Abstract
Interleukin-6 (IL-6) is a growth and antiapoptotic factor for human myeloma cells. The autocrine loop and increased expression of the growth factor receptors have been postulated as the mechanisms of tumorigenesis. Here we show that IL-6 stimulation induced the phosphorylation of insulin-like growth factor-I (IGF-I) receptors in a human myeloma cell line, NOP2, highly expressing IL-6 receptor α (IL-6Rα) and in the IL-6Rα–transfected U266 cell line. IL-6–dependent complex formation of IL-6Rα with IGF-I receptor β was found in NOP2 where IL-6Rα colocalized with IGF-I receptors at lipid rafts. Moreover, the IL-6–induced phosphorylation of IGF-I receptor β was not blocked by a Janus kinase 2 (Jak2) inhibitor. In addition to the activation of the signal transducer and activator of transcription 3 and extracellular signal-regulated kinase 1/2, IL-6 stimulation led to the activation of Akt, presumably following the phosphorylation of IGF-I receptors. Thus, our results suggest that in NOP2, IL-6Rα and IGF-I receptors exist on the plasma membrane in close proximity, facilitating the efficient assembly of 2 receptors in response to IL-6. The synergistic effects of highly expressed IL-6Rα on IGF-I receptor–mediated signals provide a novel insight into the Jak-independent IL-6 signaling mechanism of receptor cross-talk in human myeloma cells.
Introduction
Multiple myeloma (MM) is a hematopoietic tumor characterized as the monoclonal accumulation of malignant plasma cells. The growth of myeloma cells is mediated by the autocrine and paracrine secretion of interleukin-6 (IL-6),1,2 and IL-6 production from myeloma cells is enhanced by IL-13 or CD40 stimulation,4,5 leading to accelerated autocrine growth. IL-6 also has an antiapoptotic effect on myeloma cells,6,7 and thus, IL-6 supports the survival and expansion of myeloma cells by both stimulating cell proliferation and preventing apoptosis.
IL-6 belongs to a family of IL-6 proteins, including IL-11, leukemia inhibitory factor, oncostatin M, cardiotrophin-1, and ciliary neurotrophic factor, and the receptors of these family members share gp130 as a signal transducing molecule.8 Accordingly, some IL-6 family members can be growth factors for myeloma cells.9 The IL-6 receptor complex consists of IL-6Rα and gp130, and the latter is responsible for signal transduction.8 As both IL-6Rα and gp130 lack kinase domains, the association of gp130 with Janus kinases (Jaks) is thought to be critical for IL-6–mediated signals. The activated Jaks phosphorylate tyrosine residues in the cytoplasmic region of gp130. The phosphorylated gp130 recruits the signal transducer and activator of transcription 3 (Stat3) whose tyrosine residues are also phosphorylated by Jaks. The phosphorylated Stat3 forms dimers and translocates to the nucleus.10,11 Another major signal transduction pathway via gp130 is the Ras–mitogen activated protein kinase (MAPK) pathway by the complex formation of Src homology 2 domain containing tyrosine phosphatase (SHP-2) and growth factor receptor binding protein-2 (Grb2) with gp130.12 Activation of both the Stat and Ras-MAPK pathways is necessary for cell proliferation.12 The phosphatidylinositol-3 kinase (PI3-K) that prevents hepatocytes from undergoing apoptosis13 is also activated via gp130.14
Some growth factors, such as insulin,15 insulin-like growth factor-I (IGF-I),16,17 and basic fibroblast growth factor (bFGF/FGF-2),18 whose receptors have tyrosine kinase domains, have recently been reported to function as growth and survival factors for MM. Most receptor tyrosine kinases (RTKs) are composed of a single polypeptide chain and are monomeric in the absence of their ligands. Ligand binding to the extracellular portion of RTKs leads to the dimerization of receptors, resulting in autophosphorylation of specific tyrosine residues in the cytoplasmic portion.19 Tyrosine autophosphorylation either stimulates the intrinsic catalytic (kinase) activity of the receptor or generates recruitment sites for the downstream signaling proteins containing phosphotyrosine-recognition domains, such as Src homology 2 (SH2) domain or phosphotyrosine-binding domain.20 Insulin and IGF-I receptors consist of the disulfide-linked dimers of 2 polypeptide chains that form α-β heterodimers. Insulin or IGF-I binds to the extracellular α subunits of the receptors, subsequently activating the intracellular tyrosine kinase domain of the β subunits via rearrangement within the quaternary structure of heterotetrameric (α2-β2) receptors. Once activated, the insulin/IGF-I receptors phosphorylate a number of important proximal substrates, including members of the insulin receptor substrate 1 (IRS-1), IRS-2, IRS-3, and IRS-4, and Src homology/collagen (Shc) adapter proteins.21 Tyrosine phosphorylation of IRS protein creates recognition sites for additional effector molecules containing SH2 domains. These include the small adapter proteins Grb2, noncatalytic region of tyrosine kinase (Nck), SHP2 protein tyrosine phosphatase, and the regulatory subunit of PI3-K. The targets of PI3-K action are 2 classes of serine/threonine kinases, Akt, and atypical protein kinase C isoforms ζ and λ.22 Akt is a key mediator for cell proliferation and survival by the phosphorylation of several target molecules, such as Bad,23 forkhead transcription factor (FKHR),24 caspase-9,25 mammalian target of rapamycin (mTOR),26 IκB-α,27 p70S6 kinase,28 murine double minute 2 (MDM2),29 p21Cip1/Waf1,30 and p27Kip1.31-33
The altered structure or enhanced expression of the RTKs, the most typical being proto-oncogene products, is involved in the transformation of some cell types.34-37 Mutated receptors may become constitutive active kinases, and the increased expression of the receptors may amplify the receptor-mediated growth signals. However, the physiologic consequences of the increased expression of cytokine receptors on tumor cells remain to be understood. Although IL-6 is a growth factor for human myeloma cells, its biologic functions are quite variable in different types of cells.38 In this study we found that IL-6 was a potent growth and survival factor for NOP2 expressing high levels of IL-6Rα in the serum-free condition, and that Akt, as well as Stat3 and extracellular signal-regulated kinase 1/2 (ERK1/2), was unusually activated by IL-6 and contributed to the cell survival of NOP2. The elevated expression of IL-6Rα on NOP2 led to the Jak-independent phosphorylation of IGF-I receptors in response to IL-6, resulting in the activation of PI3-K and Akt that subsequently suppressed the functions of FKHR, p27Kip1, and p53. This study provided evidence of the cross-talk between a cytokine receptor, IL-6Rα, and a growth factor receptor, IGF-I receptor β, on cell membranes harboring increased IL-6Rα.
Materials and methods
Myeloma cells from the patients, myeloma cell lines, and cell culture
Mononuclear cell fractions were isolated from bone marrow aspirates from 3 myeloma patients (MM1: immunoglobulin A (IgA)–κ, stage II; MM2: IgG-λ, stage II; MM3: IgG-κ, stage III), and CD38++ MPC-1- immature myeloma cells were collected by a cell sorter (Epics Elite ESP; Coulter, Hialeah, FL) as reported previously.39 These sorted myeloma cells were cultured in RPMI1640 medium (Nissui, Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS; M. A. Bioproducts, Walkersville, MD) with a cell-culture insert well (1.0-μm pore size; Becton Dickinson, Franklin Lakes, NJ) at 37°C with 5% CO2. All samples were obtained from the patients with appropriate informed consent. This study was performed according to the guidelines of the Ethical Review Committee of Gene Analysis Research for the Japanese government. ILKM240 and ILKM3,40 as IL-6–dependent myeloma cell lines, and NOP241 and U266, as IL-6–independent myeloma cell lines, were cultured with or without 10% FBS. ILKM2 and ILKM3 were maintained in the presence of IL-6 (2 ng/mL; Sawady Technology, Tokyo, Japan). A synthetic medium composed of RPMI1640 with 200 μg/mL AlbuMax (GibcoBRL, Grand Island, NY) and 2 μg/mL transferrin (GibcoBRL) was used as a serum-free culture.
Growth factors and cell survival assay
Myeloma cells isolated from the MM patients were cultured in a cell-culture insert well containing complete medium with or without IL-6 (2 ng/mL) or IGF-I (100 ng/mL) for 14 days. Myeloma cell lines were cultured in a serum-free medium with either IL-6 (1 ng/mL), insulin (3 or 30 ng/mL), IGF-I (3 or 30 ng/mL), epidermal growth factor (100 ng/mL; EGF), bFGF (100 ng/mL), vascular EGF (100 ng/mL; VEGF), stem cell factor (100 ng/mL; SCF), macrophage colony-stimulating factor (100 ng/mL; M-CSF), or platelet-derived growth factor (100 ng/mL; PDGF) for 2 to 10 days. Cell viability was examined by flow cytometry with forward and side scatters (Epics Elite ESP). Insulin, IGF-I, EGF, SCF, M-CSF, and PDGF were purchased from Sigma (St Louis, MO), and bFGF and VEGF were obtained from Techne (Minneapolis, MN).
Cell surface antigen expression
Cells (1 × 105) were stained with either the phycoerythrin (PE)–conjugated antihuman CD126 (IL-6Rα) antibody (Coulter) or the fluorescein isothiocyanate (FITC)–labeled antihuman IGF-I receptor antibody (R&D Systems, Minneapolis, MN) at 4°C for 30 minutes followed by flow cytometry (Epics Elite ESP) as described previously.42
Nuclear and cytoplasmic extracts
Nuclear and cytoplasmic fractions were isolated by the modified method of Schreiber et al.43 Briefly, 2 to 4 × 106 cells were suspended in 400 μL buffer A (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH7.9], 10 mM KCl, 0.1 mM EDTA [ethylenediaminetetraacetic acid], 0.1 mM EGTA [ethylene glycol tetraacetic acid], 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.1 mM Vanadate, 10 μg/mL aprotinin, 10 μg/mL pepstatin, and 10 μg/mL leupeptin), and incubated on ice for 15 minutes. Following incubation, 10% nonidet P-40 (25 μL) was added, and the samples were then vigorously vortexed. After centrifugation, supernatants were used as cytoplasmic fractions. Pellets were resuspended in 50 μL ice-cold buffer B (20 mM HEPES, pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, and phosphatase inhibitors) and vigorously shaken. After centrifugation, supernatants were used as nuclear fractions. Protein concentrations of cytoplasmic and nuclear extracts were evaluated by the Bradford assay as described.42
Western blot analysis
Cells cultured without serum for 3 hours were stimulated with either IL-6 (2 ng/mL), IGF-1 (100 ng/mL), or insulin (100 ng/mL) for 0, 10, and 30 minutes, and cell lysates were prepared by adding lysis buffer followed by Western blot analysis as described.42 Specific antibodies for Stat3, ERK1/2, Akt, FKHR, Bad, IκB-α, caspase-9, phosphorylated Stat3 (Tyr705), phosphorylated ERK1/2 (Thr202/Thr204), phosphorylated IκB-α (Ser32), phosphorylated Bad (Ser112) or Bad (Ser136), phosphorylated Akt (Ser473) or Akt (Thr308), and phosphorylated FKHR (Ser256) or FKHR (Thr24)/FKHRL1 (Thr32) were purchased from Cell Signaling Technology (Beverly, MA). Antibodies recognizing p27Kip1 or p53 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Immunoprecipitation and Western blot analysis
Cells incubated in a serum-free medium for 3 hours (ILKM2 and ILKM3) or for 9 hours (NOP2) were treated with or without reagents for 5 or 10 minutes. Cell lysis, immunoprecipitation, and Western blot analysis were described previously.42 Phosphorylation of tyrosine residues of IGF-I receptor β and IRS-1 was detected by an antiphosphotyrosine antibody 4G10 (Upstate Biotechnology, Lake Placid, NY). Anti–IGF-I receptor β, insulin receptor β, IRS-1, IRS-2, IL-6Rα, and gp130 antibodies were bought from Santa Cruz Biotechnology. The activation of Jak2 was detected by a phospho-specific Jak2 (Tyr1007/1008) antibody (Cell Signaling).
RNA extraction and reverse transcription–polymerase chain reaction (RT-PCR)
After the stimulation of cells with reagents for several time periods, total RNA was extracted using TRIZOL (GibcoBRL) followed by first-strand cDNA synthesis using 200 U Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. For conventional PCR, 1 μL cDNA was amplified using 2.5 U Taq DNA polymerase (Takara Bio, Kusatsu, Japan), and for real-time PCR to detect the gene expression quantitatively, LightCycler reagents and instruments (Roche, Mannheim, Germany) were used according to the manufacturer's instructions. The sequences of the used primers for p21Cip1/Waf1 (Ookawa et al44 ) and bax45 were described elsewhere. The primers for Fas ligand gene (sense, 5′-CCT CCA GGC ACA GTT CTT CC-3′; antisense, 5′-TAC CAA GGC AAC CAG AAC CA-3′) and for p27Kip1 gene (sense, 5′-GGG GCT CCG GCT AAC TCT GA-3′; antisense, 5′-GGC TTC TTG GGC GTC TGC TC-3′) were also used for PCR.
Pharmacologic experiments
Cells were treated with 5 μM U0126 (Cell Signaling), 5 μM LY294002 (Cell Signaling), or 20 μM AG490 (Calbiochem, San Diego, CA). For the viable cell assay, these inhibitors were added 30 minutes prior to stimulation with IL-6, IGF-I, or insulin, and then incubated for 48 hours. For the signaling assay, AG490 was added 30 minutes before IL-6 treatment followed by further incubation for 10 minutes.
Confocal laser microscopy
Cells (1 × 105) treated with IL-6 (2 ng/mL) for 10 minutes were washed with culture medium and suspended in 50 μL medium. To detect the colocalization of IL-6Rα with IGF-I receptor on cell membranes, cells were incubated with a PE-labeled anti–IL-6Rα antibody and an FITC-labeled anti–IGF-I receptor antibody for 30 minutes on ice in the dark. To examine the localization of IL-6Rα and IGF-I receptor on plasma membranes, cells were incubated with cholera toxin B (CTXB) conjugated with FITC (Sigma) as a marker of lipid raft portion together with either PE-labeled anti–IL-6Rα or anti–IGF-I receptor antibodies (R&D Systems). After washing, cells were suspended in 50 μL medium, and 10 μL cell suspension was used to detect colocalization and lipid raft localization of IL-6Rα and IGF-I receptors by a confocal laser microscope (LSM 510; Carl Zeiss, Jena, Germany).
Transfection of IL-6Rα expression plasmid
Human IL-6Rα cDNA was amplified by PCR from pIR-2 (kindly provided by Dr T. Hirano, Osaka University, Osaka, Japan) using KOD-PLUS DNA polymerase (TOYOBO, Osaka, Japan). The primers used were 5′-TTGGATCCGGGGCACAAGGTGGCAGGATGCTGGCCGTCGGCTGCGC-3′ for sense strand and 5′-TTGGATCCGCTATCTGGGGAAGAAGTAGTC-3′ for the antisense strand. The sense strand primer includes human Stat1 5′–untranslated region (UTR) sequence (from -18 base to -1 base before ATG, underlined) instead of the original 5′-UTR that results in enhanced cell surface expression of the protein (N.T., unpublished data, January 2003). The amplified cDNA was cloned into TOPO TA cloning vector (Invitrogen), and the sequence was verified. The IL-6Rα expression vectors were constructed by subcloning the human IL-6Rα cDNA into the EcoRI site of the retroviral vector pRep (kindly provided by Dr H. Suzuki Yamaguchi University, Ube, Japan) with correct orientation. pRep was generated by the insertion of woodchuck PRE (posttranscriptional regulatory element; 595 bp) between an internal ribosomal entry site (IRES)/green fluorescent protein (GFP) and simian virus 40 (SV40)–driven puromycin-resistant cassette of the pMX vector. The pRep-(Stat1) IL-6Rα or empty pRep was transfected into PT-67 packaging cells (Clontech, Palo Alto, CA) by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At 4 days after transfection, the supernatant containing the infectious replication-incompetent retroviruses encoding the IL-6Rα gene was collected, filtered, and either used to infect the U266 myeloma cell line or stored at -70°C. U266 was incubated in the virus-containing medium for 2 days, and then cultured in fresh media containing 0.5 μg/mL puromycin (Sigma) for 2 or 3 weeks. Surviving cells were subjected to flow cytometry to confirm the IL-6Rα expression by enhanced GFP (EGFP) expression and by staining with an anti–IL-6Rα antibody.
Results
IL-6 together with IGF-I supports the in vitro proliferation of IL-6–dependent myeloma cell lines and myeloma cells from the patients
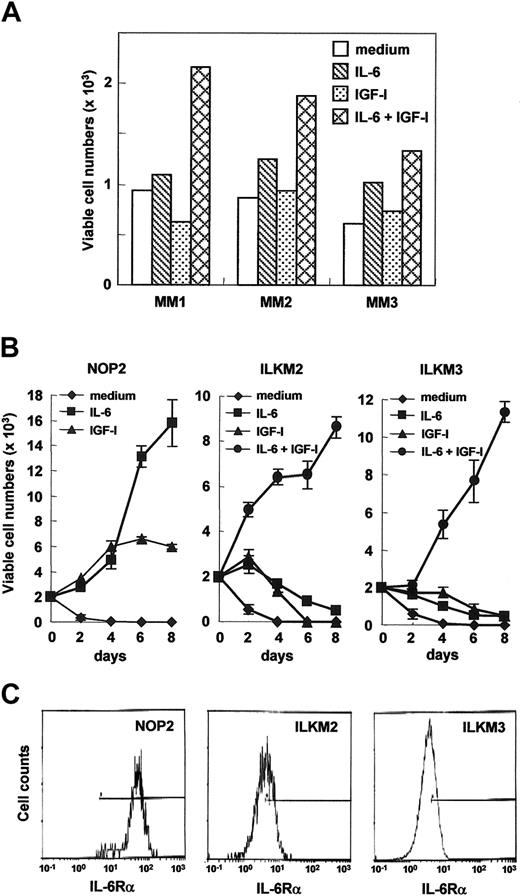
In primary myeloma cells isolated from 3 MM patients, IL-6 together with IGF-I, but not IL-6 or IGF-I alone, markedly supported their viability in serum-containing medium (Figure 1A) in which IL-6–dependent myeloma cell lines, ILKM2 and ILKM3, proliferated with IL-6.42 However, IL-6 was unable to maintain these cell lines in a RPMI1640-based synthetic medium containing AlbuMax (lipid-rich bovine serum albumin) and transferrin but lacking FBS and insulin (hereafter called serum-free medium; Figure 1B). Both ILKM2 and ILKM3 rapidly lost their cell viability with IL-6 or IGF-I alone (nearly 90% of cells were apoptotic by 2 days), but proliferated in response to IL-6 together with IGF-I at least up to 10 days in this serum-free condition (Figure 1B and data not shown). Among the several growth factors described in “Materials and methods,” only insulin or IGF-I could trigger the cell proliferation of ILKM2 and ILKM3 with IL-6 in serum-free media, whereas the effect of insulin was weaker than that of IGF-I (data not shown). These data suggest that, as reported previously,46 IL-6 together with IGF-I but not IL-6 or IGF-I alone have growth and antiapoptotic effects on ILKM2 and ILKM3. The combinatory effect of IL-6 with IGF-I on myeloma cell growth seemed to be synergistic rather than additive because neither IL-6 nor IGF-I alone could support cell viability. Moreover, this synergistic effect is not due to the up-regulation of receptor expression because the expression of IL-6Rα, gp130, and IGF-I receptors was not altered by either IL-6 or IGF-I stimulations (data not shown).
Effects of IL-6 or IGF-I on the cell proliferation of ILKM2, ILKM3, and NOP2 cell lines that differ in IL-6Rα expression levels. (A) Myeloma cells isolated from myeloma patients were viably cultured with IL-6 (2 ng/mL) and IGF-I (100 ng/mL). Myeloma cells from 3 myeloma patients were cultured with or without IL-6 and IGF-I in serum-containing complete media for 2 weeks, and the relative viable cell numbers were counted by flow cytometry with forward and side scatters at a constant flow rate for one minute. (B) In serum-free media, IL-6 (1 ng/mL) could support NOP2 growth, whereas the cell proliferation of ILKM2 and ILKM3 was promoted by IL-6 together with IGF-I (3 ng/mL). The growth curves for 8 days cultured with medium alone (♦), with IL-6 (▪), with IGF-I (▴), or with IL-6 and IGF-I (•) are indicated. Values of relative viable cell numbers are indicated by the means and SDs obtained from 3 independent experiments. Statistical analysis by Student t test indicated that viable cell numbers of NOP2 after day 2 between medium alone and medium with IL-6 were significantly different (P < .001). Differences of viable cell numbers of NOP2 after day 6 between IGF-I and IL-6 were also significant (P < .001). Viable cell numbers of ILKM2 and ILKM3 after day 2 between medium alone and medium with IL-6 and IGF-I were significantly different (P < .001). Differences of viable cell numbers of ILKM2 after day 2 and of ILKM3 after day 4 between IGF-I or IL-6 alone and IL-6 together with IGF-I were also significant (P < .001). (C) The cell surface expression of IL-6Rα of NOP2, ILKM2, and ILKM3 is shown by flow cytometry. Compared with ILKM2 and ILKM3, NOP2 exhibited a markedly increased expression of IL-6Rα.
Effects of IL-6 or IGF-I on the cell proliferation of ILKM2, ILKM3, and NOP2 cell lines that differ in IL-6Rα expression levels. (A) Myeloma cells isolated from myeloma patients were viably cultured with IL-6 (2 ng/mL) and IGF-I (100 ng/mL). Myeloma cells from 3 myeloma patients were cultured with or without IL-6 and IGF-I in serum-containing complete media for 2 weeks, and the relative viable cell numbers were counted by flow cytometry with forward and side scatters at a constant flow rate for one minute. (B) In serum-free media, IL-6 (1 ng/mL) could support NOP2 growth, whereas the cell proliferation of ILKM2 and ILKM3 was promoted by IL-6 together with IGF-I (3 ng/mL). The growth curves for 8 days cultured with medium alone (♦), with IL-6 (▪), with IGF-I (▴), or with IL-6 and IGF-I (•) are indicated. Values of relative viable cell numbers are indicated by the means and SDs obtained from 3 independent experiments. Statistical analysis by Student t test indicated that viable cell numbers of NOP2 after day 2 between medium alone and medium with IL-6 were significantly different (P < .001). Differences of viable cell numbers of NOP2 after day 6 between IGF-I and IL-6 were also significant (P < .001). Viable cell numbers of ILKM2 and ILKM3 after day 2 between medium alone and medium with IL-6 and IGF-I were significantly different (P < .001). Differences of viable cell numbers of ILKM2 after day 2 and of ILKM3 after day 4 between IGF-I or IL-6 alone and IL-6 together with IGF-I were also significant (P < .001). (C) The cell surface expression of IL-6Rα of NOP2, ILKM2, and ILKM3 is shown by flow cytometry. Compared with ILKM2 and ILKM3, NOP2 exhibited a markedly increased expression of IL-6Rα.
IL-6 is sufficient as a growth and survival factor for NOP2 cells highly expressing IL-6Rα
Interestingly, although in serum-containing media IL-6 had no augmenting effect on the proliferation of an IL-6–nonproducing myeloma cell line, NOP2,42 cell viability and the proliferation of NOP2 clearly depend on IL-6 in serum-free conditions (Figure 1B). The effect of IL-6 on NOP2 seemed comparable with that of IL-6 together with IGF-I on ILKM2 and ILKM3 (Figure 1B). Flow cytometry showed that NOP2 expressed IL-6Rα 10 times more than ILKM2 or ILKM3 (Figure 1C), implying that the increased expression of IL-6Rα may play a critical role, such as anti–cell death and proliferation, in the biology of myeloma cells in response to IL-6.
Akt is activated by IL-6 in NOP2 but not in ILKM2 or ILKM3 cells
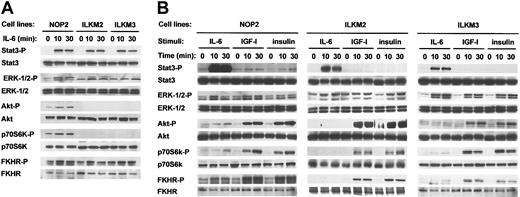
To understand the striking biologic effect of IL-6 on NOP2, we investigated the IL-6–induced activation of intracellular signaling molecules in NOP2 as well as ILKM2 and ILKM3 in serum-free conditions. As shown in Figure 2A, IL-6 stimulation could induce the phosphorylation of Stat3 and ERK1/2 similarly in 3 cell lines. In contrast, Akt, FKHR, and p70S6 kinase was activated by IL-6 only in NOP2 but not in ILKM2 or ILKM3. Upon IL-6 stimulation, the phosphorylation of FKHR and p70S6K observed in NOP2 may follow Akt activation, whereas the phosphorylation of other molecules postulated as downstream targets of Akt, such as Bad, IκB-α, and caspase-9, was not detected (data not shown). IGF-I or insulin stimulation induced the phosphorylation of ERK1/2, Akt, p70S6K, and FKHR in 3 cell lines, whereas Stat3 was phosphorylated very weakly in NOP2 but not in ILKM2 and ILKM3 (Figure 2B). Hence, the activated signaling molecules by IGF-I or insulin were almost equivalent to those by IL-6 in NOP2 (Figure 2B). These results, together with the biologic results shown in Figure 1, indicate that the IL-6–induced activation of the Stat and Ras-MAPK pathways is important but not sufficient and the activation of the Akt pathway is further required for myeloma cell survival.
IL-6 stimulation induced the phosphorylation of Akt, p70S6K, and FKHR in NOP2 but not in ILKM2 and ILKM3. Phosphorylation of Stat3, ERK1/2, Akt, p70S6K, and FKHR in myeloma cell lines stimulated with IL-6 (2 ng/mL), IGF-I (100 ng/mL), or insulin (100 ng/mL) for 0, 10, or 30 minutes was determined by Western blot analysis using phospho-specific antibodies. Western blots of 3 cell lines by IL-6 stimulation on the same blot (A) and of each cell line by 3 reagents (B) are shown. IL-6 stimulation induced the phosphorylation of Akt, p70S6K, and FKHR in NOP2 but not in ILKM2 and ILKM3, whereas the phosphorylation of ERK1/2, Akt, p70S6K, and FKHR but not of Stat3 in NOP2, ILKM2, and ILKM3 stimulated with IGF-I or insulin was comparable with NOP2 with IL-6.
IL-6 stimulation induced the phosphorylation of Akt, p70S6K, and FKHR in NOP2 but not in ILKM2 and ILKM3. Phosphorylation of Stat3, ERK1/2, Akt, p70S6K, and FKHR in myeloma cell lines stimulated with IL-6 (2 ng/mL), IGF-I (100 ng/mL), or insulin (100 ng/mL) for 0, 10, or 30 minutes was determined by Western blot analysis using phospho-specific antibodies. Western blots of 3 cell lines by IL-6 stimulation on the same blot (A) and of each cell line by 3 reagents (B) are shown. IL-6 stimulation induced the phosphorylation of Akt, p70S6K, and FKHR in NOP2 but not in ILKM2 and ILKM3, whereas the phosphorylation of ERK1/2, Akt, p70S6K, and FKHR but not of Stat3 in NOP2, ILKM2, and ILKM3 stimulated with IGF-I or insulin was comparable with NOP2 with IL-6.
ERK1/2 and PI3-K pathways are necessary for myeloma cell survival
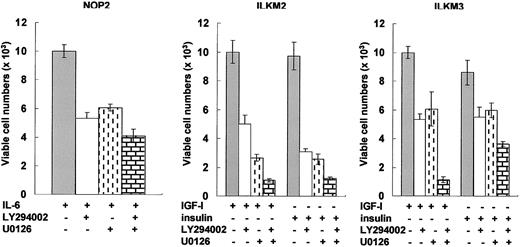
To clarify the physiologic relevance of the ERK1/2 (downstream of MEK1/2) and Akt (downstream of PI3-kinase) pathways, we investigated the viability of 3 myeloma cell lines treated with IL-6, IGF-I, or insulin by using pharmacologic inhibitors. The efficacy of inhibitors was evaluated for short-term culture because, even in serum-free conditions, ILKM2 and ILKM3 remained viable for 2 days with IGF-I or insulin but without factors (Figure 1B and data not shown). Either a MEK1/2 inhibitor, U0126, or a PI3-K inhibitor, LY294002, partially inhibited the effect of IGF-I or insulin, and both together completely blocked the effect of these growth factors on the cell viability of ILKM2 and ILKM3 (Figure 3). The effect of IL-6 on NOP2 cell survival was similarly suppressed by U0126 or LY294002 and the combination of U0126 with LY294002 exhibited the most striking inhibitory effect (Figure 3). These results confirm that both the ERK1/2 and PI3-K pathways are important for the survival of myeloma cell lines.
Both the ERK1/2 and PI3-K pathways were important for the viability of myeloma cell lines. Flow cytometry was used to evaluate the effects of the indicated inhibitors and their combinations on the cell viability of ILKM2 and ILKM3 treated with 30 ng/mL IGF-I or insulin and of NOP2 treated with 1 ng/mL IL-6 for 2 days. The MEK1/2 inhibitor, U0126 (5 μM), and the PI3-K inhibitor, LY294002 (5 μM), similarly reduced the cell viability of myeloma cell lines (P < .01), and both together completely blocked the effects of IGF-I, insulin, or IL-6 (P < .001). The values of relative viable cell numbers are indicated by the means and SDs from 3 independent experiments. Statistical analysis by Student t test indicated significant differences of viable cell numbers between inhibitors that were or were not treated.
Both the ERK1/2 and PI3-K pathways were important for the viability of myeloma cell lines. Flow cytometry was used to evaluate the effects of the indicated inhibitors and their combinations on the cell viability of ILKM2 and ILKM3 treated with 30 ng/mL IGF-I or insulin and of NOP2 treated with 1 ng/mL IL-6 for 2 days. The MEK1/2 inhibitor, U0126 (5 μM), and the PI3-K inhibitor, LY294002 (5 μM), similarly reduced the cell viability of myeloma cell lines (P < .01), and both together completely blocked the effects of IGF-I, insulin, or IL-6 (P < .001). The values of relative viable cell numbers are indicated by the means and SDs from 3 independent experiments. Statistical analysis by Student t test indicated significant differences of viable cell numbers between inhibitors that were or were not treated.
IL-6 stimulation affects both the expression of Fas ligand, p27Kip1, p21Cip1/Waf1, and bax genes and subcellular localization of p27Kip1 and p53 in NOP2 cells
As FKHR is an Akt target molecule, its phosphorylation was well correlated with Akt activation in myeloma cell lines (Figure 2). The phosphorylated FKHR by Akt is located in the cytoplasm as an inactive transcription factor via binding to 14-3-3. The expression of Fas ligand (FasL) and p27Kip1 genes is controlled by FKHR, which is constitutively active without survival signals. The elevated FasL expression induces apoptosis via binding to Fas on cell membranes, and the increase in a cyclin-dependent kinase (CDK) inhibitor, p27Kip1, causes cell cycle arrest. In NOP2, the expression of FasL and p27Kip1 genes was induced by serum depletion, whereas IL-6 almost completely blocked the increase in both FasL and p27Kip1 gene expression (Figure 4A).
IL-6 induced the cytoplasmic accumulation of p27Kip1 and p53 proteins and blocked the expression of Fas ligand, p27Kip1, p21Cip1/Waf1, and bax genes in NOP2. In serum-free media, NOP2 cultured with or without IL-6 (2 ng/mL) for 24 hours (A) or for 12 hours (B) was harvested and cellular RNA was isolated. Subsequent real-time PCR analysis using cDNA synthesized from total RNA evaluated quantitatively the expression of Fas ligand, p27Kip1 (A), p21Cip1/Waf1, and bax (B) genes. The expression of Fas ligand, p27Kip1, p21Cip1/Waf1, and bax genes was blocked by IL-6 treatment in NOP2. Values of relative gene expressions are indicated by the ratio to glyceraldehyde-3-phosphate dehydrogenase (G3PDH) when the value of time 0 was 1. The values are indicated by the means and SDs obtained from 2 independent experiments. Differences of relative gene expression between cells cultured without and with IL-6 were all statistically significant (P < .01). (C) Western blot showed that both p27Kip1 and p53 proteins remained in the cytoplasm of NOP2 treated with IL-6. Cytoplasmic (indicated as C) and nuclear (indicated as N) extracts were probed with an anti-p53 antibody in Western blot analysis. Values of the relative amounts of proteins were determined as densities of bands by Scion image software (Scion, Frederick, MA) and shown as a ratio to time 0.
IL-6 induced the cytoplasmic accumulation of p27Kip1 and p53 proteins and blocked the expression of Fas ligand, p27Kip1, p21Cip1/Waf1, and bax genes in NOP2. In serum-free media, NOP2 cultured with or without IL-6 (2 ng/mL) for 24 hours (A) or for 12 hours (B) was harvested and cellular RNA was isolated. Subsequent real-time PCR analysis using cDNA synthesized from total RNA evaluated quantitatively the expression of Fas ligand, p27Kip1 (A), p21Cip1/Waf1, and bax (B) genes. The expression of Fas ligand, p27Kip1, p21Cip1/Waf1, and bax genes was blocked by IL-6 treatment in NOP2. Values of relative gene expressions are indicated by the ratio to glyceraldehyde-3-phosphate dehydrogenase (G3PDH) when the value of time 0 was 1. The values are indicated by the means and SDs obtained from 2 independent experiments. Differences of relative gene expression between cells cultured without and with IL-6 were all statistically significant (P < .01). (C) Western blot showed that both p27Kip1 and p53 proteins remained in the cytoplasm of NOP2 treated with IL-6. Cytoplasmic (indicated as C) and nuclear (indicated as N) extracts were probed with an anti-p53 antibody in Western blot analysis. Values of the relative amounts of proteins were determined as densities of bands by Scion image software (Scion, Frederick, MA) and shown as a ratio to time 0.
Since 3 papers have recently reported that Akt also regulated the subcellular localization of p27Kip1 via the phosphorylation of a threonine residue,31-33 we examined whether the subcellular localization of p27Kip1 in NOP2 was altered by IL-6 stimulation. Figure 4C shows that IL-6 induced the cytoplasmic accumulation of p27Kip1 in NOP2, indicating that the net activity of p27Kip1 was suppressed by IL-6 via not only the reduced expression but also cytoplasmic localization of p27Kip1. Akt controls the p53-dependent apoptotic pathways because MDM2, a ubiquitin ligase, when phosphorylated by Akt, induces the degradation of p53. In NOP2, IL-6 blocked the serum depletion–induced nuclear localization of p53 (Figure 4C), and moreover, the serum depletion–induced expression of bax and p21Cip1/Waf1, target genes of p53,47 was blocked by IL-6 stimulation (Figure 4B). These results indicate that the antiapoptotic effect of IL-6 on NOP2 presumably results from the activation of Akt, which inhibits the functions of FKHR, p27Kip1, and p53.
IL-6 stimulation induces the phosphorylation of IGF-I receptors in NOP2 but not in ILKM3 cells
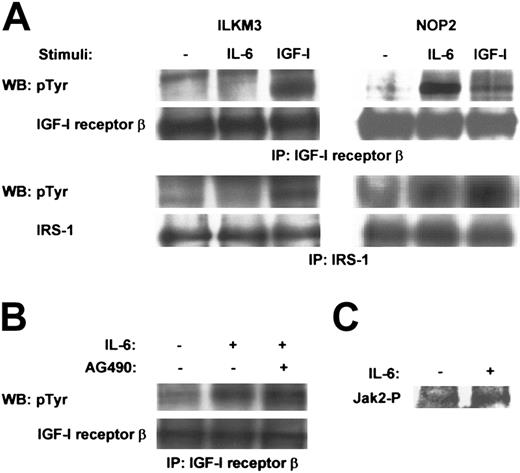
As similar intracellular signaling cascades were activated in NOP2 by IL-6 and in 3 cell lines by IGF-I or insulin, we investigated how IL-6 mimics the IGF-I/insulin signaling in NOP2. Interestingly, in a sharp contrast to ILKM3, in NOP2, IL-6 stimulation induced the phosphorylation of IGF-I receptor β and IRS-1, which was comparable with that by IGF-I stimulation (Figure 5A), thereby indicating that the phosphorylation of IGF-I receptors by IL-6 is likely to trigger the activation of the PI3-K–Akt pathway in NOP2. Although Jak2 was activated in response to IL-6 in NOP2 (Figure 5C), a Jak2 inhibitor, AG490, did not suppress the IL-6–induced phosphorylation of IGF-I receptor β (Figure 5B). Thus, the IL-6–activated Jak2 does not participate in the phosphorylation of IGF-I receptors in NOP2.
IL-6 induced the tyrosine phosphorylation of IGF-I receptor β in NOP2 but not in ILKM3. (A) Immunoprecipitation (IP) and Western blot (WB) analysis with the indicated antibodies showed tyrosine phosphorylation of IGF-I receptor β and IRS-1 in ILKM3 and NOP2 treated with or without IL-6 (2 ng/mL) or IGF-I (100 ng/mL) for 5 minutes. (B) A Jak2 inhibitor, AG490 (20 μM), had no effect on the IL-6–induced phosphorylation of IGF-I receptor β in NOP2. (C) IL-6 (2 ng/mL) activated Jak2 in NOP2, as shown by IP-Western analysis using a phospho-specific Jak2 antibody.
IL-6 induced the tyrosine phosphorylation of IGF-I receptor β in NOP2 but not in ILKM3. (A) Immunoprecipitation (IP) and Western blot (WB) analysis with the indicated antibodies showed tyrosine phosphorylation of IGF-I receptor β and IRS-1 in ILKM3 and NOP2 treated with or without IL-6 (2 ng/mL) or IGF-I (100 ng/mL) for 5 minutes. (B) A Jak2 inhibitor, AG490 (20 μM), had no effect on the IL-6–induced phosphorylation of IGF-I receptor β in NOP2. (C) IL-6 (2 ng/mL) activated Jak2 in NOP2, as shown by IP-Western analysis using a phospho-specific Jak2 antibody.
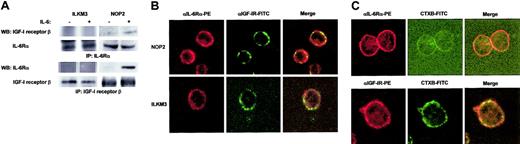
Complex formation of IL-6Rα with IGF-I receptor β on NOP2 cells
Immunoprecipitation followed by Western blot analysis using IL-6Rα and IGF-I receptor β antibodies or vice versa gave evidence of the interaction between IL-6Rα and IGF-I receptor β in NOP2 but not in ILKM3, and importantly, these interactions were dependent on IL-6 stimulation (Figure 6A). In addition, colocalization of IL-6Rα with IGF-I receptors was observed on the plasma membranes of NOP2 but not of ILKM3 by confocal laser microscopy (Figure 6B). In NOP2, some IL-6Rα and IGF-I receptors were colocalized at the raft portions of plasma membranes marked with a raft marker, cholera toxin B subunit (CTXB; Figure 6C). Taken together, these results suggest that IL-6Rα could in part be codistributed with IGF-I receptors at lipid rafts of NOP2 cell membranes. Thus, this may suggest the possibility of the IL-6–induced complex formation of IL-6Rα, gp130, and/or IGF-I receptor β, followed by the subsequent autophosphorylation of IGF-I receptors.
IL-6Rα and IGF-I receptor formed a complex on plasma membranes of NOP2. (A) IP-Western analysis showed IL-6–dependent coprecipitation of IL-6Rα and IGF-I receptor β in NOP2 but not in ILKM3. (B) Confocal laser microscopy showed the colocalization of IL-6Rα labeled with PE (red) and IGF-I receptor labeled with FITC (green) in NOP2 but not in ILKM3. Colocalization is shown in yellow in the merged pictures. (C) Some IL-6Rα and IGF-I receptors were located at lipid rafts in NOP2. IL-6Rα or IGF-I receptor was recognized by specific antibodies labeled with PE (red), while lipid rafts were indicated in green with FITC-conjugated cholera toxin B subunits (CTXB). Fluorescent imaging at an initial magnification of × 630.
IL-6Rα and IGF-I receptor formed a complex on plasma membranes of NOP2. (A) IP-Western analysis showed IL-6–dependent coprecipitation of IL-6Rα and IGF-I receptor β in NOP2 but not in ILKM3. (B) Confocal laser microscopy showed the colocalization of IL-6Rα labeled with PE (red) and IGF-I receptor labeled with FITC (green) in NOP2 but not in ILKM3. Colocalization is shown in yellow in the merged pictures. (C) Some IL-6Rα and IGF-I receptors were located at lipid rafts in NOP2. IL-6Rα or IGF-I receptor was recognized by specific antibodies labeled with PE (red), while lipid rafts were indicated in green with FITC-conjugated cholera toxin B subunits (CTXB). Fluorescent imaging at an initial magnification of × 630.
IL-6–induced phosphorylation of IGF-I receptors in IL-6Rα–transfected U266 cells
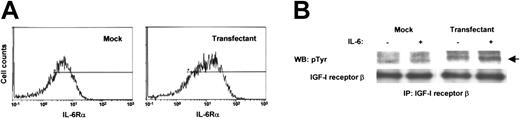
To demonstrate that the findings in NOP2 are actually due to the increased expression of IL-6Rα, we introduced the IL-6Rα gene into another myeloma cell line, U266, and established a stable transfectant, IL-6Rα-U266. Compared with the mock-transfected U266 (mock U266), the expression of IL-6Rα was found to be increased in IL-6Rα–U266 (Figure 7A). IL-6 stimulation induced the phosphorylation of IGF-I receptor β in IL-6Rα–U266 but not in mock U266 (Figure 7B), consistent with the results obtained from NOP2. Hence, we confirmed that the increased levels of IL-6Rα expression could induce the phosphorylation of IGF-I receptors on myeloma cell lines in response to IL-6.
IGF-I receptors were phosphorylated by IL-6 in IL-6Rα–transfected U266. (A) Flow cytometry showed the elevated expression of IL-6Rα on IL-6Rα–U266. (B) IP-Western analysis showed the phosphorylation of IGF-I receptor β by IL-6 (2 ng/mL) in IL-6Rα–U266 but not in mock U266.
IGF-I receptors were phosphorylated by IL-6 in IL-6Rα–transfected U266. (A) Flow cytometry showed the elevated expression of IL-6Rα on IL-6Rα–U266. (B) IP-Western analysis showed the phosphorylation of IGF-I receptor β by IL-6 (2 ng/mL) in IL-6Rα–U266 but not in mock U266.
Discussion
In this study, we found that IGF-I receptors were phosphorylated by IL-6 stimulation in IL-6Rα highly expressing NOP2 and IL-6Rα–transfected U266 (Figures 1C, 5, and 7). The mechanism appeared to be a complex formation of IL-6Rα with IGF-I receptor β by IL-6 stimulation but was likely to be independent of Jaks in NOP2 (Figures 5, 6). In addition to the Stat3 and ERK1/2 pathways, IL-6 activated Akt downstream of PI3-K via the phosphorylation of IGF-I receptors in NOP2 (Figure 2), and the PI3-K–Akt pathway was further necessary for the IL-6–promoted proliferation of NOP2 in serum-free conditions (Figure 3). The activated Akt could contribute to the antiapoptosis of cells presumably due to the inactivation of FKHR, p27Kip1, and p53 (Figures 2 and 4). Hence, in myeloma cell lines, the IL-6–induced activation of Stat3 and ERK1/2 is important but not sufficient for cell survival and proliferation, which further require the activation of PI3-K–Akt–mediated pathways. Accordingly, under the cellular context of the elevated expression of IL-6Rα on myeloma cells, it is possible that IL-6Rα molecules are located close to IGF-I receptors at lipid rafts, and IL-6 stimulation triggers the complex formation of IL-6Rα not only with gp130 but also with IGF-I receptors, leading to the autophosphorylation of IGF-I receptor β and subsequent activation of PI3-K–Akt pathways.
Numerous biologic responses of different cell types are induced by IL-6, which activates Stat3 and Ras-ERK1/2 via Jaks, and the balance of activation of both pathways is considered to direct the cell fate in response to IL-6.48 IL-6 is the most potent growth factor for myelomas in vitro and in vivo, while IGF-I, a member of the so-called growth factors whose receptors possess tyrosine kinase domains, is a potent growth and survival factor for a wide variety of cells. We showed here that IGF-I cooperating with IL-6 played a pivotal role in the growth and survival in vitro of myeloma cell lines and myeloma cells isolated from MM patients (Figures 1A-B). Moreover, a surprising finding was revealed in this study that IGF-I receptors were phosphorylated in NOP2 but not in ILKM3 in response to IL-6 (Figure 5A). The phosphorylation of IGF-I receptor β by IL-6 stimulation was further confirmed in IL-6Rα–transfected U266 (Figure 7). Although the cross-talk of signals mediated by a cytokine and growth factor has been previously reported in the case of the phosphorylation of EGF receptors by the growth hormone–activated Jak2,49 a Jak2 inhibitor could not suppress the phosphorylation of IGF-I receptor β in NOP2 (Figure 5B). This suggests that the IL-6–induced activation of Jak2 is not involved in the activation of IGF-I receptor–mediated signals in NOP2. It has been reported that the PI3-K and Akt pathways were activated via gp130 recruiting some adaptor molecules to create binding sites to the SH2 domain of the p85 subunit of PI3-K.50 Although we cannot completely rule out this possibility, Gab-1, IRS-1, and IRS-2 could not be coprecipitated with gp130 in NOP2 (data not shown), suggesting that the activation of PI3-K and Akt resulted from the phosphorylation of IGF-I receptors.
A previous paper using simple immunoprecipitation showed the detection of protein complexes of transmembrane molecules, such as gp130 and ErbB2, on a prostate carcinoma cell line,51 and we also detected complexes composed of IL-6Rα and IGF-I receptor β in NOP2 (Figure 6A). Insulin also weakly supported the IL-6–induced proliferation of ILKM2 and ILKM3, whereas, unlike IGF-I receptors, insulin receptor β was not coprecipitated with IL-6Rα in NOP2 (data not shown). These results indicate that IGF-I receptors rather than insulin receptors seem to be relevant for the IL-6–induced activation of PI3-K–Akt pathways in NOP2. Although both IGF-I receptor β and gp130 were coprecipitated with IL-6Rα in NOP2 (Figure 6A and data not shown), it is currently unclear whether some or all IL-6Rα/IGF-I receptor β complexes contain gp130 (IL-6Rα/gp130/IGF-I receptor β). Since the interaction of IL-6Rα with IGF-I receptor β was observed in only NOP2 highly expressing IL-6Rα, some free IL-6Rα not associated with gp130 may interact with IGF-I receptors (IL-6Rα/IGF-I receptor β) on NOP2 in response to IL-6. Unlike IL-6–stimulated NOP2, we failed to detect complexes consisting of IGF-I receptor β and IL-6Rα or gp130 in NOP2 stimulated with IGF-I by immunoprecipitation (data not shown), whereas it is intriguing to note that IGF-I stimulation induced the tyrosine phosphorylation of Stat3 in NOP2 but not in ILKM2 and ILKM3 (Figure 2B). Nevertheless, the IGF-I–induced phosphorylation of Stat3 was very weak (Figure 2B) and the biologic effect of IGF-I was much weaker than that of IL-6 in NOP2 (Figure 1B). These data suggest that IGF-I may induce only an inefficient complex formation of IGF-I receptors with gp130 or with IL-6Rα/gp130 in NOP2 even though these complexes could be induced by IGF-I stimulation.
Furthermore, our finding of the colocalization of IL-6Rα and IGF-I receptors at lipid rafts in NOP2 (Figure 6B-C) is consistent with a recent report that revealed the blockade of IL-6 and IGF-I signaling by a raft inhibitor.52 This suggests that IL-6 receptors and IGF-I receptors seem to be closely linked, and therefore the highly expressed IL-6Rα likely enhances the accessibility of IL-6Rα with IGF-I receptor β on cell surface membranes. In the case of interferon (IFN) signaling, although IFN-α/β and IFN-γ transmit signals through distinct receptor complexes, the IFN-α/β receptor component, IFNAR-1, facilitates the efficient assembly of IFN-γ–activated transcription factors. This cross-talk is contingent on constitutive subthreshold IFN-α/β signaling and the association between the 2 nonligand-binding receptor components, IFNAR-1 and IFNGR-2, in the caveolar membrane domains.53 More recently, cross-talk of IFN-α/β signals with gp130 has also reported that IFNAR-1 and gp130 exist in close proximity.54 These results suggest the assembly of cytokine receptor subunits, which may represent a “receptosome”-like structure, allowing the unique signaling cross-talks to occur.54 Taken together, it is tempting to speculate that the elevated expression of IL-6Rα could increase the frequency of the complex formation of IL-6Rα with IGF-I receptors at lipid rafts in response to IL-6, presumably resulting in the altered conformation of IGF-I receptors that triggers their autophosphorylation independently of both Jaks and IGF-I.
IGF-I–mediated growth and anti–cell death signals are widely conserved in worms, flies, and mammals. Initially, strains of Caenorhabditis elegans with mutations in the dauer formation (Daf) pathway were found to have unusually long lives.55 In mammals, PI3-K homologous to AGE-1 is activated by the IGF-I receptor, a counterpart of DAF-2. PI3-K activates Akt that inhibits the function of FKHR, a mammalian DAF-16. In contrast to NOP2, the IGF-I receptor–Akt pathway was not activated by IL-6 in ILKM2 and ILKM3, with the result that IL-6 alone failed to support the cell viability of both cell lines in the serum-free condition (Figures 1B and 2). The indispensable roles of PI3-K in antiapoptosis were demonstrated by experiments using a PI3-K inhibitor, LY294002 (Figure 3). In agreement with a genetic study in C elegans, these results indicate that IGF-I receptor, PI3-K, and Akt are also involved in the survival mechanism of myeloma cells.
A transcription factor, FKHR, becomes inactive via phosphorylation by Akt. Among a number of target molecules as substrates for Akt, FKHR was phosphorylated in NOP2 by IL-6, and in all cell lines by insulin or IGF-I (Figure 2). In NOP2, IL-6 suppressed the induction of FKHR target genes, such as CDK inhibitor, p27Kip1, and proapoptotic FasL genes (Figure 4A), and insulin or IGF-I had the same effect on 3 cell lines (data not shown). The inhibited expression of FasL on the cell surface of these myeloma cell lines expressing Fas/CD95 was also confirmed by flow cytometry (data not shown). In addition, in NOP2, IL-6 induced the cytoplasmic accumulation of p27Kip1 and blocked the nuclear accumulation of apoptosis-inducing factor p53 and the expression of p53-target genes,47 such as proapoptotic bcl-2 family member, bax, and a CDK inhibitor, p21Cip1/Waf1 (Figures 4B-C). The cytoplasmic localization of p27Kip1 may result from the phosphorylation of p27Kip1 (Yamazaki et al35 ; Humphrey et al36 ; and Roussel et al37 ), and the localization of p53 from the phosphorylation of MDM2 by Akt. A ubiquitin ligase, MDM2 phosphorylated by Akt, moves to the cytoplasm with p53 from the nucleus.29 As the growth factor–induced activation of Akt is considered to be an important mechanism of the antiapoptotic effect on a wide variety of cells, these results indicate that in myeloma cells, Akt seems to play a central role in the antiapoptotic pathways via the inhibition of FKHR, p27Kip1, and p53 activities.
Finally, as the overexpression of growth factor receptors has been postulated as a mechanism of tumorigenesis or advantage of tumor growth, the overexpression of the growth factor receptors that harbor kinase activity (RTKs) shows constitutive active kinases due to the ligand-independent dimerization of the receptors or the enhanced signals mediated by receptors, suggesting that the levels of growth factor receptors regulate the strength of intracellular signals and consequently influence the biologic outcomes. In MM, myeloma cells use cytokine receptors lacking kinase activity, such as IL-6 receptors, as the main receptors for promoting their growth, while they also express RTKs, such as IGF-I receptors, as well as other types of receptors. This study shows that increased IL-6Rα could interact with IGF-I receptors at lipid rafts in response to IL-6 and induce signal divergence from IL-6 receptor complexes to downstream interacting receptors. This report therefore provides a novel insight into the IL-6–induced growth mechanism of IL-6Rα highly expressing myeloma cells of the Jak-independent synergy between IL-6 receptors and IGF-I receptors.
Prepublished online as Blood First Edition Paper, October 30, 2003; DOI 10.1182/blood-2003-07-2187.
Supported in part by grants from the Ministry of Education, Science, Sports and Culture of Japan; the Ministry of Health, Labour and Welfare of Japan; the Japan Society for the Promotion of Science; and the Public Trust Japan Leukaemia Research Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr H. Asaoku (Hiroshima Red Cross Hospital) for bone marrow aspirates from MM patients, Dr H. Suzuki for the generous gift of the retrovirus vector, pRep, and Dr T. Hirano and Dr M. Hibi for human IL-6Rα cDNA. We also thank the Center for Gene Research, Yamaguchi University for DNA sequencing.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal