Abstract
Recently there has been interest in developing cell and gene therapies with adult stem cells from human bone marrow referred to as mesenchymal stem cells or marrow stromal cells (hMSCs). We incubated early-passage hMSCs in serum-free medium without cytokines or other supplements for 2 to 4 weeks. Surprisingly, a subpopulation of the cells survived serum deprivation and then began to proliferate in serum-containing medium. The cells selected by serum deprivation had longer telomeres than control cells. Also, the patterns of gene expression revealed by reverse transcriptase–polymerase chain reaction (RT-PCR) assays and microarray data indicated that the cells selected by serum deprivation were a subpopulation of very early progenitor cells with enhanced expression of octomer-binding transcription factor 4 (OCT-4) and several other genes characteristically expressed in embryonic cells.
Introduction
Human mesenchymal stem cells or marrow stromal cells (hMSCs) can differentiate into osteoblasts, chondrocytes, adipocytes, myocytes, and other cell types.1-9 They are readily isolated from bone marrow by their adherence to tissue culture plastic and can be expanded extensively in medium containing high concentrations of fetal calf serum (FCS). The cells are readily cloned as single-cell–derived colonies, but both the colonies and the cells within a colony are heterogeneous in morphology, rates of proliferation, and efficacy with which they differentiate.3,10,11 Two morphologically distinct cell phenotypes are seen in early-passage, low-density cultures: small, spindle-shaped cells that are rapidly self renewing (RS cells) and large, flat cells that replicate slowly and appear more mature.3,10,12 The proportion of RS cells remains high for several passages if the cultures are maintained at low density, but the larger cells predominate in later passages and the cells cease proliferation at about the Hayflick limit of 50 population doublings.9,10,12,13, Several investigators10,13-15 identified distinctive surface epitopes on hMSCs but none of the epitopes have been shown to distinguish early progenitors from more mature cells in cultures of hMSCs. Marrow cells that may or may not be related to hMSCs were recently isolated by first selecting Lin– and glycophorin– marrow cells and then culturing the cells on fibronectin-coated plated cells in medium containing 2% FCS and a series of growth factors.16,17 After about 25 population doublings, cultures of multipotent adult precursor cells (MAPCs) were obtained that expressed telomerase, were multipotential for differentiation, and continued to expand for well over 50 population doublings. Recently, we found that a subpopulation of hMSCs with distinctive features can be selected from early-passage cultures of hMSCs by incubation of the cells for 2 to 4 weeks without serum, growth factors, or other supplements.
Materials and methods
Human MSCs were prepared as described previously.10,12 In brief, nucleated cells were isolated with a density gradient (Ficoll-Paque; Pharmacia, Uppsala, Sweden) from 2 mL human bone marrow aspirated from the iliac crests of healthy volunteers under a protocol approved by the Institutional Review Board of Tulane University Health Sciences Center. All the nucleated cells (30 to 70 million) were plated in a 145-cm2 dish in 20 mL complete culture medium: α–modified minimum essential media (αMEM; GIBCO BRL, Carlsbad, CA); 17% fetal bovine serum ([FBS] lot-selected for rapid growth of MSCs; Atlanta Biologicals, Norcross, GA); 100 units/mL penicillin; 100 μg/mL streptomycin; and 2 mM l-glutamine (GIBCO BRL). After 24 hours at 37°C in 5% CO2, nonadherent cells were discarded and the adherent cells were incubated in fresh medium for 4 days. The cells were lifted with 0.25% trypsin and 1 mM EDTA (ethylenediaminetetraacetic acid) for 5 minutes at 37°C and replated at 50 cells/cm2 in an interconnecting system of culture flasks (6320 cm2; Cell Factory; Nunc, Wiesbaden, Germany). After 7 to 9 days, the cells were lifted with trypsin/EDTA, suspended at about 106 cells/mL in 5% dimethyl sulfoxide (DMSO) and 30% FCS in αMEM and frozen in 1-mL aliquots in liquid nitrogen as passage 1 cells. The vials of passage 1 cells were thawed, plated in a 60-cm2 dish, incubated for 4 days, and lifted with trypsin/EDTA to recover viable cells. The cells were then plated in complete medium at 50 to 500 cells/cm2, incubated for 4 to 7 days, and lifted with trypsin/EDTA to recover passage 2 cells. Later-passage cells were obtained by replating the cells at 50 to 500 cells/cm2, incubating them for 4 to 7 days, and recovering the cells with trypsin/EDTA.
To prepare serum-deprived (SD) cells and controls, passage 2 or later-passage cells were plated at 50 to 500 cells/cm2 in 15-cm diameter plates. One set of plates was washed with phosphate-buffered saline (PBS) and incubated with αMEM without serum or growth factors to prepare SD cells. The second set was incubated with complete culture medium with FCS as a parallel control set. The medium was replaced every 4 days for 2 to 4 weeks. After serum deprivation, both control and SD cells were recovered by lifting with trypsin/EDTA and replated with complete culture medium with 17% FCS. Both controls and SD cultures were expanded in complete culture medium containing FCS.
Telomere length assay
In assays of telomere lengths, the day-0 sample was prepared by plating passage 2 hMSCs at 100 cells/cm2 in a 15-cm diameter dish and incubating in complete medium for 5 days. The SD sample was prepared by incubation of the day-0 sample in medium without FCS for 3 weeks, then replating all the surviving cells in a 15-cm diameter dish, and incubation in complete medium for 5 days. The control sample was prepared by incubating the day-0 sample in complete medium for 3 weeks, then replating at 100 cells/cm2, and incubation in complete medium for 5 days. Genomic DNA was isolated from 1 × 106 cells (MagNA Pure LC DNA Isolation Kit I; Roche Molecular Biochemicals, Indianapolis, IN) and telomere length was assayed with a commercial kit (Telo Tagg; Roche Molecular Biochemicals). In brief, 10 μg genomic DNA was digested with RsaI and Southern blotted into nylon membrane. Telomere lengths were determined using chemiluminescent assay to detect digoxigenin (DIG)–labeled probe.
Western blot analysis
Cells were prepared as for the assays of telomere length and lysed in buffer (Lysis Buffer; Roche Molecular Biochemicals) supplemented with protease inhibitor cocktail (Sigma Biochemicals, St Louis, MO) and protein concentration was determined (Micro BCA Kit; Pierce Biotechnology, Rockford, IL). The cell lysate (50 to 100 μg of protein) was fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; Novex 12% gels; Invitrogen, Carlsbad, CA). The sample was transferred to a filter (Immobilon P; Millipore, Bedford, MA) by electro-blotting (Immunoblotting Apparatus; Invitrogen). The filter was blocked for 30 minutes with PBS containing 5% nonfat dry milk and 0.1% Tween 20 and then incubated for one hour with the primary antibody. For detection of p21WAF1, the filter was incubated with 1:500 dilution of anti-p21 antibody (Pharmingen, San Diego, CA). The p53 was detected by incubating with a 1:500 monoclonal antibody (DO-1; Pharmingen). The filter was washed 4 times for 15 minutes each with PBS containing 0.1% Tween 20. Bound primary antibody was detected by incubating for 1 hour with horseradish peroxidase goat antimouse immunoglobulin G (IgG; Pharmingen) diluted 1:10 000 in PBS containing 5% nonfat dry milk. The filter was washed with PBS containing 0.1% Tween 20 and developed using a chemiluminescence assay (West-Femto Detection Kit; Pierce Biotechnology).
RT-PCR analysis
RNA was isolated from 0.5 × 106 cells (RNeasy RNA Isolation Kit; Qiagen, Valencia, CA) and 50 ng total RNA was used to perform one-step reverse transcriptase–polymerase chain reaction (RT-PCR; Titan One Step RT-PCR Kit; Roche Biochemical). Five microliters of the product was loaded for agarose gel electrophoresis. For semiquantitative RT-PCR analysis, 100, 20, and 4 ng of total RNA was used to perform one-step RT-PCR. The following primer sets were used: catalytic subunit of telomere replicating telomerase (hTERT): 5′-cgctggtggcccagtgcctg and 5′-ctcgcacccggggctggcag; OCT-4: 5′-cgctccggcccacaaatctc and 5′-ccgcacgacaaccgcaccat; ornithine decarboxylase (ODC) antizyme: 5′-ccgcacgacaaccgcaccat and 5′-cgctccggcccacaaatctc; activating transcription factor 5 (ATF-5): 5′-aaggagctggaacagatggaagac and 5′-ttgtaaacctcgatgagcaggtcc; fibroblast growth factor-2 (FGF2): 5′-gtgtgctaaccgttacctggctat and 5′-aggtaagcttcactgggtaacagc; FGF2 receptor (FGF2R): 5′-tgtgctaaccgttacctggctatg and 5′-aggtaagcttcactgggtaacagc; glutathione-S–transferase (GST): 5′-tgggaagaacaagatcacccagag and 5′-gttgtccaggtagctcttccaagt; keratin-associated protein 1 (KAP1): 5′-acccaaccttcagatcaactcctg and 5′-ccggttgagaagctaggaaatcca; lysyl oxidase (LOX): 5′-ttacccagccgaccaagatattcc and 5′-tcataacagccaggactcaatccc; sine oculis homeobox homolog-2 (SIX2): 5′-actgagtcttgaaccacagaaggg and 5′-acagaaggagagaatgaacggtgg; homeobox C6 (HOXC6): 5′-tcaattccaccgcctatgatccag and 5′-aatcctgagcgattgaggtctgtg; and β-actin: 5′-tcatgaagtgtgacgttgacatccgt and 5′-cttagaagcatttgcggtgcacgatg.
Clonogenicity, growth curve, and differentiation assays
For the clonogenicity assay, cells were plated at 1 cell/well into a 96-well plate using an automated instrument (Clonecyte Accessory and FACSvantage; BD Biosciences, San Jose, CA). The cells were incubated with complete culture medium for 10 days, stained with Crystal Violet, and colonies with diameters of 2 mm or greater were counted. For the differentiation assay, the cells were plated at 1 cell/well in a 96-well plate and incubated in complete culture medium for 9 days and medium was changed to either osteogenic medium (10–8 M dexamethasone/0.2 mM ascorbic acid/10 mM β-glycerolphosphate; Sigma) or adipogenic medium (0.5 μM hydrocortisone/0.5 mM isobutylmethylxanthine/60 μM indomethacin). The incubation was continued for 3 weeks with a change of medium every 4 days. The cells were fixed with 10% formalin and stained with Alizarin Red (Sigma) or Oil Red O (Fisher Scientific, Hampton, NH). For chondrocyte differentiation, a pellet culture system18 was used. Approximately 200 000 MSCs were placed in a 15-mL polypropylene tube (Falcon, Heidelberg, Germany) and centrifuged to pellet. The pellet was cultured at 37°C with 5% CO2 in 500 μL chondrogenic media that contained 500 ng/mL bone morphogenetic protein-6 (BMP-6; R&D Systems, Minneapolis, MN) in addition to high-glucose Dulbecco modified Eagle medium (DMEM) supplemented with 10 ng/mL transforming growth factor-β3 (TGF-β3), 107 M dexamethasone, 50 μg/mL ascorbate-2-phosphate, 40 μg/mL proline, 100 μg/mL pyruvate, and 50 mg/mL ITS+ Premix (6.25 μg/mL insulin, 6.25 μg/mL transferrin, 6.25 ng/mL selenious acid, 1.25 mg/mL bovine serum albumin [BSA], and 5.35 mg/mL linoleic acid; Becton Dickinson, San Jose, CA). The medium was replaced every 3 to 4 days for 21 days. For immunostaining, paraffin-embedded sections were deparaffinized using xylene and rehydrated though graded alcohols. The pellet was pretreated with 25 mg/mL hyaluronidase (Sigma) in PBS for 30 minutes at 37°C for optimal antigen retrieval. After washing in PBS 3 times at room temperature, nonspecific staining was blocked with PBS containing 10% normal goat serum for 1 hour at room temperature. Rabbit antibody against type II collagen (1:500; Cosmo Bio, Tokyo, Japan) was placed on section for 72 hours at 4°C. After washing with PBS, the sections were incubated with biotinylated goat antirabbit (1:500; Vector Laboratories, Braintree, MA) for 1 hour at room temperature, washed, and then incubated with streptavidin-conjugated Texas Red (1:400; Vector Laboratories) for 1 hour at room temperature. The slides were washed in PBS, dried, and coverslipped with antifade mounting medium containing DAPI (4,6 diamidino-2-phenylindole; Vectashield; Vector Laboratories). For growth curve assay, 105 cells were plated into 3 wells of a 6-well plate. The cells were trypsinized and counted after 5 days of incubation. The data presented here are a mean of 2 donor samples.
Microarray analysis
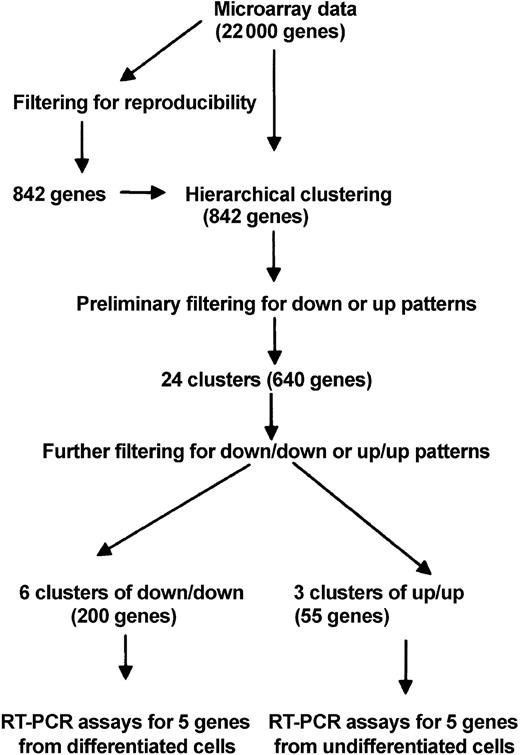
Total RNA was extracted (RNeasy Kit; Qiagen) from 1 × 106 cells of 5 samples from each of 2 donors as described in Figure 4A. The RNA expression was assayed with a chip containing oligonucleotide probes for about 22 000 human genes (HG-U133A array; Affymetrix, Santa Clara, CA). For the initial filtering for reproducibility of the data, the Microarray Suite 5.0 program (Affymetrix) was used to obtain signal intensities. The data were then filtered in the following steps: (1) genes that were not consistently scored as absent or present in the 3wkS (cells incubated with serum for 3 weeks) and 3wkSD (cells incubated without serum for 3 weeks) samples from both donors (Figure 4A) were eliminated; (2) genes scored as absent in all 4 samples were eliminated; (3) steps 1 and 2 were combined to reduce the number of genes to about 8000; (4) genes in the 4 samples that did not show significant change from day 0 (Figure 4A) were eliminated; (5) genes that did not show consistent scores of increase or decrease in the 4 samples were eliminated; (6) steps 4 and 5 were combined to reduce the number of genes to 915; and (7) steps 5 and 6 were repeated for the 4 samples of +5dSDS (3wkSD samples incubated for 5 days in medium containing 17% FCS) and +5dSS (3wkS samples incubated for 5 days in medium containing 17% FCS) and redundancies were eliminated to reduce the number of genes to 842. The hierarchical cluster analysis was carried out on the 842 genes with the dChip 1.3+ program.19 Adjacent genes were merged if the cluster of merged genes maintained the same pattern of expression.
RNA and RT-PCR. (A) Schemes for the preparation of cells for microarray and RT-PCR analyses. Day 0 indicates initial samples of passage 2 hMSCs; 3wkSD and 3wkS, cells incubated without or with serum for 3 weeks; and +5dSDSS and +5dS, 3wkSD and 3wkS samples incubated for 5 days in medium containing 17% FCS. (B) RT-PCR analyses of RNA obtained from the samples described in panel A showing enhanced expression of genes that are expressed in very early progenitor cells.
RNA and RT-PCR. (A) Schemes for the preparation of cells for microarray and RT-PCR analyses. Day 0 indicates initial samples of passage 2 hMSCs; 3wkSD and 3wkS, cells incubated without or with serum for 3 weeks; and +5dSDSS and +5dS, 3wkSD and 3wkS samples incubated for 5 days in medium containing 17% FCS. (B) RT-PCR analyses of RNA obtained from the samples described in panel A showing enhanced expression of genes that are expressed in very early progenitor cells.
Results
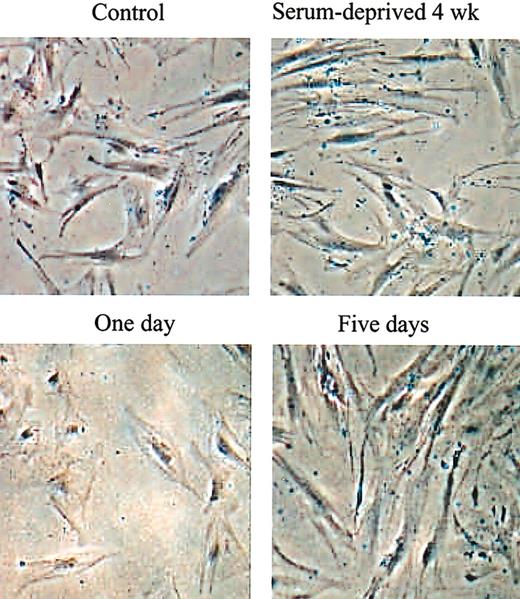
The experiments were carried out with frozen vials of passage 2 hMSCs that were previously shown to expand rapidly in cultures.12,13 The vials were thawed and viable cells were recovered by incubating them on the plates for 2 days in complete medium containing FCS. The adherent viable cells were lifted with trypsin/EDTA, plated at 100 cells/cm2, and incubated in complete medium containing FCS for 5 days so that the cultures expanded into the log phase of growth and about half the cells consisted of RS cells.10,12 The medium was then replaced with the same medium devoid of serum, growth factors, or other supplements, and the incubation continued for 2 to 4 weeks. Initially, many of the cells in the serum-free medium appeared apoptotic and necrotic (Figure 1). Control cultures incubated in medium containing FCS became confluent. The SD cells were lifted with trypsin/EDTA, plated at 100 cells/cm2, and incubated in medium containing FCS. After 5 days, the morphology of the SD cells changed from large, apparently senescent cells to the spindle-shaped cells characteristic of early-passage hMSCs (Figure 1).
Phase contrast micrographs of hMSCs during and after selection for SD cells. (Top row) Before recovery of SD cells. (Bottom row) After recovery of SD cells. Passage 2 hMSCs were incubated in complete medium containing 17% FCS for 28 days with a change of medium every 4 days to obtain control cells. Parallel hMSC cultures were incubated under the same conditions in medium without serum to obtain SD cells. The SD cells were then recovered by incubation in medium with 17% FCS for 1 to 5 days. Original magnification, × 10.
Phase contrast micrographs of hMSCs during and after selection for SD cells. (Top row) Before recovery of SD cells. (Bottom row) After recovery of SD cells. Passage 2 hMSCs were incubated in complete medium containing 17% FCS for 28 days with a change of medium every 4 days to obtain control cells. Parallel hMSC cultures were incubated under the same conditions in medium without serum to obtain SD cells. The SD cells were then recovered by incubation in medium with 17% FCS for 1 to 5 days. Original magnification, × 10.
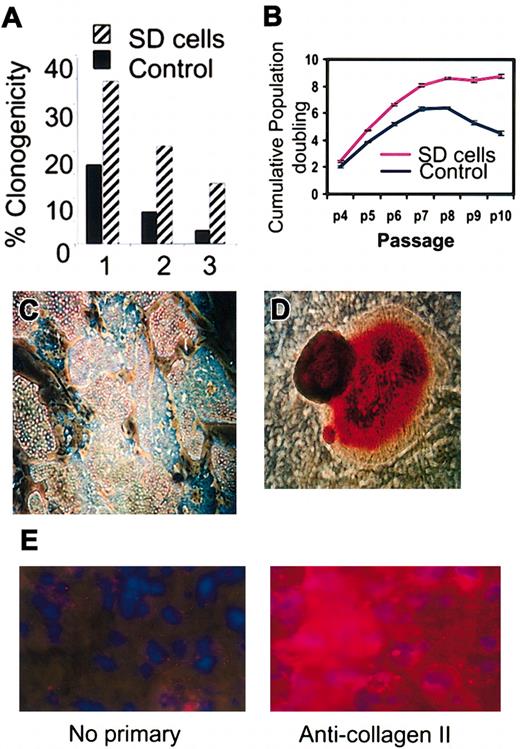
The replated SD cells (not shown) displayed a lag period of 4 to 5 days similar to the lag period seen when standard cultures of hMSCs are replated.10,12 Thereafter, the SD cells grew rapidly with a doubling time of about 24 hours for 4 to 5 days and until the cultures approached confluence. The SD cells were more clonogenic than parallel samples incubated for 4 weeks at the same densities in medium containing FCS (Figure 2A). The colonies formed were smaller than colonies formed by controls (not shown), but the SD cells retained their potential to differentiate into adipocytes and mineralizing cells (Figure 2C-D). The cells also differentiated into chondrocytes that expressed type II collagen (Figure 2E). However, the micropellets formed stained only weakly for proteoglycans, indicating that the differentiation into chondrocytes was not robust.18 Although SD cells continued to propagate longer than the control cells, they underwent senescence after passage 10 (Figure 2B), suggesting that they are not immortal. MSCs from 34 donors were incubated without serum and SD cells were obtained from 28 donors. However, the yields varied more than 10-fold, an observation consistent with previous observations indicating that preparations of MSCs varied in their content of early progenitors.9,10 Only 4% to 7% of SD cells and control cultures of MSCs were positive in assays for stromal antigen 1 (Stro-1; not shown) that was used to characterized some MSCs.14
Clonogenicity, expansion, and differentiation of SD cells. (A) Percent of cells that gave colonies after plating single cells into separate wells (1-3) of a 96-well microtiter plate. (B) Cumulative population doubling in medium containing 17% FCS as a function of passage number (p4 to p10). (C) Differentiation of SD cells into adipocytes (original magnification, × 10). Cells stained with Oil Red O. (D) Differentiation of SD cells into mineralizing cells (original magnification, × 10). Cells stained with Alizarin Red. (E) Differentiation of SD cells into chondrocytes (original magnification, × 40).
Clonogenicity, expansion, and differentiation of SD cells. (A) Percent of cells that gave colonies after plating single cells into separate wells (1-3) of a 96-well microtiter plate. (B) Cumulative population doubling in medium containing 17% FCS as a function of passage number (p4 to p10). (C) Differentiation of SD cells into adipocytes (original magnification, × 10). Cells stained with Oil Red O. (D) Differentiation of SD cells into mineralizing cells (original magnification, × 10). Cells stained with Alizarin Red. (E) Differentiation of SD cells into chondrocytes (original magnification, × 40).
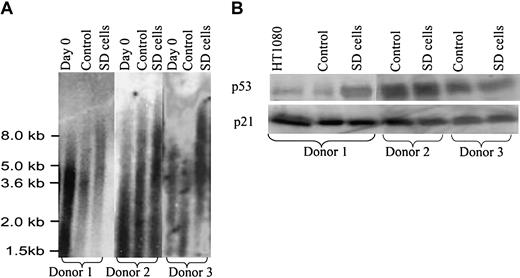
In assays of SD cells prepared from 15 different donors of bone marrow aspirates, the average telomere lengths were consistently longer than in the same cultures before serum deprivation (Figure 3A). Also, the average telomere lengths in the SD cells were longer than in control cells from the same hMSC preparations that were incubated in parallel in serum-containing medium. Assays for telomerase activity gave low and variable values for both SD cells and controls (not shown). RT-PCR (not shown) and Western blot assays (Figure 3B) indicated that SD cells expressed p53 and p21, genes not usually expressed in transformed cells.20,21 Soft agar assays to test for the loss of contact inhibition did not show any colonies with either control or SD cells, and no tumor growths were apparent 5 weeks after immunodeficient mice were subcutaneously injected with 2 million SD cells per mouse (n = 10). Therefore, there was no evidence that the cells were transformed.
Telomere length assay and Western blots. (A) Telomere length assay. Passage 2 cells were plated at 100 cells/cm2 for 5 days and then incubated in medium with and without 17% FCS for 28 days as in Figure 1. Day-0 cells were assayed before initial plating. The control and SD cells were allowed to recover in medium with FCS for 5 days. (B) Western blot for p53 and p21 with hMSCs from 3 different donors. The hMSCs were incubated with and without serum as for panel A. A human fibrosarcoma cell line, HT1080, was used as a positive control.
Telomere length assay and Western blots. (A) Telomere length assay. Passage 2 cells were plated at 100 cells/cm2 for 5 days and then incubated in medium with and without 17% FCS for 28 days as in Figure 1. Day-0 cells were assayed before initial plating. The control and SD cells were allowed to recover in medium with FCS for 5 days. (B) Western blot for p53 and p21 with hMSCs from 3 different donors. The hMSCs were incubated with and without serum as for panel A. A human fibrosarcoma cell line, HT1080, was used as a positive control.
On the basis of these observations, we tested the hypothesis that the SD cells expressed a profile of genes more characteristic of early progenitors than the other cells in cultures of hMSCs. Cells from 2 different donors were assayed under 5 different conditions: (1) initial plating of passage 2 hMSCs at low density (100 cells/cm2) and incubation for 5 days (day-0 cells in Figure 4) so that the cultures contained about equal proportions of RS cells and more mature cells10,12 ; (2) incubation of the day-0 samples in serum-free medium for 3 weeks (3wkSD cells in Figure 4A); (3) incubation of parallel day-0 samples in serum-containing complete medium for 3 weeks (3wkS); (4) replating and incubation of 3wkSD samples in serum-containing medium for 5 days (+5dSDS) so that the cells regained their original spindle-shaped morphology (Figure 1); (5) replating and incubation of the 3wkS samples in serum-containing medium for 5 days (+5dSS).
RT-PCR assays (Figure 4A) indicated that mRNA levels were higher in SD cells than in controls for Oct-4, hTERT, and ODC antizyme, 3 genes characteristically expressed in embryonic cells.22-25 The same RNA samples were assayed by microarrays and the data were analyzed by hierarchical clustering of the genes by their patterns of expression.26 In brief (Figure 5), the data from a chip containing about 22 000 genes were first filtered, and differentially expressed genes and 842 filtered genes were assigned to hierarchical clusters with the dChip 1.3+ program19 (Figure 6).
Hierarchical cluster analyses of 842 genes expressed in SD and control cells. The data on samples are presented in the order (left to right) day 0, 3wkSD, +5dSDS, 3wkS, + 5dSS (see Figure 4A).
Hierarchical cluster analyses of 842 genes expressed in SD and control cells. The data on samples are presented in the order (left to right) day 0, 3wkSD, +5dSDS, 3wkS, + 5dSS (see Figure 4A).
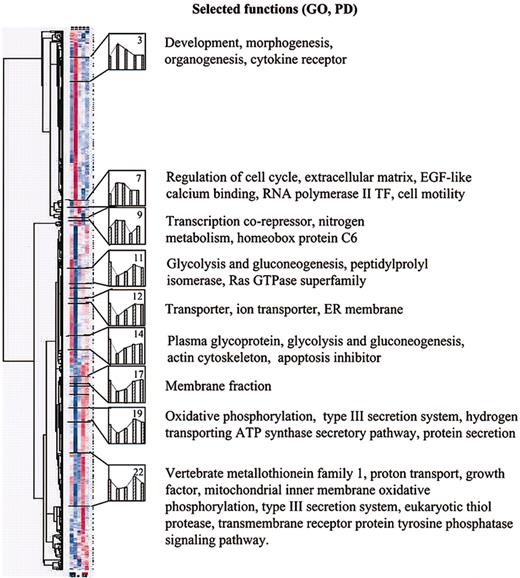
The 24 clusters were further filtered to identify (1) clusters in which genes were down-regulated in response to serum deprivation and remained down-regulated when the cells were returned to medium containing FCS (down/down pattern); and (2) clusters in which genes were up-regulated and remained up-regulated (up/up pattern). Six down/down clusters (arbitrarily numbered 11, 12, 14, 17, 19, and 22), and 3 up/up clusters (numbers 3, 7, and 9) were identified. The functional annotations assigned to 5 of the 6 down/down clusters by the dChip program19 (Figure 6) included genes encoding membrane fractions and membrane-associated receptors or transporters. Two of 6 down/down clusters (clusters 11 and 14) also included genes for intermediary metabolism. One down/down cluster (cluster 14) contained a gene for an apoptosis inhibitor. The 3 up/up clusters included genes involved in development, morphogenesis, and organogenesis (cluster 3); genes involved in regulation of cell cycle, genes for an endothelial growth factor–like (EGF–like) calcium binding protein, RNA polymerase II transcription factor, and cell motility (cluster 7); and genes for a transcription corepressor, nitrogen metabolism, and homeobox protein C6 (cluster 9).
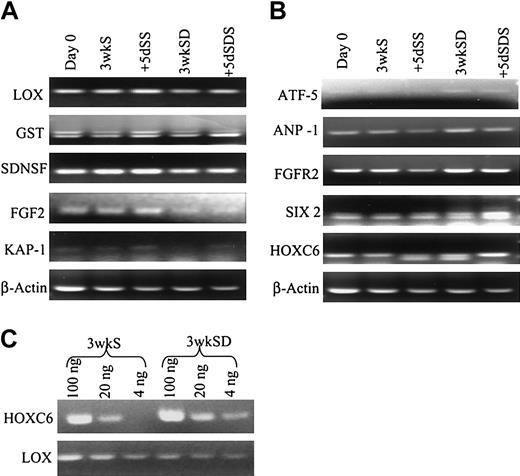
In the next step of analysis of the microarray data (Figure 5), we examined in greater detail 5 individual genes from the down/down clusters that are expressed in differentiated cells and 5 genes from the up/up clusters that are expressed in uncommitted cells. The 5 down/down genes (Figure 7) included a tumor suppressor gene also referred to as LOX because it encodes an enzyme that is required for the extracellular cross-linking of collagen and elastin; GST that is involved in the blood barrier in brain and testes; neural stem cell–derived neuronal survival protein (SDNSF); FGF2; and KAP1. The up/up genes (Figure 7) included ATF-5 that binds to the cyclic adenosine monophosphate (cAMP) response elements in many promoters; angiopoietin-1 (ANP-1) that promotes sprouting of endothelial cells; FGF2R; SIX2; and HOXC6 that belongs to the family of homeobox D4 genes involved in early development.
Genes showing prominent down/down and up/up patterns.LOX indicates lysyl oxidase (accession no. NM_002317); GST, glutathione-S–transferase (AL527430); SDNSF, neural stem cell–derived neuronal survival protein (BE_880828); FGF2, fibroblast growth factor-2 (M27968); KAP-1, keratin associated protein-1 (NM_030967); ATF-5, activating transcription factor 5 (NM_012068); ANP-1, angiopoietin-1 (U83508); FGFR2, fibroblast growth factor receptor-2 (NM_022969); SIX2, sine oculis homeobox homolog-2 (AF3332197); HOXC6, homeobox C6 (NM_004503). The black line and diamonds indicate SD cells; red line and squares, control cells; 0, 21, and +5 samples prepared as in Figure 4A.
Genes showing prominent down/down and up/up patterns.LOX indicates lysyl oxidase (accession no. NM_002317); GST, glutathione-S–transferase (AL527430); SDNSF, neural stem cell–derived neuronal survival protein (BE_880828); FGF2, fibroblast growth factor-2 (M27968); KAP-1, keratin associated protein-1 (NM_030967); ATF-5, activating transcription factor 5 (NM_012068); ANP-1, angiopoietin-1 (U83508); FGFR2, fibroblast growth factor receptor-2 (NM_022969); SIX2, sine oculis homeobox homolog-2 (AF3332197); HOXC6, homeobox C6 (NM_004503). The black line and diamonds indicate SD cells; red line and squares, control cells; 0, 21, and +5 samples prepared as in Figure 4A.
RT-PCR assays (Figure 8) confirmed the expression patterns of the 10 genes in the 2 donors used for microarray analysis. RT-PCR assays on samples from 3 additional donors demonstrated that 2 of 3 had similar profiles of expression for SDNSF, FGF2, FGF2R, HOXC6, KAP1, and LOX (not shown). A semiquantitative RT-PCR assay on HOXC6 and LOX (Figure 8C) confirmed the difference in expression pattern of the genes (Figure 8C).
RT-PCR analyses. (A-B) RT-PCR analyses to confirm the expression patterns of the genes shown in Figure 6. (C) Semiquantitative RT-PCR analysis of HOXC6 and LOX of 3wkS and 3wkSD samples.
RT-PCR analyses. (A-B) RT-PCR analyses to confirm the expression patterns of the genes shown in Figure 6. (C) Semiquantitative RT-PCR analysis of HOXC6 and LOX of 3wkS and 3wkSD samples.
Discussion
We also analyzed the data for expression of genes that were recently identified as signature genes for murine stem cells.27,28 About half of the genes with human homologs were present in the filtered data from one or more of the samples of SD cells and control MSCs but there were no large and persistent differences in expression that distinguished the SD cells. In addition, we examined the unfiltered data from the experiments for expression of the polycromb group gene Bmi-1 that is essential for the self renewal of hematopoietic stem cells.29,30 The gene was expressed in all 5 sets of samples and there were no significant changes in the level of expression.
The results demonstrate that subjecting early-passage hMSCs to serum deprivation for 2 to 4 weeks selects for a distinct subpopulation of cells. The SD cells are remarkable in that they survive complete serum deprivation for prolonged periods of time, have long telomeres, and enhanced expression of genes are expressed primarily in early progenitor cells. At the same time, the SD cells retained most of the characteristics of hMSCs in that they generated single-cell–derived colonies and they differentiated into osteoblasts, chondrocytes, and adipocytes. SD cells were obtained from 75% of early-passage hMSCs obtained from more than 30 separate donors of marrow aspirates. The yield of SD cells decreased markedly with passage number so that they could not be isolated from hMSCs preparations after 3 passages (not shown). Therefore, SD cells were probably not present in significant numbers in the hMSC preparations used in most previous experiments. Compared with the hierarchy of a hematopoietic system,31 RS cells that were previously identified as a rapidly self-renewing subpopulation in hMSC cultures10 are probably comparable to transitory amplifying cells. SD cells are more slowly replicating earlier progenitors and therefore more closely resemble hematopoietic stem cells or embryonic stem cells.
Prepublished online as Blood First Edition Paper, November 26, 2003; DOI 10.1182/blood-2003-06-1967.
Supported in part by National Institutes of Health (NIH) grants AR 47796 and AR 48323, the Oberkotter Foundation, HCA the Healthcare Company, and the Louisiana Gene Therapy Research Consortium (D.J.P.), NIH/National Institutes of Arthritis, Musculoskeletal, and Skin Disease fellowship (R.R.P.), and Louisiana Board of Regents Support Fund Fellowship (J.R.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Mr Alan Tucker for flow cytometer support and Dr Leena Ala-Kokko for critical reading of the manuscript.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal