Abstract
Endothelial cell adhesive interactions are mediated by both fibrinogen and fibrin, and growth is stimulated by fibroblast growth factor 2 (FGF-2). We have shown previously that FGF-2 binds specifically and with high affinity to fibrinogen and fibrin and that fibrinogen potentiates the proliferative capacity of FGF-2 and also protects it from proteolytic degradation. To further characterize this interaction we have performed FGF-2 mutagenesis to identify the interactive site. Because FGF-1 has a similar structure to FGF-2 but does not bind to fibrinogen, we used a strategy of cassette and site-directed mutagenesis, exchanging residues from FGF-1 and FGF-2 and correlating structural changes with fibrinogen binding. Two cassette interchange mutants, 2212 and 2211, contained either the third cassette or both the third and fourth cassettes from FGF-1, and neither exhibited any affinity for fibrinogen. Exchange of 5 residues (Phe95, Ser100, Asn102, Arg107, and Arg109) from FGF-2 into the corresponding sites in the third cassette of FGF-1 imparted high-affinity binding with apparent dissociation constants (Kd) of 5.3 nM and 8.6 nM, respectively, compared with 1.3 nM for wild-type FGF-2. We conclude that these 5 residues define a high-affinity binding site in FGF-2 for fibrinogen.
Introduction
The fibroblast growth factors (FGFs) form a large family of structurally related, multifunctional proteins that regulate various biologic responses including embryonic growth, development, cell proliferation, differentiation, and angiogenesis.1,2 FGF-2 and FGF-1 are 2 prototype members that share over 50% sequence identity and mediate cellular functions by binding to specific tyrosine kinase receptors that are activated by oligomerization facilitated by heparin or heparan sulfate proteoglycans acting as cofactors.3-6
Fibrinogen is a 340-kDa plasma glycoprotein composed of 2 pairs of 3 separate polypeptide chains that are converted by thrombin to fibrin that is needed for hemostasis, and it also provides the temporary matrix required for initial cell responses to injury.7 Fibrinogen interacts with endothelial cells through specific receptors including αvβ3, α5β1, and intercellular adhesion molecule-1 (ICAM-1) to support matrix or cell adhesion.8-10 Endothelial cells respond differently to fibrin, which stimulates changes in shape, migration, and protein synthesis11-16 mediated in part through interaction of the amino terminus of the fibrin β chain that is exposed following thrombin cleavage with the endothelial cell receptor, VE-cadherin.17-19
We have shown previously that FGF-2 binds to fibrinogen and fibrin with high affinity,20 whereas FGF-1 does not.21 Fibrinogen-bound FGF-2 has enhanced proliferative activity for endothelial cells,22 can sustain prolonged endothelial cell growth in the absence of the soluble growth factor,21 and is protected from proteolytic degradation.23 We have also demonstrated that vascular endothelial growth factor (VEGF), another important regulator of angiogenesis, binds with high affinity to fibrinogen.24 The specific binding of FGF-2 and VEGF to fibrin(ogen) indicates an important level of coordination at sites of injury between growth factors with critical cell regulatory functions and the fibrin matrix. The high-affinity binding also suggests that FGF-2 in blood may circulate in association with fibrinogen that is present in high concentration. Specific binding of FGF-2 may facilitate localization of this activity at sites of vessel injury, thrombosis, malignancy, and inflammation where FGF-2–directed cell responses are required. This concept is consistent with the observation that mice deficient in fibrinogen exhibit delayed wound healing, a process requiring FGF-2.25
To better understand the structural basis for the interaction between FGF-2 and fibrinogen, we have sought to characterize the binding site on FGF-2. To do this, we exploited the structural relationships between FGF-2, which binds to fibrinogen, and FGF-1, which does not. Crystal structures of both molecules show a very similar structure,26-30 and we reasoned that the binding site on FGF-2 would represent a surface exposed group of residues in physical proximity that differed from the comparable region on FGF-1. We employed a strategy of cassette and site-directed mutagenesis to identify FGF-2 residues that support high-affinity binding to fibrinogen.
Materials and methods
Construction of wild-type and mutant FGF expression plasmids
Expression vector pET-26b was purchased from Novagen (Madison, WI). All restriction enzymes, Deep Vent DNA Polymerase, and T4 ligase were purchased from New England Biolabs (Beverly, MA). DH5α-competent cells were purchased from Gibco (Rockville, MD), and the QuikChange Site-Directed Mutagenesis Kit was purchased from Stratagene (La Jolla, CA). The plasmids pHBGF-1a-pET3c and pTI70301, which contain FGF-1 cDNA and FGF-2 cDNA, respectively, were initially divided into 4 cassettes: A, B, C, and D (Figure 1A).31 Each cassette contains approximately 25% of the open reading frame. The division between the first and second cassettes of FGF-1 was determined by the location of a single BamHI site, and division of the third and fourth by the natural presence of an SphI site, a single NcoI site was introduced to divide the second and third cassette. The FGF-1 construct was used as a template for division of FGF-2 into analogous cassettes. Since FGF-1 and FGF-2 have similar 3-dimensional structures, “shuffling” of cassettes between FGF-1 and FGF-2 allows for structural or functional analysis of regions of either in the context of the structural background of the other. Using these plasmids, FGF-2, FGF-1, FGF-2 deletion, chimeric, and site-directed mutants were manipulated and subcloned into pET-26b bacterial expression vector with a C-terminal His-tag, and all constructs were confirmed by DNA sequencing. FGF-2 and FGF-1 cDNA fragments were generated by polymerase chain reaction (PCR) and subcloned by using NdeI and XhoI cloning sites. The sense and antisense FGF-2 and FGF-1 primers were as follows, FGF-2 (sense): 5′ GGA ATT CCA TAT GGC AGC CGG GAG CAT CAC C 3′; FGF-2 (antisense): 5′ GGC CAT ATG GCA GCC GGG AGC ATC ACC 3′; FGF-1 (sense): 5′ GGA ATT CCA TAT GGC TAA TTA CAA GAA GCC C 3′; FGF-1 (antisense): 5′ CCG CTC GAG ATC AGA AGA GAC TGG 3′. The FGF-2 chimeric mutants were subcloned by using the restriction enzymes indicated (Figure 1B). For example, BamHI and XhoI fragments of pET26FGF-1 were ligated into BamHI- and XhoI-digested pET26FGF-2 vector. The resulting construct was termed 1222, because residues representing the first cassette were derived from FGF-1, and those in the next 3 cassettes were from FGF-2. Other chimeric mutants were subcloned in a similar way. The deletion mutants were generated by PCR and cloned into pET26FGF-2 using appropriate restriction enzymes resulting in clones ABC, ABD, ACD, and BCD (Figure 1C). Based on the initial results of fibrinogen binding of FGF-1, FGF-2, and of FGF-2 chimeric and deletion mutants, 4 regions were selected in FGF-2 cassettes C and D for site-directed mutagenesis. Deletions and substitutions of amino acids were generated by PCR-based site-directed mutagenesis.
Cassette structure of FGF constructs and strategy of mutagenesis. (A) Full-length FGF-2 and partial-length FGF-1 cDNA fragments were initially synthesized in 4 cassettes (A, B, C, and D) and ligated as indicated. The amino acid changes (Glu → Lys [E → K], His → Tyr [H → Y], Ala → Thr [A → T]) are shown above the FGF-2 sequence and were inserted to generate specific cleavage sites for the restriction enzymes indicated in parentheses. Numbering of residues is based on that used in the published crystal structures of FGF-1 and FGF-2.26-30 (B) Construction of FGF chimeric mutants. The mutants were subcloned by using indicated restriction enzymes. (C) Construction of FGF-2 deletion mutants. Deletion mutants were generated by PCR and cloned into pET26bFGF-2 with appropriated restriction enzyme as indicated, and the resulting constructs were ABC, BCD, ABD, and ACD. Gray shading in B and C represents FGF-2 fragments; white sections, FGF-1 fragments.
Cassette structure of FGF constructs and strategy of mutagenesis. (A) Full-length FGF-2 and partial-length FGF-1 cDNA fragments were initially synthesized in 4 cassettes (A, B, C, and D) and ligated as indicated. The amino acid changes (Glu → Lys [E → K], His → Tyr [H → Y], Ala → Thr [A → T]) are shown above the FGF-2 sequence and were inserted to generate specific cleavage sites for the restriction enzymes indicated in parentheses. Numbering of residues is based on that used in the published crystal structures of FGF-1 and FGF-2.26-30 (B) Construction of FGF chimeric mutants. The mutants were subcloned by using indicated restriction enzymes. (C) Construction of FGF-2 deletion mutants. Deletion mutants were generated by PCR and cloned into pET26bFGF-2 with appropriated restriction enzyme as indicated, and the resulting constructs were ABC, BCD, ABD, and ACD. Gray shading in B and C represents FGF-2 fragments; white sections, FGF-1 fragments.
Production of the His-tagged wild-type and mutant FGF proteins
Ni-NTA agarose was purchased from Qiagen (Valencia, CA), Halt protease inhibitor cocktail and GelCode Blue Stain Reagent were from Pierce (Rockford, IL), isopropyl-1-thio-β-D-galactopyranoside (IPTG) and BL21(DE3)plysS-competent cells were from Novagen, BL21-codonplus–competent cells were from Stratagene, and commercial FGF-2 and FGF-1 were from PeproTech (Rocky Hill, NJ). The FGF-1 constructs were transformed into Escherichia coli (E coli) strain BL21(DE3)plysS, and FGF-2 and FGF-2 mutant constructs were transformed into E coli strain BL21-codonplus. FGF expression was induced with 1 mM IPTG for 4 hours, after which the cultures were centrifuged and the pellet lysed in buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 1% Halt protease inhibitor cocktail, pH 8.0). The lysate was then incubated with lysozyme (1 mg/mL) for 30 minutes, sonicated on ice and centrifuged at 10 000g for 20 minutes. The supernatant was removed, incubated with 50% Ni-NTA agarose slurry at 4°C for 1 hour, and then the His-tagged protein was eluted with elution buffer. Commercially available and recombinant FGF-1 and FGF-2 were characterized by electrophoresis on 15% sodium dodecyl sulfate (SDS)–polyacrylamide gels followed by staining with GelCode Blue Stain Reagent.
Cell culture
Primary endothelial cells were obtained from human umbilical veins as described previously,22 seeded on 0.2% wt/vol gelatin-coated, 25-cm, tissue-culture flasks, and cultured in McCoy 5A medium (Flow Laboratories, McLean, VA) containing 20% fetal bovine serum (FBS), 50 μg/mL endothelial cell growth supplement (ECGS; Collaborative Research, Bedford, MA), and 100 μg/mL heparin (Sigma, St Louis, MO) until they reached confluence. The cells were passaged up to 2 times before use and then placed in suspension by trypsinization. Cells were suspended by rinsing in Hanks balanced salt solution followed by brief incubation with trypsin-EDTA (ethylenediaminetetraacetic acid; Gibco). The cells were pelleted by centrifugation for 10 minutes at 500g and resuspended in McCoy 5A medium.
[3H]thymidine incorporation
Cell proliferation was quantitated using [3H]thymidine incorporation as described previously.22 Briefly, approximately 2 × 104 endothelial cells were suspended in McCoy 5A medium supplemented with 20% FBS, 50 μg/mL ECGS, and 100 μg/mL heparin, were plated in gelatin-coated, 12-well, tissue-culture plates (Becton Dickinson, Franklin Lakes, NJ) and allowed to adhere for 4 to 5 hours. The medium was then removed, and the cells were washed twice with serum-free McCoy 5A medium. Serum-free medium was then added containing 1% Nutridoma (Roche Molecular Biochemicals, Indianapolis, IN), 1 ng/mL to 25 ng/mL commercial FGF-2, FGF-1 (PeproTech), or recombinant forms, and 1 μCi/mL (0.037 MBq) [3H]thymidine (NEN Life Science Products, Boston, MA). After incubation at 37°C for 24 hours, nonadherent cells were removed by washing twice with ice-cold phosphate-buffered saline (PBS). To each well, 500 mL of 10% ice-cold trichloroacetic acid was then added, and precipitates were collected on a filter using a manifold. Filters were washed twice with ice-cold 5% trichloroacetic acid, followed by 95% ethanol, allowed to air-dry, and then suspended in scintillation fluid overnight. Acid precipitable counts were quantitated using a scintillation counter. Each experiment was performed at least 3 times, and triplicate wells were used in each experiment. The significance of differences in means was determined using a 2-tailed Student t test. Variance is described as plus or minus SE.
Fibrinogen binding assay
FGF-2 recombinants were immobilized on Sepharose beads as described previously.20 Briefly, polyclonal anti–FGF-2 antibody (100 μg/mL) was incubated with 1 mL AFFI-gel-15 (Bio-Rad, Hercules, CA) in 0.1 M sodium phosphate buffer, pH 7.4, containing 0.5 M NaCl and gently mixed at 25°C for 2 hours, and over 97% of antibody was bound to the beads. Residual active ester sites were then blocked by the addition of 1 M ethanolamine, pH 8.0, and the suspension was washed several times with 0.1 M sodium phosphate buffer, pH 7.4, containing 0.5 M NaCl. Then, various FGF-2 recombinants (50 μg/mL) were added to this suspension and gently mixed at 25°C for one hour, after which unbound FGF was removed by washing with 0.1 M sodium phosphate buffer, pH 7.4, containing 0.25 M NaCl. For screening of FGF-2 recombinants, 3 nM 125I-fibrinogen was incubated at 37°C with a 0.04 mL suspension of immobilized FGF-2 recombinants in a final volume of 0.1 mL. Following incubation, the beads were separated by centrifugation at 3000g for 10 minutes, after which the supernatant was removed, and the beads were then washed rapidly twice with cold 0.1 M sodium phosphate buffer, pH 7.4, containing 0.5 M NaCl to minimize nonspecific association. The amount of bound fibrinogen was calculated from the radioactivity associated with the beads. Total binding was defined as the percent of radioactivity added that was bound, normalized to recombinant FGF-2 as 100%. Nonspecific binding was determined in parallel experiments in the presence of a 20-fold molar excess of unlabeled fibrinogen. Specific binding was total binding minus nonspecific binding. The same method was used for quantitative binding analysis of selected FGF-2 recombinants using 125I-fibrinogen at concentrations from 0.5 nM to 300 nM. Scatchard analysis was performed using the Ligand program from Biosoft (Ferguson, MO). Because a nonequilibrium binding assay was used, all dissociation constant (Kd) values represent apparent Kd. Each experiment was performed at least 3 times, and either triplicate or quadruplicate wells were used in each experiment.
Results
Recombinant FGF-1 and FGF-2 had appropriate mobilities on SDS–polyacrylamide gel electrophoresis (PAGE) that were identical to FGF-1 and FGF-2 peptides purchased commercially (Figure 2). Differences in structure from the wild-type forms include alterations of 3 residues in FGF-2 to create restriction sites at cassette junctions (Figure 1A) and also the addition of 6 histidine residues at the carboxyl terminus to allow affinity purification on Ni-NTA agarose. Additionally, FGF-1 lacks the first 20 residues coded by the cDNA because the form containing these residues cannot be expressed in sufficient amount in E coli.1 Recombinant and commercially available forms of FGF-1 and FGF-2 were equivalent in their capacity to support endothelial cell proliferation at concentration up to 25 ng/mL (Figure 3A-B). These results indicate that recombinant FGF-1 and FGF-2 are fully active and undegraded with identical electrophoretic migration and similar activities to peptides purchased commercially.
SDS-PAGE of commercial and recombinant FGF-1 and FGF-2. Lane 1, molecular weight markers; lane 2, commercial FGF-1; lane 3, recombinant FGF-1; lane 4, commercial FGF-2; and lane 5, recombinant FGF-2. kDa indicates kilodaltons.
SDS-PAGE of commercial and recombinant FGF-1 and FGF-2. Lane 1, molecular weight markers; lane 2, commercial FGF-1; lane 3, recombinant FGF-1; lane 4, commercial FGF-2; and lane 5, recombinant FGF-2. kDa indicates kilodaltons.
Proliferative activities of commercial and recombinant FGFs. [3H]Thymidine incorporation into DNA of endothelial cells. Cpm indicates counts per minute. (A) Commercial FGF-1 ( ) and recombinant FGF-1 (▪). (B) Commercial FGF-2 (
) and recombinant FGF-1 (▪). (B) Commercial FGF-2 ( ) and recombinant FGF-2 (▪). The data are mean ± SE.
) and recombinant FGF-2 (▪). The data are mean ± SE.
Proliferative activities of commercial and recombinant FGFs. [3H]Thymidine incorporation into DNA of endothelial cells. Cpm indicates counts per minute. (A) Commercial FGF-1 ( ) and recombinant FGF-1 (▪). (B) Commercial FGF-2 (
) and recombinant FGF-1 (▪). (B) Commercial FGF-2 ( ) and recombinant FGF-2 (▪). The data are mean ± SE.
) and recombinant FGF-2 (▪). The data are mean ± SE.
We next constructed a series of FGF-2 deletion mutants and FGF-1/FGF-2 chimeric forms by interchanging complementary cassettes. These were expressed, purified, and screened for fibrinogen binding (Table 1). The data are normalized to that observed with recombinant FGF-2 as 100% for total binding. None of the FGF-2 mutants with deletion of an entire cassette representing between 34 and 42 residues showed any affinity for fibrinogen, probably reflecting major structural disruptions. Analysis of binding results with the FGF-1/FGF-2 chimeric mutants showed that fibrinogen binding was comparable to intact FGF-2 for the forms 1222, 1122, and 2122. All other chimeric constructs were comparable to FGF-1 and showed no binding. This result suggested that residues within cassette C and D of FGF-2 were responsible for fibrinogen binding since substitution of the FGF-1 sequence for either the third or fourth FGF-2 cassette abolished binding.
Results of screening assays for binding of recombinant FGFs to fibrinogen
Constructs . | Total bound, %* . | Specific bound, %† . |
|---|---|---|
| FGF-2 (2222) | 100 | 100 |
| FGF-1 (1111) | 3 ± 2 | 1 ± 1 |
| 2212 | 2 ± 1 | 0 |
| 2211 | 5 ± 3 | 0 |
| 2221 | 9 ± 4 | 0 |
| 1121 | 6 ± 2 | 0 |
| 1122 | 45 ± 9 | 99 ± 15 |
| 2122 | 64 ± 21 | 99 ± 21 |
| 1112 | 5 ± 3 | 0 |
| 1222 | 38 ± 12 | 87 ± 18 |
| 2111 | 6 ± 2 | 0 |
| ACD‡ | 3 ± 1 | 8 ± 4 |
| BCD‡ | 4 ± 1 | 0 |
| ABD‡ | 1 ± 1 | 1 ± 1 |
| ABC‡ | 1 ± 1 | 0 |
| FGF-2_ID§∥ | 86 ± 6 | 100 ± 9 |
| FGF-2_IS∥ | 94 ± 8 | 100 ± 7 |
| FGF-2_2D∥ | 77 ± 17 | 79 ± 26 |
| FGF-2_2S∥ | 77 ± 14 | 83 ± 19 |
| FGF-2_3D∥ | 5 ± 1 | 23 ± 9 |
| FGF-2_3S∥ | 84 ± 6 | 100 ± 13 |
Constructs . | Total bound, %* . | Specific bound, %† . |
|---|---|---|
| FGF-2 (2222) | 100 | 100 |
| FGF-1 (1111) | 3 ± 2 | 1 ± 1 |
| 2212 | 2 ± 1 | 0 |
| 2211 | 5 ± 3 | 0 |
| 2221 | 9 ± 4 | 0 |
| 1121 | 6 ± 2 | 0 |
| 1122 | 45 ± 9 | 99 ± 15 |
| 2122 | 64 ± 21 | 99 ± 21 |
| 1112 | 5 ± 3 | 0 |
| 1222 | 38 ± 12 | 87 ± 18 |
| 2111 | 6 ± 2 | 0 |
| ACD‡ | 3 ± 1 | 8 ± 4 |
| BCD‡ | 4 ± 1 | 0 |
| ABD‡ | 1 ± 1 | 1 ± 1 |
| ABC‡ | 1 ± 1 | 0 |
| FGF-2_ID§∥ | 86 ± 6 | 100 ± 9 |
| FGF-2_IS∥ | 94 ± 8 | 100 ± 7 |
| FGF-2_2D∥ | 77 ± 17 | 79 ± 26 |
| FGF-2_2S∥ | 77 ± 14 | 83 ± 19 |
| FGF-2_3D∥ | 5 ± 1 | 23 ± 9 |
| FGF-2_3S∥ | 84 ± 6 | 100 ± 13 |
Total binding was measured in the absence of cold fibrinogen and is expressed as a percentage of recombinant FGF-2.
Specific binding = (total binding - nonspecific binding) / (total binding × 100).
A, B, C, and D refer to cassettes in FGF-2.
The numbers 1, 2, and 3 refer to areas of site-directed mutagenesis in Figure 2A.
D indicates the deletion of the sequence indicated in Figure 2A, and S indicates substitution of FGF-2 sequence for the corresponding residues in FGF-1.
To identify specific residues that could contribute to a unique binding site on FGF-2, we next compared the sequences of FGF-1 and FGF-2 within cassettes C and D (Figure 4A) and focused attention on the 4 regions indicated for site-directed mutagenesis. Deletion mutants were constructed for regions 1, 2, and 3 in which the indicated residues in FGF-2 were removed, and substitution mutants were also made in which the residues from FGF-2 were substituted for those present in FGF-1 (Figure 4B). These mutants were expressed, purified, and screened for binding. We expected that deletion of the binding site on FGF-2 would eliminate binding activity and that substitution of the residues representing the binding site in FGF-2 into the FGF-1 backbone would impart new binding activity. The deletion and substitution mutants involving sites 1 and 2 exhibited no reduction in binding activity (Table 1) indicating that they did not involve the binding site.
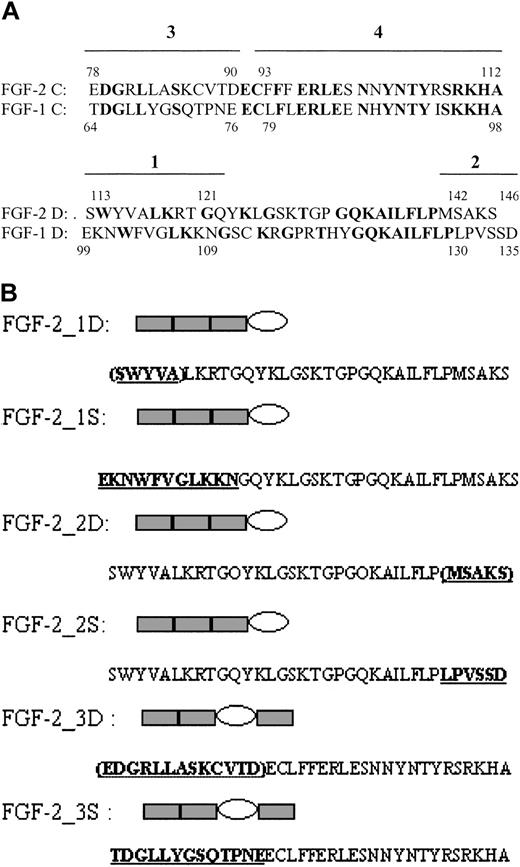
Location and structure of site-directed mutants. (A) Amino acid sequence of cassettes C and D of FGF-2 and FGF-1. Bold characters represent identical residues between FGF-1 and FGF-2. The 4 regions that were selected for site-directed mutagenesis are indicated by lines above the sequence. (B) Structures of recombinant FGF forms made by site-directed mutagenesis. The ovals indicate the cassettes in which changes in sequence were made. FGF-2_1D, _2D, _3D represent deletion mutants in which the residues in parentheses were removed. FGF-2_1S, _2S, _3S represent substitution mutants in which the residues in bold derived from FGF-1 were substituted into the FGF-2 cassette. Residues in bold with underlining are derived from FGF-1.
Location and structure of site-directed mutants. (A) Amino acid sequence of cassettes C and D of FGF-2 and FGF-1. Bold characters represent identical residues between FGF-1 and FGF-2. The 4 regions that were selected for site-directed mutagenesis are indicated by lines above the sequence. (B) Structures of recombinant FGF forms made by site-directed mutagenesis. The ovals indicate the cassettes in which changes in sequence were made. FGF-2_1D, _2D, _3D represent deletion mutants in which the residues in parentheses were removed. FGF-2_1S, _2S, _3S represent substitution mutants in which the residues in bold derived from FGF-1 were substituted into the FGF-2 cassette. Residues in bold with underlining are derived from FGF-1.
The deletion mutant involving region 3 showed absent binding activity, but the mutant with FGF-1 residues substituted for those in FGF-2 exhibited binding comparable to that of intact FGF-2. These findings were interpreted as indicating that the large deletion of 13 residues disrupted the conformation in a similar way to that observed with deletion of a whole cassette, and that this abolished binding. However, the high binding observed with the substitution mutant indicated that the FGF-2–specific binding site was not contained within this region.
In region 4, comparisons of the sequences of FGF-2 from residues 93-112 with the corresponding residues 79-98 in FGF-1 showed that only 5 differed (Figure 5).
Specific residues involved in fibrinogen binding. The sequences of FGF-2 and FGF-1 in cassette C are shown. These are compared with the sequence of cassette C of 2 recombinant forms termed 221*2 and 221*1. The cassette C sequence in both of these mutants is derived from FGF-1 with the exception of the 5 residues indicated by bold letters and underlining that represent the corresponding, different residues derived from the FGF-2 sequence.
Specific residues involved in fibrinogen binding. The sequences of FGF-2 and FGF-1 in cassette C are shown. These are compared with the sequence of cassette C of 2 recombinant forms termed 221*2 and 221*1. The cassette C sequence in both of these mutants is derived from FGF-1 with the exception of the 5 residues indicated by bold letters and underlining that represent the corresponding, different residues derived from the FGF-2 sequence.
We, therefore, constructed a new substitution mutant replacing these residues in cassette C of FGF-1 with the 5 corresponding residues from FGF-2 and expressed 2 new mutants. The chimeric form 2212, with the C cassette representing the wild-type sequence from FGF-1, showed no binding. The new mutant, 221*2, altered only by the substitution of the 5 residues indicated, showed high-affinity fibrinogen binding (Figure 6A). Scatchard analysis suggested the presence of 2 binding sites with Kd1 of 5.3 nM and Kd2 of 67 nM (Figure 6B). A second construct was made with the same 5 residues from FGF-2 substituted into the 2211 chimeric form, which also showed no appreciable binding. The new construct, 221*1, showed high-affinity binding (Figure 6C), demonstrating a single binding site with Kd of 8.6 nM (Figure 6D). These affinities were close to those of the wild-type FGF-2 which showed 2 sites of Kd1 of 1.3 nM and Kd2 of 260 nM.20 Thermodynamic stability of the complexes was determined from Kd values obtained from the binding experiments using the equation: ΔG0 = –RT lnKd. Comparisons of the binding energies attributed to the high-affinity sites in construct 221*1 (11.0 kcal/mol) and 221*2 (11.3 kcal/mol) with wild-type FGF-2 (12.2 kcal/mol) indicated that this 5-residue substitution contributed over 90% of the thermodynamic stability to the respective interactions with fibrinogen.
Binding of recombinant 221*2 and 221*1 to fibrinogen. [125I]-fibrinogen was incubated with 221*2 (A) and 221*1 (C) immobilized on Sepharose beads, and the amount of bound protein was determined as radioactivity associated with the beads following centrifugation and washing. Nonspecific binding was determined in the same way in the presence of a 20-fold molar excess of unlabeled fibrinogen. Specific binding is shown and was calculated by subtracting the nonspecific from the total bound. Each point represents the mean ± SD of 3 separate experiments. Scatchard plot of the data for 221*2 (B) and 221*1 (D). The best fit of the data was determined using the ligand program.32 The correlation coefficients were 0.99.
Binding of recombinant 221*2 and 221*1 to fibrinogen. [125I]-fibrinogen was incubated with 221*2 (A) and 221*1 (C) immobilized on Sepharose beads, and the amount of bound protein was determined as radioactivity associated with the beads following centrifugation and washing. Nonspecific binding was determined in the same way in the presence of a 20-fold molar excess of unlabeled fibrinogen. Specific binding is shown and was calculated by subtracting the nonspecific from the total bound. Each point represents the mean ± SD of 3 separate experiments. Scatchard plot of the data for 221*2 (B) and 221*1 (D). The best fit of the data was determined using the ligand program.32 The correlation coefficients were 0.99.
Discussion
The results demonstrate that binding of FGF-2 to fibrinogen is mediated by residues including Phe95, Ser100, Asn102, Arg107, and Arg109. These residues each differ from those at comparable sites in FGF-1, which does not bind to fibrinogen, and substituting them into the FGF-1 sequence converts it from a nonbinding to a binding form. This area represents a high-affinity site with Kd of 5.3 nM for one construct and Kd of 8.6 nM for the second construct. These affinities are close to the Kd for the high-affinity site of native FGF-2 of 1.3 nM and together are responsible for over 90% of the binding energy. These 5 residues are conserved in the FGF-2 sequence of 2 mammals, rabbit and mouse, but are absent in Drosophila melanogaster and Caenorhabditis elegans that lack fibrinogen.33,34 This suggests an evolutionary conservation of an important functional site. The location of the lower-affinity site is uncertain, but analysis of fibrinogen binding of the construct 221*2 suggests a second, lower-affinity site, whereas a single site was found for 221*1. Therefore, the lower-affinity binding site may be located within the fourth cassette.
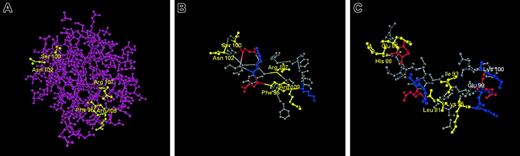
The location within FGF-2 of the 5 residues involved in fibrinogen binding is shown in Figure 7A. The structures of FGF-1 and FGF-2 are similar in the region containing the 5 residues that impart fibrinogen binding.26-30 Within FGF-2 (Figure 7B), Phe95, Arg107, and Arg109 form a cluster, and Ser100 and Asn102 are close, separated by 0.54 nM. The structural arrangement in FGF-1 is similar, but the difference in residues results in a significant change in charge distribution. Thus, the Phe95, Arg107, Arg109 “cluster” has a greater basic electrostatic character than the comparable Leu81, Ile93, Lys95 group of FGF-1, where a hydrophobic Ile replaces a basic Arg residue. Furthermore, the Ser100 and Asn102 of FGF-2 likely possess less electronegative character than Glu86 and His88 of FGF-1. Based on these modest sequence differences, we speculate that a more basic electrostatic potential within this region makes a significant contribution to the affinity of FGF-2 for fibrinogen. Alternatively, the enhanced affinity may result from favorable long-range interactions facilitated by the replacement of FGF-1 residues with those from FGF-2.
Location of the binding site. (A) The 5 residues involved in fibrinogen binding to FGF-2 are shown in yellow. (B) Structure of FGF-2 in the fibrinogen binding region. Portions of the crystal structure of FGF-2 are shown with the 5 residues in FGF-2 related to fibrinogen binding indicated in yellow. Positively charged residues are shown in blue and negatively charged residues in red. Approximate distances are: Ser100 to Agn102, 0.54 nM; Ser100 to Arg109, 2.06 nM. (C) Comparison of structure of FGF-1 with that of FGF-2 in the fibrinogen binding region. The residues in FGF-1 corresponding to the 5 residues in FGF-2 involved in fibrinogen binding are shown in yellow. Other positively charged residues are shown in blue and negatively charged residues in red. The location of 2 additional residues in FGF-1 with no corresponding counterparts in FGF-2 (Glu99 and Lys100) are also indicated. Based on published crystal structures.26-30
Location of the binding site. (A) The 5 residues involved in fibrinogen binding to FGF-2 are shown in yellow. (B) Structure of FGF-2 in the fibrinogen binding region. Portions of the crystal structure of FGF-2 are shown with the 5 residues in FGF-2 related to fibrinogen binding indicated in yellow. Positively charged residues are shown in blue and negatively charged residues in red. Approximate distances are: Ser100 to Agn102, 0.54 nM; Ser100 to Arg109, 2.06 nM. (C) Comparison of structure of FGF-1 with that of FGF-2 in the fibrinogen binding region. The residues in FGF-1 corresponding to the 5 residues in FGF-2 involved in fibrinogen binding are shown in yellow. Other positively charged residues are shown in blue and negatively charged residues in red. The location of 2 additional residues in FGF-1 with no corresponding counterparts in FGF-2 (Glu99 and Lys100) are also indicated. Based on published crystal structures.26-30
Structural differences in this area may also explain the failure of the 2221 recombinant to bind even though the putative 5-residue binding site is intact. We note that the fourth cassette of FGF-1 contains an extra 2 residues not present in FGF-2. These are Glu99 and Lys100 and are located close to the binding site in FGF-2 (Figure 7C). It is possible that the addition of these charged residues to the FGF-2 site interferes with binding.
The residues identified as important in fibrinogen binding are close to those previously shown to be involved in receptor interactions. Plotnikov et al35 identified FGF-2 sites that interact with the FGF receptor using crystallography and identified Glu99 to Tyr103 as an important sequence. Residues Asn102 and Tyr103 were part of a primary interactive site, and Glu99, Ser100, and Asn101 were involved in the secondary binding site. Using site-directed mutagenesis, Springer et al36 identified Tyr103 as part of the primary receptor interactive site and Lys110, Tyr111, and Trp114 within the secondary receptor binding site. Venkataraman et al37 suggested that the primary receptor binding site involved Glu96, Asn101, and Tyr103, whereas Lys110, Tyr111, and Trp113 were part of the secondary binding site for the receptor based on molecular modeling. Therefore, the fibrinogen binding site described herein appears to overlap the receptor binding site, and residues Ser100 and Asn102 may be common to both. Binding to fibrinogen apparently does not prevent receptor binding, but rather, the proliferative activity of FGF-2 is increased by fibrinogen.21 Specific effects of fibrinogen on binding and activation of FGF receptors by FGF-2 will require additional study.
Interactions with heparin primarily involve residues carboxyl terminal to the fibrinogen binding region.6,37,38 However, Asn101 has been identified as interacting with heparin based on cocrystal structures of FGF-2 and tetra- and hexasaccharides.38 Therefore, the binding of FGF-2 with fibrinogen may also modify heparin interactions. We have found that fibrinogen can substitute for cell surface heparan sulfate proteoglycans as a cofactor in endothelial cell proliferation (A.S. and C.F., unpublished observations, November 2003), and the proximity of binding sites suggests that fibrinogen binding may alter interactions with heparin and heparan sulfate proteolycans. VEGF, another angiogenic growth factor, also binds fibrinogen but does not compete for binding with FGF-2,22 consistent with its marked differences in structure. Interleukin-1 α (IL-1α) and IL-1β, however, share structural features with FGF-1 and FGF-2,39,40 including similar secondary structure in the comparable region to the FGF-2 fibrinogen binding site, suggesting that they may also interact with fibrinogen. Binding of critical cell regulatory peptides to fibrin(ogen) indicates that it plays in an important role in localizing and organizing the cellular responses to injury.
Prepublished online as Blood First Edition Paper, November 20, 2003; DOI 10.1182/blood-2003-08-2638.
Supported in part by grant nos. HL-30616 (C.W.F.), and HL-35627, HL-34328, and RR15555 (T.M.) from the National Institutes of Health, Bethesda, MD.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 1. Cassette structure of FGF constructs and strategy of mutagenesis. (A) Full-length FGF-2 and partial-length FGF-1 cDNA fragments were initially synthesized in 4 cassettes (A, B, C, and D) and ligated as indicated. The amino acid changes (Glu → Lys [E → K], His → Tyr [H → Y], Ala → Thr [A → T]) are shown above the FGF-2 sequence and were inserted to generate specific cleavage sites for the restriction enzymes indicated in parentheses. Numbering of residues is based on that used in the published crystal structures of FGF-1 and FGF-2.26-30 (B) Construction of FGF chimeric mutants. The mutants were subcloned by using indicated restriction enzymes. (C) Construction of FGF-2 deletion mutants. Deletion mutants were generated by PCR and cloned into pET26bFGF-2 with appropriated restriction enzyme as indicated, and the resulting constructs were ABC, BCD, ABD, and ACD. Gray shading in B and C represents FGF-2 fragments; white sections, FGF-1 fragments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/6/10.1182_blood-2003-08-2638/6/m_zh80060458530001.jpeg?Expires=1765925207&Signature=SqKSS~dtWO1ww2sHO59RzwJXbyGwFTqaNqwuoQDZvlZUX3vzEwXCqovazMMhDbn77usMz3zmU6md-uJkPiTMoszbX4zNvlUR4lNUjKvFIWubA-GDctaSUsxQNkBOdUM5xFctBDncbU-WPdrpbk2Pr~OobqGbXTJSJq8w3HZRDollrJ8u7nFQt4XP0jQYBSGdqowLHzZH5u~zWpOUJG222MfTqNAdwgTMuXuQAe2hJt55czAm7riIKzJ~RIPCw1nICE-hIWvtcg9Pe3c~MgCBIg3hzr~OMePWeP7VJ3H4AhMiWqQaZs4r-B3PH0JX9oGOghG1aZ6psW47WLDp4a2kmQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 3. Proliferative activities of commercial and recombinant FGFs. [3H]Thymidine incorporation into DNA of endothelial cells. Cpm indicates counts per minute. (A) Commercial FGF-1 () and recombinant FGF-1 (▪). (B) Commercial FGF-2 () and recombinant FGF-2 (▪). The data are mean ± SE.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/6/10.1182_blood-2003-08-2638/6/m_zh80060458530003.jpeg?Expires=1765925207&Signature=0E8wQ9HvngHsHMHcemrv2-KO7NtKecULqSqi12yVWfBPVi92HFGyUWkFJ2w04bBcAKDOI-38TT96eZDVk-oYWNlF4hms-tJhzDBgHAPhKml-kEGurKTyjTzr9450mG-0lgdJId3UCWqVH9qhnMXbUulaHOxw7tm-zEL2ThlDwhikCb~dcNL1rpX5-W4XKgpD~zi7kemnRH-Xbhveyz7JqlWt7IoLF-w1HSR1c8ad7AsssG-49-6~g~HnW4O3GgODUEdG-KZWpNZR2QC3m0av1ahlAO4wZghlVNOgQR0jwJ23zNuU0fYN2nniFhcSYx0zcCCGlXHfw65kgoIlSW~ngQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 6. Binding of recombinant 221*2 and 221*1 to fibrinogen. [125I]-fibrinogen was incubated with 221*2 (A) and 221*1 (C) immobilized on Sepharose beads, and the amount of bound protein was determined as radioactivity associated with the beads following centrifugation and washing. Nonspecific binding was determined in the same way in the presence of a 20-fold molar excess of unlabeled fibrinogen. Specific binding is shown and was calculated by subtracting the nonspecific from the total bound. Each point represents the mean ± SD of 3 separate experiments. Scatchard plot of the data for 221*2 (B) and 221*1 (D). The best fit of the data was determined using the ligand program.32 The correlation coefficients were 0.99.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/6/10.1182_blood-2003-08-2638/6/m_zh80060458530006.jpeg?Expires=1765925207&Signature=StB2Bgn4pfiR1RrGvfQiyrHmFjPMZM2cW793sKS-FO5NeBTjEcmVffd4JBe-1sZNTqDTIqc61f1JZa8GzoiscgT0bdwk0Q2wRaLx8LJ040XpAnibKQkbI9b9eIay-aKEKBh69q0-ZxsryVv1N81JPmWTuRsisXZ1NNODqGJxoK-bBObMde3zDGkr~23ouuxe65MQ44s5cZNN1hYP8QU8rtooJRzNoKplpKhbl64xAeGIQl1V-0-~D~QPz15l7NpsgcA3njKJd1o2KadX2XcBcecCFpg8FZPHJVkFMRcbpYYXsmOGhNUFoDV~NAyX94YjVYXVJNxIiluxluUJ0Anzzg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal