (40) | 48,XY,+21,+8[11] | 317 del 13 | ND | Wild-type band and deletion band | 317 del 13 (5/9) | 317 del 13 - exon 2 splice mutant | CCR/55 mo |
| L11 | M | AMKL | 21 | Diagnosis BM | 63.7 (65) | 47,XY,+21(c)[8] | 299 ins 1 | Wild-type (1/4); 299 ins 1 (3/4) | Wild-type band | ND | 299 ins 1 - stop codon before Met 84 | Died of sepsis during treatment |
| T03 | F | TMD | Birth | PB | 200 (10) | 47,XY,+21(c)[10] | 161C>T | ND | Wild-type band | ND | 161 C>T - stop codon before Met 84 | Died at day +4; liver failure secondary to fibrosis |
PB indicates peripheral blood sample; BM, bone marrow; CCR, complete continuous remission; and ND, not done.
Sequence obtained from direct sequencing of genomic DNA
Nucleotide position 1 is taken from the submitted GenBank sequence of human GATA1 (NM_002049)
GATA1 mutations in TMD/AMKL patients with multiple samples
Pt and diagnosis . | Sex . | Age at diagnosis (mo) . | DNA source . | WBC count, × 109/L (% blast) . | Karyotype . | Sequence in genomic DNA*† . | Cloned sequence from genomic DNA (no. mutant clones/no. total clones)† . | Radiolabeled GATA1 exon 2 PCR products . | Cloned sequence of bands cut from gels (no. mutant clones/no. total clones)† . | Consequence of mutation† . | Clinical status/time since diagnosis . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| L12 | F | CCR/48 mo | |||||||||
| Normal | Birth | BG | — | ND | Wild type | Wild type (0/6) | Wild-type band and faint deletion band | 269 del 23 (5/10) | 269 del 23 - Stop codon before Met 84 | ||
| AMKL | 13 | Diagnosis BM | 8.3 (44) | 48,XX,del (6) (q?13q? 21), +21c,+21[5] | 269 del 23 | 269 del 23 (3/15) | Wild-type band and prominent deletion band 1; faint deletion band 1 | 269 del 23 (8/15); NA | 269 del 23 - Stop codon before Met 84 | ||
| 48, idem, dup (1) (q? 31q 42)[6] | |||||||||||
| 47, XX, +21c[6] | |||||||||||
| Remission | — | CR BM | (3) | 47, XX, +21c[11] | Wild type | ND | Wild-type band | ND | No mutation detected | ||
| L13 | M | CCR/51 mo | |||||||||
| Normal | Birth | BG | — | ND | Wild type | Wild type (0/12) | Wild-type band and faint deletion band 1 | 273 del 17 (1/6) | 273 del 17 - Stop codon before Met 84 | ||
| AMKL | 26 | Diagnosis BM | 3.95 (39) | 47,XY,21+(c)del(6)(?q) [4] | 273 del 17 | ND | Wild-type band and prominent deletion band 1; faint deletion band 1; faint deletion band 2 | 273 del 17 (4/4); NA; NA | 273 del 17 - Stop codon before Met 84 | ||
| 47,XY,21 +(c) in [12] | |||||||||||
| Remission | CR BM | (2) | 47, XY, +21(c)[14] | Wild type | ND | Wild-type band | ND | No mutation detected | |||
| L14 | F | CCR/60 mo | |||||||||
| TMD | Birth | PB smear | 58 (50) | 47, XY, +21(c) | 235 del 132 | 235 del 132 (3/6) | Wild-type band and prominent deletion band | 235 del 132 (3/9) | 235 del 132 - Exon 2 splice mutant | ||
| AMKL | 21 | Diagnosis BM | 5.4 (46) | Hyperdiploid 52 chromosomes | 235 del 132 | 235 del 132 (3/20) | Wild-type band and prominent deletion band | ND | 235 del 132 - Exon 2 splice mutant | ||
| 52 XX, +21(c), +11, +13,+19,+21, +X[2] | |||||||||||
| 52 idem, der3t(1:3)(2?;p2.6) [2] | |||||||||||
| Remission | CR BM | (4) | 47, XX, +21c[8] | Wild type | ND | Wild-type band | ND | No mutation detected | |||
| L15 | M | CCR/4 mo | |||||||||
| Normal | Birth | BG | — | ND | 202 del 2 | 202 del 2 (9/28); 262 ins 1 (2/28); 331A>G (2/28) | Wild-type band | ND | 202 del 2 - Stop codon before Met84; 262 ins 1 - Stop codon before Met 84; 331A>G - Exon 2 splice mutant | ||
| AMKL | 12 | Diagnosis BM | 7.3 (50) | 50,XY,+19,+21c,+21,+22[6] | 202 del 2 | 202 del 2 (6/27) | Wild-type band | ND | 202 del 2 - Stop codon before Met 84 | ||
| 47,XY,+21c[13] | |||||||||||
| Remission | CR BM | (<5) | 47,XY,+21c[10] | Wild type | ND | Wild-type band | ND | No mutation detected | |||
| L17 | M | CCR/42 mo | |||||||||
| Normal | Birth | BG | — | ND | Wild type | Wild type (0/58) | Wild-type band | ND | No mutation detected | ||
| MDS/AMKL | 22 | Diagnosis BM | 7.7 (27) | 48,XY,dup(3) (q2? 5q2? 6.2), der (10) | 332G>A | 332G>A (7/15) | Wild-type band | ND | 332G>A - Exon 2 splice mutant | ||
| t(1:10)(q21;q26), +21c,+21[11] 47,XY,+21c[2] | |||||||||||
| Remission | CR BM | (4) | 47, XX, +21c[11] | Wild type | ND | Wild-type band | ND | No mutation detected | |||
| L16 | M | 2 mo into treatment | |||||||||
| AMKL | 1 | Diagnosis BM | 23.1 (50) | 47,XY,+21c[14] | 161 C>T | 161 C>T(2/10) | Wild-type band | 161 C>T(2/4) | 161 C>T - Stop codon before Met 84 | ||
| Remission | Post-Rx BM | (2) | ND | Wild type | ND | Wild-type band | ND | No mutation detected | |||
| T01 | F | Alive and well at 30 mo | |||||||||
| TMD | Birth | PB smear | 80 (50) | 47, XX, +21(c)[7] | 286 ins 16 (dup270-285) | ND | Wild-type band and insertion band 1 | 286 ins 16 (dup270-285) (5/7) | 286 ins 16 (dup 270-285) - Stop codon before Met 84 | ||
| Remission | 18 | CR PB | 8.6 (0) | ND | Wild type | ND | Wild-type band | ND | No mutation detected | ||
| T02 | M | CCR/85 mo | |||||||||
| TMD | 10 days | PB smear | 250 (44) | 47, XY, +21(c) Mosaic Down[3] | 235 del 132 | 235 del 132 (13/15) | Wild-type band and prominent deletion band | 235 del 132 (3/5) | 235 del 132 - Exon 2 splice mutant | ||
| Remission | 84 | PB | 4.8 (0) | ND | Wild type | ND | Wild-type band | ND | No mutation detected |
Pt and diagnosis . | Sex . | Age at diagnosis (mo) . | DNA source . | WBC count, × 109/L (% blast) . | Karyotype . | Sequence in genomic DNA*† . | Cloned sequence from genomic DNA (no. mutant clones/no. total clones)† . | Radiolabeled GATA1 exon 2 PCR products . | Cloned sequence of bands cut from gels (no. mutant clones/no. total clones)† . | Consequence of mutation† . | Clinical status/time since diagnosis . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| L12 | F | CCR/48 mo | |||||||||
| Normal | Birth | BG | — | ND | Wild type | Wild type (0/6) | Wild-type band and faint deletion band | 269 del 23 (5/10) | 269 del 23 - Stop codon before Met 84 | ||
| AMKL | 13 | Diagnosis BM | 8.3 (44) | 48,XX,del (6) (q?13q? 21), +21c,+21[5] | 269 del 23 | 269 del 23 (3/15) | Wild-type band and prominent deletion band 1; faint deletion band 1 | 269 del 23 (8/15); NA | 269 del 23 - Stop codon before Met 84 | ||
| 48, idem, dup (1) (q? 31q 42)[6] | |||||||||||
| 47, XX, +21c[6] | |||||||||||
| Remission | — | CR BM | (3) | 47, XX, +21c[11] | Wild type | ND | Wild-type band | ND | No mutation detected | ||
| L13 | M | CCR/51 mo | |||||||||
| Normal | Birth | BG | — | ND | Wild type | Wild type (0/12) | Wild-type band and faint deletion band 1 | 273 del 17 (1/6) | 273 del 17 - Stop codon before Met 84 | ||
| AMKL | 26 | Diagnosis BM | 3.95 (39) | 47,XY,21+(c)del(6)(?q) [4] | 273 del 17 | ND | Wild-type band and prominent deletion band 1; faint deletion band 1; faint deletion band 2 | 273 del 17 (4/4); NA; NA | 273 del 17 - Stop codon before Met 84 | ||
| 47,XY,21 +(c) in [12] | |||||||||||
| Remission | CR BM | (2) | 47, XY, +21(c)[14] | Wild type | ND | Wild-type band | ND | No mutation detected | |||
| L14 | F | CCR/60 mo | |||||||||
| TMD | Birth | PB smear | 58 (50) | 47, XY, +21(c) | 235 del 132 | 235 del 132 (3/6) | Wild-type band and prominent deletion band | 235 del 132 (3/9) | 235 del 132 - Exon 2 splice mutant | ||
| AMKL | 21 | Diagnosis BM | 5.4 (46) | Hyperdiploid 52 chromosomes | 235 del 132 | 235 del 132 (3/20) | Wild-type band and prominent deletion band | ND | 235 del 132 - Exon 2 splice mutant | ||
| 52 XX, +21(c), +11, +13,+19,+21, +X[2] | |||||||||||
| 52 idem, der3t(1:3)(2?;p2.6) [2] | |||||||||||
| Remission | CR BM | (4) | 47, XX, +21c[8] | Wild type | ND | Wild-type band | ND | No mutation detected | |||
| L15 | M | CCR/4 mo | |||||||||
| Normal | Birth | BG | — | ND | 202 del 2 | 202 del 2 (9/28); 262 ins 1 (2/28); 331A>G (2/28) | Wild-type band | ND | 202 del 2 - Stop codon before Met84; 262 ins 1 - Stop codon before Met 84; 331A>G - Exon 2 splice mutant | ||
| AMKL | 12 | Diagnosis BM | 7.3 (50) | 50,XY,+19,+21c,+21,+22[6] | 202 del 2 | 202 del 2 (6/27) | Wild-type band | ND | 202 del 2 - Stop codon before Met 84 | ||
| 47,XY,+21c[13] | |||||||||||
| Remission | CR BM | (<5) | 47,XY,+21c[10] | Wild type | ND | Wild-type band | ND | No mutation detected | |||
| L17 | M | CCR/42 mo | |||||||||
| Normal | Birth | BG | — | ND | Wild type | Wild type (0/58) | Wild-type band | ND | No mutation detected | ||
| MDS/AMKL | 22 | Diagnosis BM | 7.7 (27) | 48,XY,dup(3) (q2? 5q2? 6.2), der (10) | 332G>A | 332G>A (7/15) | Wild-type band | ND | 332G>A - Exon 2 splice mutant | ||
| t(1:10)(q21;q26), +21c,+21[11] 47,XY,+21c[2] | |||||||||||
| Remission | CR BM | (4) | 47, XX, +21c[11] | Wild type | ND | Wild-type band | ND | No mutation detected | |||
| L16 | M | 2 mo into treatment | |||||||||
| AMKL | 1 | Diagnosis BM | 23.1 (50) | 47,XY,+21c[14] | 161 C>T | 161 C>T(2/10) | Wild-type band | 161 C>T(2/4) | 161 C>T - Stop codon before Met 84 | ||
| Remission | Post-Rx BM | (2) | ND | Wild type | ND | Wild-type band | ND | No mutation detected | |||
| T01 | F | Alive and well at 30 mo | |||||||||
| TMD | Birth | PB smear | 80 (50) | 47, XX, +21(c)[7] | 286 ins 16 (dup270-285) | ND | Wild-type band and insertion band 1 | 286 ins 16 (dup270-285) (5/7) | 286 ins 16 (dup 270-285) - Stop codon before Met 84 | ||
| Remission | 18 | CR PB | 8.6 (0) | ND | Wild type | ND | Wild-type band | ND | No mutation detected | ||
| T02 | M | CCR/85 mo | |||||||||
| TMD | 10 days | PB smear | 250 (44) | 47, XY, +21(c) Mosaic Down[3] | 235 del 132 | 235 del 132 (13/15) | Wild-type band and prominent deletion band | 235 del 132 (3/5) | 235 del 132 - Exon 2 splice mutant | ||
| Remission | 84 | PB | 4.8 (0) | ND | Wild type | ND | Wild-type band | ND | No mutation detected |
BG indicates Birth Guthrie DNA (neonatal blood spot) sample; PB, peripheral blood sample; BM, bone marrow; Post-Rx, after treatment; CR, complete remission; CCR, complete continuous remission; ND, not done; NA, not available.
Sequence obtained from direct sequencing of genomic DNA
Nucleotide position 1 is taken from the submitted GenBank sequence of human GATA-1 (NM_002049)
Isolation of DNA
DNA was extracted from a number of different sources: cord blood, neonatal blood spots (Guthrie cards), diagnostic bone marrow slides, fresh bone marrow, and peripheral blood samples (QIAamp DNA Mini Kit; Qiagen, Crawley, United Kingdom). The Guthrie cards used had been stored in a hospital laboratory at ambient temperature in a sealed cabinet. They were handled with gloves at all times. Spots used were carefully cut (being careful to avoid the margins of the spot) and individually put into sterile plastic containers. It is unlikely that any 2 Down patients were filed together or that the Down cards selected were allowed to come into contact with each other. However, because they are filed like pieces of paper, they would have come into contact with other, almost certainly non-Down, Guthrie cards. Once in our lab the spots were stored at room temperature in individual sterile universal tubes until the DNA was extracted. For each individual where neonatal blood spots were available, DNA from more than one spot was extracted on separate occasions. The rationale was to verify results obtained in independently processed samples.
GATA1 mutation detection
GATA1 mutations were ascertained in genomic DNA by multiple methods. In all cases GATA1 exon 2 was directly sequenced. Many mutations were nucleotide insertions or deletions that resulted in a double sequence chromatogram trace. To accurately determine the nature of the mutation, GATA1 exon 2 PCR products were cloned and sequenced. To detect minor populations of cells containing GATA1 mutations, radiolabeled PCR reaction products were separated on a polyacrylamide gel (see next paragraph).
PCR, cloning, and sequencing
Polymerase chain reaction (PCR) of GATA1 exon 2 was carried out using 2 distinct sets of previously published primers and conditions.16,19 This was done to ensure results were consistent and not artefactual. No-template and normal-template controls were included. PCR products were purified (Qiagen Minielute Gel Extraction Kit and Minielute PCR Purification Kit; Qiagen) and directly sequenced. Mutations were confirmed by sequencing cloned PCR products. Purified PCR products were cloned into a TA vector (pGEM-T Easy vector; Promega, Southhampton, United Kingdom). For all mutations, enough clones were sequenced to confirm the mutation in 2 independent clones. Bidirectional sequencing was carried out using ABI BigDye terminator sequencing kit v3.1 (Perkin Elmer, Southampton, United Kingdom) and an ABI 3100 Capillary Array sequencer (Perkin Elmer). Sequences were analyzed using MacVector (Accelrys, Cambridge, United Kingdom) and Sequencher v3.1.1 (Gene Codes Corporation, Ann Arbor, MI) software packages.
To generate radiolabeled GATA1 exon 2 PCR products, the forward PCR primer was end labeled with 32Pγ-ATP using standard techniques and then used in a 1:12 ratio of labeled-to-unlabeled primer in the PCR reaction. Five microliters of radiolabeled PCR products were added to 10 μL denaturing solution (95% formamide, 20 mM EDTA [ethylenediaminetetraacetic acid], 0.05% bromophenol blue, and 0.05% xylenecyanol). Samples were denatured by heating at 95°C for 10 minutes and then immediately placed on ice prior to loading on a 6% polyacrylamide (acrylamide-bisacrylamide ratio 19:1) denaturing gel (7M Urea/1X TBE). In some cases after electrophoresis, mutant GATA1 exon 2 PCR products were excised and DNA eluted. Eluted DNA was reamplified and cloned. On other occasions, gels were dried and exposed to Kodak LS film (Kodak, Crawley, United Kingdom) for 4-8 hours at -20°C.
Precautions taken to avoid contamination
Precaution was taken to avoid DNA contamination of DNA samples and PCR products generated. Three separate laboratories were used for DNA extraction, setting up PCR reactions, and cloning PCR products, respectively. Dedicated reagents, Gilson pipettes, filtered barrier tips, and plasticware were used for each of these 3 activities and were kept physically apart. After extraction, DNA samples were immediately aliquoted for single use only.
Results
All patients with AMKL and TMD have mutations in GATA1, which disappear on remission of disease.
All 12 patients with AMKL and 4 patients with TMD had genomic DNA mutations in exon 2 of the GATA1 gene (Tables 1, 2): either insertions and deletions or substitutions within exon 2. The mutations would be predicted either to abrogate splicing of exon 2 or generate a stop codon prior to the alternative translational start codon at position 84 (Tables 1, 2). In either case, the predicted GATA1 protein would start at codon 84 and lack the N-terminal domain. These mutations disappeared with resolution or remission of disease. These results concur with published data on the spectrum of mutations, their predicted functional effect, and association with the disease.
During the course of mutation analysis in AMKL and TMD patients we developed an alternative technique to detect GATA1 mutations compared to those previously reported. As many mutations result in a length change of GATA1 exon 2, radiolabeled GATA1 exon 2 PCR products were separated on polyacrylamide gels. In Figure 1A, examples of this analysis are shown in diagnostic samples from 4 patients with AMKL (L02, L13, L03, and L11). Patients L02, L13, and L03 have an extra mutant band in addition to the wild-type GATA1 exon 2 product. In all 3 patients, the ratio of wild type to mutant band varies. Depending on the ratio of wild type and mutant PCR product, direct sequencing of genomic DNA revealed either a double sequence trace (corresponding to both mutant and wild-type sequences) or mainly a mutant trace (Figure 1B). The mutant bands were excised, cloned, and sequenced (Figure 1C). The sequence of the mutant band was in agreement with that of the mutation inferred by direct sequencing of genomic DNA. By contrast, amplification of exon 2 from DNA of patient L11 only resulted in a wild-type-sized band. Both direct sequence analysis and analysis of cloned products from the excised band identified the mutation as a one-base pair insertion. This patient illustrates that not all mutations will be detected by electrophoresis of radiolabeled PCR products on a polyacrylamide gel. In our study 12 of 21 mutations were detected on polyacrylamide gels.
GATA1 exon 2 mutation detection in AMKL patients. (A) A representative polyacrylamide gel illustrates positions of radiolabeled mutant GATA1 exon 2 PCR products with respect to wild-type product (Wt). Genomic DNA from AMKL patients (L02, L13, L03, and L11; Tables 1-2) and a normal sample (N) was used as template. Positions of DNA size markers (in base pairs) are indicated on the left. (B) Direct sequence analysis of GATA1 exon 2 PCR products from genomic DNA from patients L02, L13, L03, and L11 is shown. Note that in patients L02, L13, and L11, there are 2 superimposed sequence traces in portions of the chromatogram indicative of more than one GATA1 sequence in the template. The predominant sequence trace is wild type, while the mutant sequence is a minor trace. Only the mutant sequence trace is seen for patient L03. This reflects the amount of mutant product (relative to wild type) as seen in the polyacrylamide gel in panel A of this figure. (C) Amplified GATA1 exon 2 PCR products from patients L02, L13, L03, and L11 were cloned and sequenced. Sequence chromatograms from mutant and wild-type clones are shown. Nucleotide position 1 is taken from the submitted GenBank sequence of human GATA1 (NM_002049).
GATA1 exon 2 mutation detection in AMKL patients. (A) A representative polyacrylamide gel illustrates positions of radiolabeled mutant GATA1 exon 2 PCR products with respect to wild-type product (Wt). Genomic DNA from AMKL patients (L02, L13, L03, and L11; Tables 1-2) and a normal sample (N) was used as template. Positions of DNA size markers (in base pairs) are indicated on the left. (B) Direct sequence analysis of GATA1 exon 2 PCR products from genomic DNA from patients L02, L13, L03, and L11 is shown. Note that in patients L02, L13, and L11, there are 2 superimposed sequence traces in portions of the chromatogram indicative of more than one GATA1 sequence in the template. The predominant sequence trace is wild type, while the mutant sequence is a minor trace. Only the mutant sequence trace is seen for patient L03. This reflects the amount of mutant product (relative to wild type) as seen in the polyacrylamide gel in panel A of this figure. (C) Amplified GATA1 exon 2 PCR products from patients L02, L13, L03, and L11 were cloned and sequenced. Sequence chromatograms from mutant and wild-type clones are shown. Nucleotide position 1 is taken from the submitted GenBank sequence of human GATA1 (NM_002049).
GATA1 mutations are present at birth in patients with AMKL
Given that TMD patients have GATA1 mutations at birth, we asked if DS AMKL patients without a known clinical history of antecedent TMD had detectable GATA1 mutations at birth. In 4 such patients matching neonatal blood spots were available (L12, L13, L15, and L17; Tables 1, 2). In 3 of 4 patients (L12, L13, and L15) the GATA1 mutation present at diagnosis was identified in DNA extracted from neonatal blood spots (Tables 1, 2; Figure 2A). By contrast, in the fourth patient L17, GATA1 mutation was detected at diagnosis of AMKL in 7 of 15 clones but not detected in sequence of 58 clones of GATA1 exon 2 PCR products in the neonatal blood spot. In this patient it was not possible to identify the mutation by radiolabeled PCR, as it was a single nucleotide substitution. Unfortunately, none of these 4 babies had a blood count at birth and, therefore, it is not possible to know whether the diagnosis of TMD was missed because the abnormalities were subtle or because there was no TMD.
GATA1 mutations can be clinically silent at birth. Analysis of GATA1 mutations in sequential genomic DNA samples in AMKL patients and healthy DS neonates. (A-B) Sequential genomic DNA samples from patients L12 and L13 (A), and L14 (B) were used as a template to amplify radiolabeled GATA1 exon 2. PCR products were then separated on a polyacrylamide gel. Wt indicates the position of the wild-type GATA1 exon 2 PCR product. DNA samples used were neonatal blood spot DNA (BG); diagnostic AMKL DNA (Diag); and remission DNA sample. Patient L14 presented with TMD at birth, and this sample was used in the analysis. Positions of DNA size markers (in base pairs) are indicated on the left. (C) Genomic DNA from 21 unselected healthy DS neonates and one non-DS control (N) was used as template to amplify radiolabeled GATA1 exon 2. PCR products were then separated on polyacrylamide gel; 2 of 21 (N5 and N6) samples tested had detectable mutations (D-E). (D) Neonate N5 has a mutation in GATA1 exon 2, 218 ins 1, detected by direct sequencing of GATA1 exon 2 PCR product amplified from genomic DNA. Note that there are 2 sequences in the chromatogram indicative of 2 GATA1 PCR products, wild type and mutant. The minor trace sequence corresponds to the mutant product, reflecting its lower abundance relative to wild type. (E) Neonate N6 has a mutation in GATA1 exon 2, 302 ins 11 (dup291-301). The mutant-sized GATA1 exon 2 product (C) was excised from the gel and cloned, and sequence of the cloned product is shown. Wild-type GATA1 exon 2 sequence is illustrated for comparison. The predicted consequences of mutations in both N5 and N6 are shown. In N5, mutation leads to a stop codon before codon 84. In N6, mutation is predicted to terminate at codon 140.
GATA1 mutations can be clinically silent at birth. Analysis of GATA1 mutations in sequential genomic DNA samples in AMKL patients and healthy DS neonates. (A-B) Sequential genomic DNA samples from patients L12 and L13 (A), and L14 (B) were used as a template to amplify radiolabeled GATA1 exon 2. PCR products were then separated on a polyacrylamide gel. Wt indicates the position of the wild-type GATA1 exon 2 PCR product. DNA samples used were neonatal blood spot DNA (BG); diagnostic AMKL DNA (Diag); and remission DNA sample. Patient L14 presented with TMD at birth, and this sample was used in the analysis. Positions of DNA size markers (in base pairs) are indicated on the left. (C) Genomic DNA from 21 unselected healthy DS neonates and one non-DS control (N) was used as template to amplify radiolabeled GATA1 exon 2. PCR products were then separated on polyacrylamide gel; 2 of 21 (N5 and N6) samples tested had detectable mutations (D-E). (D) Neonate N5 has a mutation in GATA1 exon 2, 218 ins 1, detected by direct sequencing of GATA1 exon 2 PCR product amplified from genomic DNA. Note that there are 2 sequences in the chromatogram indicative of 2 GATA1 PCR products, wild type and mutant. The minor trace sequence corresponds to the mutant product, reflecting its lower abundance relative to wild type. (E) Neonate N6 has a mutation in GATA1 exon 2, 302 ins 11 (dup291-301). The mutant-sized GATA1 exon 2 product (C) was excised from the gel and cloned, and sequence of the cloned product is shown. Wild-type GATA1 exon 2 sequence is illustrated for comparison. The predicted consequences of mutations in both N5 and N6 are shown. In N5, mutation leads to a stop codon before codon 84. In N6, mutation is predicted to terminate at codon 140.
Two aspects of our data suggest, perhaps not surprisingly, that the clone containing a GATA1 mutation is smaller at birth than at diagnosis of AMKL in patients without a clinical history of antecedent TMD. First, in 2 of 3 patients (L12 and L13) we could not detect mutant GATA1 exon 2 PCR product in neonatal genomic DNA samples, either by direct sequencing or by sequencing cloned PCR product (Tables 1, 2); GATA1 mutations were detected only by the radiolabeled PCR method. Second, the ratio of mutant to wild-type GATA1 PCR products is greater in the AMKL sample compared to the neonatal sample in the cases where AMKL was not preceded by clinical TMD, in contrast with the one case where AMKL was preceded by clinical TMD (Figure 2B, compare panel B with panel A).
GATA1 mutations are found in otherwise healthy DS neonates at birth
To determine whether GATA1 mutations were detectable at birth in otherwise healthy DS neonates, we obtained neonatal blood spots from a randomly selected group of 21 DS neonates with no clinical findings from their clinical records to suggest TMD. At the time of analysis for GATA1 mutations, the age range of these DS children was between 9 and 53 months and none had AMKL. In view of our data and that previously reported, we confined our analysis to exon 2 of GATA1. GATA1 mutations were found in 2 of 21 samples (Figure 2C-E). Direct sequencing of genomic DNA revealed one sample with a mutant GATA1 sequence chromatogram (N5, age 31 months, Figure 2D). The second mutation was detected as an abnormal-sized radiolabeled PCR product (N6, age 26 months, Figure 2C,E). Sequence analysis revealed that the GATA1 mutation in N5 would be predicted to result in a stop codon before codon 84 (Figure 2D), whereas the mutation in N6 would result in production of a divergent protein from the site of insertion at Tyr 63 till a stop codon occurs at position 140. Once again, these data raise the question of whether the diagnosis of TMD was missed in the 2 neonates with GATA1 mutations as blood films and counts were not performed.
We also studied a control group of 62 cord blood samples from non-DS neonates by direct sequencing and separating radiolabeled GATA1 exon 2 PCR products on polyacrylamide gels. GATA1 exon 2 mutations were not detected in any of these samples (data not shown).
Some patients have multiple GATA1 mutations
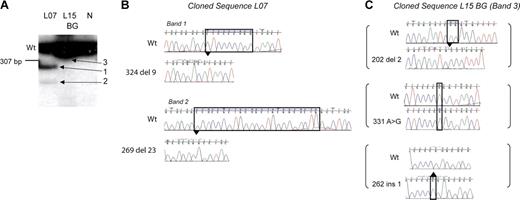
The use of radiolabeled PCR along with sequencing of mutant bands showed an unexpected novel observation: 4 of 12 patients who developed DS AMKL had multiple independent GATA1 mutations. In 3 patients (L07, L12, and L13; Tables 1, 2) multiple radiolabeled GATA1 exon 2 PCR products were seen in the diagnostic AMKL sample, suggesting multiple mutant GATA1 containing clones (Figure 2A, Figure 3). This observation was confirmed using 2 independent primer pairs (see “Patients, materials, and methods”). Sequence analysis of the excised cloned mutant PCR products in one patient, L07, also substantiated this finding (Figure 3B). Each band contained one distinct GATA1 mutation. Importantly, in all 3 patients, only the sequence corresponding to the major mutant band was observed on direct genomic sequencing. In the fourth patient (L15), sequence of DNA from neonatal blood showed 3 independent mutations: each mutant GATA1 clone contained one mutation, and there were 3 mutations in all (Figure 3C). Interestingly, in the diagnostic AMKL sample only 1 of the 3 mutations was present (Table 1). The presence of multiple mutations was confirmed in 3 patients in fresh DNA samples obtained subsequently either from archival slides (L12 and L13) or a neonatal blood spot (L15).
Multiple independent GATA1 mutations are present. (A) Radiolabeled GATA1 exon 2 PCR products generated from genomic template of patient L07 and L15's neonatal blood spot (BG) and a healthy control (N) were separated on a polyacrylamide gel. The wild-type GATA1 product is marked (Wt). Patient L07 has a single wild-type-sized band and 2 extra bands (bands 1 and 2). L15 has only one wild-type-sized product (panel C, band 3). (B-C) Radiolabeled GATA1 exon 2 bands from patient L07 and L15's BG shown in panel A were excised, cloned, and sequenced. Sequence chromatograms of clones from patient L07 (B) and L15 (C) are shown. Mutant traces are illustrated, and wild-type (Wt) traces are depicted for comparison.
Multiple independent GATA1 mutations are present. (A) Radiolabeled GATA1 exon 2 PCR products generated from genomic template of patient L07 and L15's neonatal blood spot (BG) and a healthy control (N) were separated on a polyacrylamide gel. The wild-type GATA1 product is marked (Wt). Patient L07 has a single wild-type-sized band and 2 extra bands (bands 1 and 2). L15 has only one wild-type-sized product (panel C, band 3). (B-C) Radiolabeled GATA1 exon 2 bands from patient L07 and L15's BG shown in panel A were excised, cloned, and sequenced. Sequence chromatograms of clones from patient L07 (B) and L15 (C) are shown. Mutant traces are illustrated, and wild-type (Wt) traces are depicted for comparison.
Discussion
GATA1 mutations were identified in all 12 DS patients with AMKL and 4 patients with TMD, confirming similar findings in published studies. All mutations were in exon 2, and the spectrum was similar to those previously reported.16-20 GATA1 mutations were not detectable in samples taken from patients in remission, using the assays employed. This observation suggests that GATA1 mutations are disease specific.
Three novel findings arise from our study. First, GATA1 mutations were present at birth in 3 of 4 patients who developed AMKL even though they had no clinical history of TMD after careful review of their clinical records. Second, in a cohort of 21 randomly selected and otherwise healthy DS neonates, 2 babies had GATA1 mutations and are currently aged 26 and 31 months. While our study has small sample numbers, the latter 2 findings taken together are consistent with each other and suggest that GATA1 mutations may occur without clinical signs of disease, although these babies may have had unsuspected hematologic abnormalities. Our data are consistent with GATA1 mutations occurring in a minor clone within a mixed cell population. Third, in 4 of 12 DS AMKL patients multiple GATA1 mutations were detected.
Our data, together with those from other investigators suggesting acquisition of GATA1 mutations in utero by most (if not all) DS AMKL patients, has striking parallels with studies demonstrating that other pediatric leukemia-initiating mutations also originate in utero.33-36 Our failure to detect a GATA1 mutation in the neonatal blood spot from 1 of 4 patients with AMKL without an antecedent clinical history of TMD (L17) may mean that it was absent at birth; on the other hand it seems more likely that the result reflects the rarity of the mutant clone (ie, the mutation was present in the neonatal blood spot sample but was not detected as we were unable to exploit the radiolabeled PCR technique or because there were insufficient nucleated cells in the neonatal blood spot). Analysis of neonatal blood spots for other common pediatric leukemiaassociated mutations (MLL-AF4, TEL/AML-1, and AML-1/ETO) also fails to detect all mutations at birth.33,34 TEL/AML-1 and AML-1/ETO mutations are present at birth at a frequency of 1 in 104 to 1 in 105 cells in cord blood. Neonatal blood spots contain 30 000 nucleated cells on average.33
Further studies will have to be performed to determine the method of choice to detect GATA1 mutations that combines sensitivity and ability to ascertain all mutations. There are at least 2 issues here. The first is the ability to detect minor mutant GATA1 populations, and incorporating either a radiolabel or a fluorescent tag to the DNA would be helpful in this regard. The second issue is to be able to detect both point mutations and mutations that result in length changes. SSCP (single strand conformation polymorphism) and the technique we employed may have similar sensitivities as both involve separating radiolabeled products on native acrylamide gels. SSCP has the advantage of detecting point mutations, though not all point mutations are detected. Denaturing highpressure liquid chromatography (DHPLC) combines ability to detect point mutations, length changes, and ease of use. It can be used to separate radiolabeled PCR products to increase sensitivity of the assay.
Our findings confirm that GATA1 mutations are present at high frequency in DS neonates. Indeed, for the first time multiple GATA1 mutations were detected, suggesting multiple GATA1 mutant clones coexist in an individual. There are at least 2 broad explanations for this. First, cells with trisomy 21 may have a mutator phenotype comparable to those with defective DNA repair machinery. However, we think this unlikely because, if this were the case, trisomy 21 cells should accumulate genomewide mutations in all cells, and DS individuals should be prone to a variety of types of cancers and not just hematologic malignancies, and this is not the case.10,37,38 Second, when N-terminal truncation of GATA1 protein occurs specifically in fetal trisomy 21 hematopoietic cells, it could confer a selective advantage. This may be specific to DS cells because of altered expression of any number of chromosome 21 genes. One of these includes RUNX-1, a key hematopoietic gene often deregulated in leukemogenesis. The competitive advantage afforded by such GATA1 mutation is remarkable as mutations occur as multiple independent events in 4 patients. The selection hypothesis is credible as loss of GATA1 activity leads to accumulation of immature megakaryoblasts.26,27 However, an obvious paradox raised by the selection hypothesis is that if selection were operative, why does the TMD clone extinguish, resulting in a self-limiting illness? The answer to this question may simply be that though GATA1 mutation provides a proliferative advantage, it fails to immortalize the clone. Thus, the clone eventually extinguishes. Over and above this, it may be that GATA1 mutations occur in progenitors of limited lifespan and/or that the postnatal environment fails to support the fetal hematopoietic clones with GATA1 mutations.
From the above discussion we suggest that our data have implications for the pathogenesis of TMD and DS AMKL (Figure 4). We speculate that a proportion of DS fetuses acquire GATA1 mutations during gestation. In those fetuses that acquire a GATA1 mutation, the neonatal clinical phenotype could depend on the size of the clone bearing the GATA1 mutation. Thus, a clone large enough to impair normal hematopoiesis may result in clinically evident TMD. In fetal hematopoietic cells trisomic for chromosome 21, GATA1 mutation is likely to be a required event for TMD. However, for the first time it is clear from our data that the detectable GATA1 mutations at birth do not always result in clinically overt TMD. Both clinically silent and clinically overt TMD clone(s) could then undergo clonal extinction as discussed above. However, we speculate that additional postnatal genetic (and epigenetic) events could also occur to transform the GATA1 mutant clone and result in AMKL. This model is consistent with other data on the molecular epidemiology of non-DS pediatric leukemias.36
GATA1 mutations in TMD and AMKL. A model of the relationship between GATA1 mutations and the clinical phenotypes of TMD and AMKL in DS infants and children.
GATA1 mutations in TMD and AMKL. A model of the relationship between GATA1 mutations and the clinical phenotypes of TMD and AMKL in DS infants and children.
Observations made in this study also raise unanswered questions. First, do neonates with GATA1 mutations who do not present with clinically signs of TMD have hematological abnormalities consistent with a diagnosis of TMD? In our study, neonates will have been examined by the pediatric team at birth but, as is standard practice, will not have had a blood count and film at that time. As TMD may manifest with only the presence of circulating blasts (with or without an abnormal blood count), hematologic abnormalities consistent with TMD may have been present in those neonates with GATA1 mutations without a clinical history of TMD, but we are unable to ascertain this because no blood tests were performed at that time. If, as seems likely, acquisition of GATA1 is a required event for TMD, it raises interesting questions about how to define TMD and the relative sensitivity of detection of GATA1 mutations versus circulating blasts in making the diagnosis of TMD. It is conceivable that GATA1 mutations could be detected by PCR in neonates without circulating blasts. In these cases would the neonates be considered to have TMD? Alternatively, it is noteworthy that in our small survey of a group of newborn DS babies GATA1 mutations were present in approximately 9.5% (2 of 21), similar to the frequency (10.4%) with which TMD blasts were detected in a prospective study of 77 blood films of DS newborns.6
Second, what is the risk that neonate with a GATA1 mutation will develop AMKL? Our work, together with previous work, confirms that most if not all cases of TMD have GATA1 mutations. Given that up to 30% of TMD patients develop AMKL, one could have argued that up to 30% of patients with GATA1 mutations develop AMKL. However, as our work demonstrates, for the first time, that GATA1 mutations can be present without any clinical signs of disease, the risk of a neonate with a GATA1 mutation developing leukemia is now unclear. Although our study has established the feasibility of screening for GATA1 mutations in a randomly selected cohort of DS babies at birth, the sample we have studied is too small (21 neonates) to determine the precise prevalence of GATA1 mutations in DS neonates. Moreover, though the 2 randomly selected neonates in whom we have identified GATA1 mutations have not yet developed AMKL, the length of follow up is short and they are still at risk of developing AMKL.
The answers to both questions we identify above needs to be addressed in a larger prospective cohort study of DS neonates with adequate follow up, in which GATA1 mutation status is assayed (with a carefully chosen experimental protocol to detect all GATA1 mutations), automated blood counts, and peripheral blood smears. Such a study may also reveal if minor GATA1 mutant clones subtly alter peripheral blood hematology. In addition to providing invaluable detail on the epidemiology and clinical and hematologic consequence of GATA1 mutation in DS, the potential clinical value from such a study could be to identify markers that define a subset of DS neonates at higher risk of developing AMKL later and, importantly, exclude those infants who will not.
M. Ahmed is funded by the Wellcome Trust. A.S. is the recipient of a Clinical Fellowship from the MRC (United Kingdom), and P.V. is funded by a Wellcome Trust Senior Clinical Fellowship.
Prepublished online as Blood First Edition Paper, December 4, 2003; DOI 10.1182/blood-2003-10-3383.
M. Ahmed and A.S. contributed equally to this work.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked ”advertisement” in accordance with 18 U.S.C. section 1734.
We thank the parents and children who took part in our study. We are grateful to Dr Ajay Vora, Dr Mary Morgan, Dr Jonathan Kay, Dr Beena Pushkaran, Ian Smith, and Jenny Britton, who helped collect samples and obtain consent. We thank Dr Catherine Porcher, Professor Andrew Wilkie, Dr Bill Wood, Professor Doug Higgs, and Professor Sir David Weatherall for fruitful discussions and Nicki Ventress, Linda Roberts, and Liz Rose for assistance in compiling the manuscript. This work is dedicated to Dr Richard Stevens.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal