Abstract
Juvenile or type 2 hemochromatosis (JH) is a genetic disease caused by increased intestinal iron absorption that leads to early massive iron overload. The main form of the disease is caused by mutations in a still unknown gene on chromosome 1q. Recently, we recognized a second type of JH with clinical features identical to the 1q-linked form, caused by mutations in the gene encoding hepcidin (HEPC). Hepcidin is a hepatic antimicrobial-like peptide whose role in iron homeostasis was first defined in animal models; deficiency of hepcidin in mice leads to iron overload, whereas its hepatic overexpression in transgenic animals causes iron deficiency. To define the prevalence of HEPC mutations in JH we screened the HEPC gene for mutation in 21 unrelated JH subjects. We identified a new mutation (C70R), which affects 1 of the 8 conserved cysteines that form the disulfide bonds and are critical for the stability of the polypeptide.
Introduction
Type 2 or juvenile hemochromatosis (JH) is a rare autosomal recessive disorder characterized by early onset and severe iron overload. The principal clinical manifestations, cardiomyopathy and hypogonadism, appear before the age of 30 years. Patients of both sexes have greatly increased transferrin saturation and serum ferritin concentrations at diagnosis and require intensive phlebotomy to achieve iron depletion.1
JH is a heterogeneous genetic disorder, related to at least 2 distinct loci. The first maps to chromosome 1q21,2 but the causal gene is still unknown. A rare subset is due to mutations in hepcidin (HAMP or HEPC)3 gene, which maps to chromosome 19q13.4 As in humans, inactivation of HEPC in mice leads to severe iron overload,5 whereas its overexpression in transgenic mice leads to iron deficiency anemia.6 Recently, it has been demonstrated that an inappropriately low expression of HEPC mRNA is constant in hemochromatosis related to the HFE gene, both in humans7,8 and in animal models.9-12 This implies that HFE is involved in HEPC regulation and further strengthens the concept that inability to maintain appropriate hepcidin levels is central to the development of iron excess.
Hepcidin protein shows high similarity with several cysteinerich antimicrobial peptides.4,13 It is prevalently expressed by the hepatocytes as a precursor protein of 84 amino acids. Three active peptides originate from the propeptide by protease cleavage, respectively 25, 22, and 20 amino acids long.4,13 These soluble forms of hepcidin have been isolated from the urine of healthy subjects. The 25– and 20–amino acid peptides represent the major forms, whereas the 22–amino acid peptide is present only at low concentration.4 A striking feature of the active peptides is the numerous cysteines,8 accounting for 32% of the total amino acid content. Analysis of soluble hepcidin species for both the 20 and the 25 residues shows that the 8 cysteines form 4 disulfide bonds, providing a rigid and tight structure to the final peptide14 (Figure 1D). Extensive promoter analysis has revealed the presence of consensus sequences for the transcription factor CCAAT/enhancer binding protein-α (CEBP/α), which confers liver tissue specificity.15 Hepcidin synthesis is increased by iron loading and inflammation and is inhibited by iron deficiency anemia and hypoxia.16,17
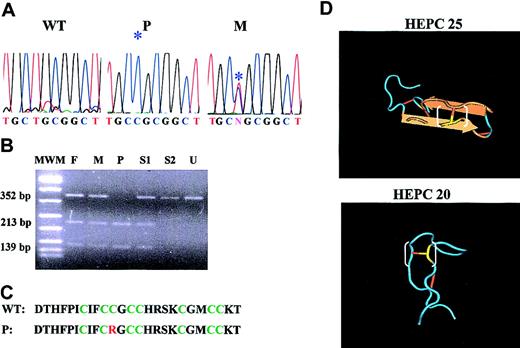
Molecular and biologic data of the C70R mutation. (A) Electrophoretograms of the HEPC genomic sequence spanning the C70R mutation. The sequence is shown for DNA of a wild-type subject (WT), proband (P), and the heterozygous mother (M). *indicates the mutation. (B) Restriction analysis of C70R mutation by SacII in the proband family. MWM indicates molecular weight marker; F, father; M, mother; P, proband; S1 and S2, siblings; and U, undigested fragment. (C) Comparison of wild-type and mutated peptide amino acid sequence. The 8 cysteines are indicated in green, the mutated arginine in red. (D) Three-dimensional structure of HEPC 25 and 20 amino acid peptides (from http://www.ncbi.nlm.nih.gov/). The mutated cysteine is indicated in yellow; a white bracket marks the S-S bond disrupted by the mutation.
Molecular and biologic data of the C70R mutation. (A) Electrophoretograms of the HEPC genomic sequence spanning the C70R mutation. The sequence is shown for DNA of a wild-type subject (WT), proband (P), and the heterozygous mother (M). *indicates the mutation. (B) Restriction analysis of C70R mutation by SacII in the proband family. MWM indicates molecular weight marker; F, father; M, mother; P, proband; S1 and S2, siblings; and U, undigested fragment. (C) Comparison of wild-type and mutated peptide amino acid sequence. The 8 cysteines are indicated in green, the mutated arginine in red. (D) Three-dimensional structure of HEPC 25 and 20 amino acid peptides (from http://www.ncbi.nlm.nih.gov/). The mutated cysteine is indicated in yellow; a white bracket marks the S-S bond disrupted by the mutation.
Here we report the results of a study aimed at identifying new HEPC mutations. During the investigation of a large series of patients with JH, we detected a homozygous nucleotide change that causes a missense (C70R) affecting one of the highly conserved cysteines. The mutation was found in a single patient of a consanguineous Italian family, previously considered affected by the 1q-linked type of JH.
Study design
Twenty-one unrelated patients with JH were studied. The clinical features of most cases have been reported previously.18,19 Six new cases were diagnosed using accepted criteria.18
DNA was prepared from peripheral blood, according to standard protocols. HEPC coding sequences (NT_011109) were amplified by polymerase chain reaction (PCR). Primers used for the amplification reaction are reported in Table 1.
Sequences of the HEPC primers used in the mutation screening
Primer . | Sequence . | PCR product length, bp . |
|---|---|---|
| Exon 1 | 206 | |
| F | 5′CAAGCTCAAGACCCAGCAGT3′ | |
| R | 5′CAGTGCCCTAGGCTGA3′ | |
| Exon 2 | 192 | |
| F | 5′CAGTCTCAGAGGTCCACT3′ | |
| R | 5′AATGTGAGCAGGGAACC3′ | |
| Exon 3 | 352 | |
| F | 5′CAGTGATGCCTTTCCTAGC3′ | |
| R | 5′AAGGCAGGGTCAGGACAA3′ |
Primer . | Sequence . | PCR product length, bp . |
|---|---|---|
| Exon 1 | 206 | |
| F | 5′CAAGCTCAAGACCCAGCAGT3′ | |
| R | 5′CAGTGCCCTAGGCTGA3′ | |
| Exon 2 | 192 | |
| F | 5′CAGTCTCAGAGGTCCACT3′ | |
| R | 5′AATGTGAGCAGGGAACC3′ | |
| Exon 3 | 352 | |
| F | 5′CAGTGATGCCTTTCCTAGC3′ | |
| R | 5′AAGGCAGGGTCAGGACAA3′ |
PCR was performed in a Gene Amp PCR System 2400 (Applied Biosystems, Foster City, CA), using 25 pmol of each primer and 50 ng template DNA, with an average protocol of 32 cycles (denaturation: 94°C 30 seconds; annealing: 56°C 30 seconds; extension: 72°C 45 seconds) and 1 U AmpliTaq DNA polymerase (Roche Applied Science, Indianapolis, IN).
For direct sequencing, PCR products were run on 1% agarose gel, purified using QIAquick PCR purification kit (Qiagen, Valencia, CA) and sequenced using Thermo Sequenase Cy5.5 dye terminator cycle sequencing kit. After purification from unincorporated dye with Autoseq G-50 columns, sequencing products were electrophoresed in an automatic sequencer (373A; Applied Biosystems) according to the manufacturer's protocols.
Restriction endonuclease digestion was carried out using 20 μL exon 3 PCR product and 10 U SacII enzyme (New England Biolabs, Beverly, MA) in a final volume of 30 μL for 2 hours.
Results and discussion
Most published information on JH families indicates that they have a genetic disorder that maps to chromosome 1q.2,20,21 Homozygous mutations in HEPC gene were identified in a rare subset of JH patients, with a phenotype indistinguishable from the 1q-linked form.3 To establish the frequency of HEPC mutations in JH, we sequenced the HEPC gene of patients from 21 families, regardless of their linkage with chromosome 1q, because linkage may be coincidental when the number of family members is small or the information from the genetic markers limited. We identified a T>C mutation at the homozygous state in exon 3 (at position 208 from the starting ATG; Figure 1A) in a young Italian patient from a consanguineous family. The proband was an 11-year-old boy with high levels of transferrin saturation, serum ferritin, and liver iron content, but without clinical complications. On the basis of the correct segregation of microsatellite alleles of chromosome 1q within the family, the disorder was previously considered 1q linked.19 The identified nucleotide change causes the substitution of the cysteine at position 70 with arginine (C70R). Because the mutation creates a restriction site for SacII enzyme, SacII digestion of HEPC exon 3 was used to demonstrate the correct segregation of the mutation within the family (Figure 1B) and to show the absence of the same mutation in 50 healthy controls (not shown). No mutations in HEPC coding region were detected in the remaining 20 patients.
C70R is a missense mutation that changes an amino acid whose role is pivotal for the final conformation of the protein. All hepcidin cysteines are highly conserved among the different species,4 indicating that their role is essential for the function of the mature protein. Also, the C70R mutation is predicted to disrupt the mature peptide, because a neutral amino acid is substituted by the basic arginine. According to the nuclear magnetic resonance (NMR) and structural analysis,14 the substituted arginine should disrupt the disulfide bond between the third and the sixth cysteine (Figure 1C) of the peptide. Taken together, these observations support the causal role of C70R mutation in the molecular pathogenesis of the disease.
C70R is the first missense mutation identified at the homozygous state in HEPC. It is of interest that an amino acid change (G71D), reported to cause hemochromatosis when associated with heterozygous C282Y,22,23 occurs adjacent to position 70. G71D occurs independently in patients of different origin.22,23 The digenic inheritance of mutations in HEPC and HFE (both G71D and C282Y at the heterozygous state) in iron-loaded patients suggests that reduced expression of hepcidin and HFE proteins synergizes in the effect on iron homeostasis. Heterozygous carriers of the proband family, as well as heterozygous carriers of the previously reported HEPC mutations,3 have normal iron parameters. However, no one had coinherited C282Y at the heterozygous state.
Our results indicate that HEPC mutations leading to JH are rare. Several lines of evidence indicate that hepcidin is a key regulator of iron homeostasis, but its mode of action is little understood. In this context, the identification of HEPC mutations is not only a new diagnostic tool for JH, but may also provide insights into the relationship between the structure of its cognate peptide and its function.
Prepublished online as Blood First Edition Paper, November 20, 2003; DOI 10.1182/blood-2003-10-3390.
Supported in part by Telethon Foundation grant no. GP00255Y01, European Community, Contract QLRT-1999-02237, and Fondo per gli Investimenti per la Ricerca di Base (FIRB) from Italian Ministry of Instruction Research and University (C.C.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal