Abstract
Human tissue factor pathway inhibitor-2 (TFPI-2) is a matrix-associated Kunitz inhibitor that inhibits the plasmin- and trypsin-mediated activation of zymogen matrix metalloproteinases involved in tumor progression, invasion, and metastasis. To directly assess its role in tumor growth and metastasis in vivo, we stably transfected HT-1080 fibrosarcoma cells expressing either fully active wild-type human TFPI-2 (WT) or inactive R24Q TFPI-2 (QT) and examined their ability to form tumors and metastasize in athymic mice in comparison to mock-transfected cells (MT). MT and QT fibrosarcoma tumors grew 2 to 3 times larger than WT tumors. Tumor metastasis was confined to the lung and was observed in 75% of mice treated with either MT or QT cells, whereas only 42% of mice treated with WT cells developed lung metastases. Real-time quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analyses of each tumor group revealed 3- to 6-fold lower levels of murine vascular endothelial growth factor gene expression in WT tumors in relation to either MT or QT tumors. Comparative tumor gene expression analysis revealed that several human genes implicated in oncogenesis, invasion, metastasis, apoptosis, and angiogenesis had significantly altered levels of expression in WT tumors. Our collective data demonstrate that secretion of inhibitory TFPI-2 by a highly metastatic tumor cell markedly inhibits its growth and metastasis in vivo by regulating pericellular extracellular matrix (ECM) remodeling and angiogenesis. (Blood. 2004;103:1069-1077)
Introduction
Proteolytic degradation of the extracellular matrix (ECM) is considered to be an essential step for malignant cells to invade and metastasize to distant tissues.1,2 Proteinase inhibitors capable of protecting the ECM from degradation by tumor-derived proteinases could potentially find utility as therapeutic agents for blocking tumor metastasis. Human tissue factor pathway inhibitor-2 (TFPI-2) is a Kunitz-type serine proteinase inhibitor synthesized and secreted into the ECM by endothelial cells, smooth muscle cells, fibroblasts, keratinocytes, and urothelium.3-7 TFPI-2 readily inhibits trypsin, plasmin, chymotrypsin, cathepsin G, plasma kallikrein, and the factor VIIa-tissue factor complex but not urokinase-type plasminogen activator (uPA), tissue-type plasminogen activator, or thrombin.8-11 TFPI-2 presumably inhibits these proteinases through an arginine residue (R24) in its first Kunitz-type domain, as an R24Q TFPI-2 mutant lost more than 90% of its inhibitory activity toward trypsin, plasmin, and the factor VIIa-tissue factor complex.12 TFPI-2 also strongly inhibited the trypsin- and plasmin-mediated activation of promatrix metalloproteinases proMMP-1 and proMMP-3 and suppressed production of active MMP-2 in HT-1080 cells stably transfected with the TFPI-2 expression vector.13,14 In addition, TFPI-2 expression is up-regulated in human atherosclerotic coronary arteries in comparison to normal, healthy arteries.15 Thus, ECM-associated TFPI-2 may play a pivotal role in the regulation of ECM remodeling, a process essential for tumor invasion and metastasis.
Given the inhibitory spectrum of TFPI-2, as well as our previous finding that TFPI-2 inhibited the degradation of fibroblast-derived ECM and Matrigel invasion by the highly invasive HT-1080 fibrosarcoma cell in a dose-dependent fashion,16 we hypothesized that expression of TFPI-2 by HT-1080 cells would markedly reduce its invasive and metastatic properties in an animal model. Since HT-1080 cells do not constitutively synthesize TFPI-2,16 we prepared stably transfected HT-1080 cells expressing high concentrations of wild-type human TFPI-2. We demonstrate that, in athymic mice, HT-1080 cells expressing wild-type TFPI-2 produce considerably smaller subcutaneous tumors and exhibited a lower metastatic rate in comparison to mock-transfected HT-1080 cells. Furthermore, HT-1080 cells stably transfected with an expression vector coding for an inactive mutant of TFPI-2, R24Q TFPI-2, produced tumors in size and metastatic rate similar to mock-transfected HT-1080 cells, providing strong evidence that the ability of TFPI-2 to reduce tumor size and metastasis correlated with its serine proteinase inhibitory activity.
Materials and methods
Materials
The murine myeloma cell line P3 × 63Ag8U.1 (P3U1) and the human fibrosarcoma cell line HT-1080 were obtained from American Type Tissue Culture Collection (Rockville, MD). Minimum essential medium Eagle (EMEM), nonessential amino acid solution, trypsin/EDTA (ethylenediaminetetraacetic acid) solution, RPMI-1640, TMBZ (3,3′,5,5′-tetramethyl-benzidine), and avidin-peroxidase were from Sigma Chemical (St Louis, MO). Lipofectamine plus reagent, sodium pyruvate, penicillin-streptomycin, and phosphate-buffered saline (PBS) were obtained from Gibco BRL Life Technologies (Rockville, MD). The pcDNA3 vector and proteinase K were obtained from Invitrogen (Carlsbad, CA). BrdU (5′-bromo-2′-deoxyuridine) and monoclonal antibody to BrdU were obtained from Amersham Pharmacia Biotech (Piscataway, NJ). Histomouse-SP bulk kit was purchased from Zymed Laboratories (South San Francisco, CA). ApopTag-peroxidase in situ apoptosis detection kit was obtained from Serological (Norcross, GA). The TaqMan reverse transcriptase (RT) reagent kit and the SYBR green master mix were obtained from Applied Biosystems (Foster City, CA). Recombinant human TFPI-2 was purified as described.8 All other reagents used were the highest quality commercially available.
Antibodies
Rabbit antihuman TFPI-2 immunoglobulin G (IgG) was prepared as described.3 Murine monoclonal antibodies against human TFPI-2 were prepared as follows. Six-week-old female Balb/c mice were injected intraperitoneally on day 0 with 50 μg recombinant TFPI-2 suspended in 50 μL of a PBS/Freund complete adjuvant emulsion. Subsequent injections containing 50 μg of TFPI-2 and Freund incomplete adjuvant were administered intraperitoneally on days 14 and 35. Mice were given an intravenous boost on days 49, 53, and 56 and killed on day 59. One mouse expressing the highest serum titer (> 105) of anti-TFPI-2 antibodies was killed and its splenocytes fused with P3 × 63Ag8U.1 myeloma cells. Fusion and hybridoma selection were optimized using standard methodology.17 Hybridomas were cultured for 7 days and their supernatants screened for antibodies to TFPI-2 by enzyme-linked immunosorbent assay (ELISA). Wells considered positive (A405 > 1) for antibody were weaned from hypoxanthine, aminopterin, and thymidine (HAT) supplement over 7 to 10 days, subcloned by limiting dilution, and grown in pristane-primed mice to generate ascites fluid. Monoclonal antibodies to human TFPI-2 (SK8, SK9) were isolated from ascites fluid using Hi Trap rProtein A affinity columns (Amersham Biosciences, Piscataway, NJ).
Cell culture and transfection
HT-1080 cells were cultured in 6% CO2 to 94% air and 96% humidity at 37°C in EMEM supplemented with 10% bovine calf serum (Hyclone, Logan, UT), sodium pyruvate, nonessential amino acids, l-glutamine, and penicillin-streptomycin. The human TFPI-28 and R24Q TFPI-212 complementary DNA (cDNA) were directionally subcloned into the EcoRI site of the pcDNA3 expression vector and the recombinant constructs transfected into HT-1080 cells using the Lipofectamine Plus reagent according to the manufacturer's instructions. Selection of transfected cells with 0.6 mg/mL G418 sulfate (Clontech, Palo Alto, CA) was initiated 48 hours after transfection and resistant colonies were cloned thrice by limiting dilution, screened for TFPI-2 expression by ELISA, and expanded. The expression levels of wild-type and R24Q TFPI-2 in stably transfected cells were assessed over a 6-week period in the presence and absence of G418.
Cell proliferation
Transfected HT-1080 cells were plated in duplicate at a density of 1 × 105 cells/well in a 6-well plate. Every 7 days, for a total of 42 days, the cells were trypsinized, counted, and replated at the same seeding density.
Human TFPI-2 ELISA
The concentration of wild-type and R24Q human TFPI-2 antigen in stably transfected HT-1080 cell supernatants was determined by ELISA using monoclonal antibody SK9 and biotinylated monoclonal antibody SK8. In this procedure, 96-well microtitration plates were coated overnight at 4°C with 100 μL/well of 50 mM carbonate buffer (pH 9.6) containing 10 μg/mL SK9. After washing the plate 3 times with Tris-buffered saline (TBS)/0.05%Tween 20, each well was blocked with 200 μL of TBS/1% gelatin/0.02% NaN3 at 37°C for 2 hours. Following 5 washes with TBS/Tween, 100-μL samples were added to each well and allowed to incubate at 37°C for 2 hours. The plate was then washed 5 times with TBS/Tween and 100 μL of biotinylated SK8 (100 ng/mL in TBS/0.1% bovine serum albumin [BSA]) was added to each well. After 2 hours incubation at 37°C, the plate was washed 5 times with TBS/Tween and subsequently treated with 100 μL of diluted peroxidase-conjugated avidin for 1 hour. After washing with TBS/Tween, each well was treated with 100 μL of tetramethyl benzidine solution. Following a suitable color development, the reaction was stopped by the addition of 1 N H2SO4 (100 μL) and the absorbance was measured at 450 nm. The concentration of TFPI-2 in test samples was interpolated from a linear standard curve (linear range 6-200 ng TFPI-2) relating A450 and known concentrations of recombinant human TFPI-2.
Tumor growth and metastasis
All animal procedures were approved by the University of New Mexico Health Sciences Center Laboratory Animal Care and Use Committee, Albuquerque. Thirty-six 6-week-old Balb/c severe combined immunodeficiency (SCID) male mice, obtained from Charles River Laboratory (Frederick, MD) through the National Cancer Institute (NCI) Contract Animal Program, were fed and watered ad libitum. The mice were divided into 3 groups of 12, and each mouse was injected with 5 × 106 HT-1080 cells subcutaneously. Group I was administered mock-transfected HT-1080 cells (MT), group II was administered wild-type TFPI-2 transfected HT-1080 cells (WT), and group III was administered R24Q TFPI-2 transfected HT-1080 cells (QT). The subcutaneous growth of tumors was readily visible, and the volume (0.5 × length × width2) of each tumor was measured externally by a tumor caliper. Five weeks after injection, each mouse received an intraperitoneal injection of BrdU (100 μg/g body mass) and was killed 3 hours later by CO2 narcosis. After being humanely killed, the mice were weighed and visible tumors and several organs were aseptically removed for further analyses.
Tissue processing
Harvested tumors and organs were formalin fixed and paraffin embedded using standard procedures. The organ tissues were visually inspected for the presence of metastatic tumors throughout the procedure. The embedded organ and tumor tissues were sectioned (5 μ) for subsequent hematoxylin and eosin (H&E) staining and further analyses.
Immunohistochemical detection of TFPI-2
Sections were deparaffinized with xylene, rehydrated in a graded series of ethanol, and washed in PBS for immunohistochemical staining. Expression of wild-type or R24Q TFPI-2 was determined using rabbit antihuman TFPI-2 IgG and detected using the Histomouse-SP Bulk kit according to the manufacturer's instructions. Briefly, rehydrated slides were incubated with 3% H2O2 to quench endogenous peroxidase activity, blocked, incubated overnight (4°C) with antibody (1:1000 dilution in PBS), incubated with a biotinylated secondary antibody, developed, and counterstained with hematoxylin.
Detection of BrdU-labeled and apoptotic cells
Tumor sections were processed as described in the previous paragraph with an additional step of trypsin (1 μg/μL) treatment after quenching. The cells were probed by incubation with primary anti-BrdU antibody (1:10 dilution) in nuclease solution. The apoptotic cells in sections were detected by TUNEL (terminal deoxynucleotidyl transferase [TdT]-mediated deoxyuridine triphosphate [dUTP] nick-end labeling) staining using the in situ apoptosis detection kit (Apoptag Peroxidase Kit) according to the manufacturer's instructions. Briefly, after deparaffinization, the sections were treated with proteinase K (20 μg/mL) for 15 minutes at room temperature and blocked. The sections were then treated with TdT enzyme for 60 minutes at 37°C. Anti-digoxigenin peroxidase conjugate was applied for 30 minutes at room temperature and was detected with the aforementioned substrate-chromogen solution. BrdU-labeled and TUNEL-positive cells were determined by counting 5 randomly chosen areas (× 100) in each section and averaged from 3 sections.
Microdissection and DNA amplification
The cells from sections were microdissected and cellular DNA purified as described.18 For each group, three 5-μ sections were deparaffinized and air dried. Under a dissecting microscope, the regions of interest were microdissected and collected. Fixed control mouse stomach and lung cells were also processed similarly. The microdissected tissue was lysed overnight at 50°C in 50 μL buffer containing 50 mM Tris-HCl (pH 8.5)/1 mM EDTA/0.5% Tween 20/200 μg/mL proteinase K followed by proteinase K inactivation at 95°C (10 minutes). The cellular DNA obtained was ethanol precipitated, washed, air dried, and redissolved in 10 μL. An aliquot (3 μL) of each was analyzed for human mitochondrial DNA (mtDNA) and the recombinant vector construct (rvcDNA) by polymerase chain reaction (PCR). The amplification of human mtDNA (1071-bp fragment) used a forward primer (GCT ATT ACC TTC TTA TTA TTT ACC) and reverse primer (GTG CGA TGA GTA GGG GAA GG). For the amplification of rvcDNA (700-bp fragment), a forward T7 primer (TAA TAC GAC TCA CTA TAG GG) and a TFPI-2-specific reverse primer (GCC TCG AGT TAA AAT TGC TTC TTC CGA TA) were employed. The thermocycling profile was set as 5 minutes of initial denaturation at 94°C, followed by 39 cycles of denaturation (94°C for 30 s), annealing for 1 minute (54°C for rvcDNA; 57°C for human mtDNA), and elongation (72°C for 2 minutes). The reaction products were electrophorized in a 1.2% agarose gel along with appropriate DNA markers.
RNA isolation
Total RNA was isolated from snap-frozen tumor samples (100-150 mg) using an RNeasy RNA extraction kit (Qiagen, Chatsworth, CA) according to the manufacturer's recommendation. Purified RNA samples were stored at -80°C in 100 μL diethyl pyrocarbonate (DEPC)-treated water. An aliquot of each RNA preparation was analyzed and quantitated using the RNA 6000 Nano assay kit in an Agilent Technologies 2100 Bioanalyzer (Palo Alto, CA).
VEGF expression: real-time quantitative RT-PCR
A 2-step real-time quantitative RT-PCR analysis was performed using a SYBR green dye-based assay in an ABI Prism 7000 Sequence Detection System according to the manufacturer's instructions (Foster City, CA). Total RNA (300 ng) from each sample was reverse transcribed using random hexamer primers (TaqMan RT reagents kit). Primers targeting the angiogenesis marker murine vascular endothelial growth factor (VEGF) and murine glyceraldehyde phosphate dehydrogenase (GAPDH), an internal control, were designed using Primer Express software (Applied Biosystems, Foster City, CA). The primers selected for sense and antisense strand, respectively, are as follows: mouse VEGF cDNA (GenBank accession no. S38083): TTACTGCTGTACCTCCACC and ACAGGACGGCTTGAAGATG; mouse GAPDH cDNA (GenBank accession no. M32599): AACGACCCCTTCATTGAC and TCCACGACATACTCAGCAC. To assure the amplicon specificity of each primer set, the PCR products were subjected to a melting curve analysis and subsequent agarose gel electrophoresis. The PCR reaction was performed in triplicates using the SYBR green master mix in a total volume of 50 μL. The reaction mixture was incubated at 95°C for 10 minutes followed by a cycling profile of 45 cycles consisting of denaturation at 95°C for 9 seconds, annealing at 57°C for 9 seconds, and extension at 72°C for 30 seconds. The efficiency for amplification of the target gene (VEGF) and the internal control gene (GAPDH) was examined using serial dilutions of cDNA with gene-specific primers. The mean difference between threshold cycle number values (ΔCT) was calculated for each cDNA dilution. The VEGF gene expression level in each sample was calculated following normalization to the GAPDH gene level and expressed as relative units.
Tumor cDNA microarray analysis
The relative abundance of messenger RNA (mRNA) in the snap-frozen tumor xenograft samples was assessed by oligonucleotide-based microarray analysis using an Affymetrix GeneChip Human Genome U133 set (Santa Clara, CA). This GeneChip consists of 2 array units of more than one million unique oligonucleotide features covering more than 39 000 transcript variants that represent 33 000 of the best characterized human genes. Additional information regarding these chips is available online at http://www.affymetrix.com/products/arrays/specific/hgu133.affx. Biotinylated cRNA probe preparation, processing, hybridization, and normalization were performed as described in the Affymetrix GeneChip Expression Analysis Manual. Fluorescence images were captured using a gene array scanner (Affymetrix) and expression analysis was performed using GeneSpring v5.1 software (Silicon Genetics, Redwood City, CA). The wild-type TFPI-2 transfected (WT) tumor ratio of medians was normalized with that obtained from mock-transfected (MT) tumors. The spots that exhibited a 2-fold or greater difference in expression levels were used to generate the gene clusters.
Results
Characterization of stably transfected HT-1080 cells
HT-1080 cells were stably transfected with the eukaryotic expression vector pcDNA3 alone (MT-1080) or vector containing cDNA constructs for either wild-type TFPI-2 (WT-1080) or R24Q TFPI-2 (QT-1080). TFPI-2-expressing stably transfected tumor cells were cloned by limiting dilution resulting in several cell lines secreting different levels of TFPI-2 that ranged from 10 to 55 ng/mL/day/106 cells determined by ELISA and consisted of 3 differential glycosylated forms of TFPI-2 (Mr of 32 kDa, 29 kDa, and 26 kDa), similar to that observed for human endothelial and smooth muscle cells.3-6 The TFPI-2-secreting HT-1080 cell lines selected for these studies secreted approximately 55 ng/mL/day/106 cells, but this number most likely underestimates the amount of TFPI-2 secreted by these cells into their ECM in vivo. In this regard, our preliminary findings indicated that the stably transfected HT-1080 cells, similar to endothelial cells and smooth muscle cells,3,4 secrete 4- to 6-fold higher levels of TFPI-2 into their ECM in cultures compared with their luminal secretion (data not shown).
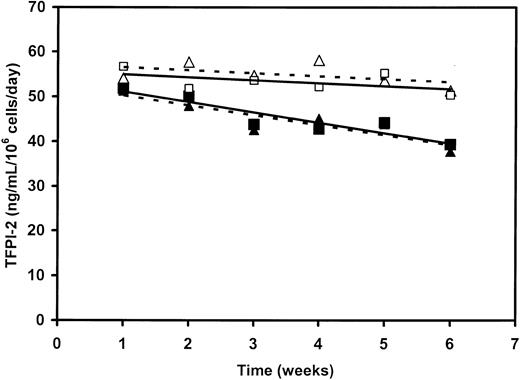
Initial in vitro studies revealed that the TFPI-2-expressing HT-1080 cells in continuous culture for 6 weeks secreted a relatively constant amount of TFPI-2 while under G418 selection (Figure 1.) In the absence of G418, these same cells continued to secrete TFPI-2, but the level declined by approximately 25% over 6 weeks of continuous culturing (Figure 1). Despite the slow and progressive loss of the expression vector in the absence of G418 (ie, under conditions that would partially mimic the in vivo growth of this cell line), the TFPI-2 expression level was still approximately 80% over a 5-week period. Accordingly, this period was selected as the time frame to evaluate the effect of TFPI-2 secretion on the in vivo growth and metastasis of this tumor cell in SCID mice.
Expression of either wild-type or R24Q human TFPI-2 by stably transfected HT-1080 cells in the presence and absence of G418. Stably transfected HT-1080 cell lines, with an initial expression level of approximately 55 ng TFPI-2/mL /day/106 cells, was continuously cultured with passaging in the presence (□, ▵) and absence (▪, ▴) of 0.6 mg/mL G418. The supernatants were assayed weekly for either TFPI-2 (□, ▪) or R24Q TFPI-2 (▵, ▴) expression by a sandwich ELISA as described in “Materials and methods.”
Expression of either wild-type or R24Q human TFPI-2 by stably transfected HT-1080 cells in the presence and absence of G418. Stably transfected HT-1080 cell lines, with an initial expression level of approximately 55 ng TFPI-2/mL /day/106 cells, was continuously cultured with passaging in the presence (□, ▵) and absence (▪, ▴) of 0.6 mg/mL G418. The supernatants were assayed weekly for either TFPI-2 (□, ▪) or R24Q TFPI-2 (▵, ▴) expression by a sandwich ELISA as described in “Materials and methods.”
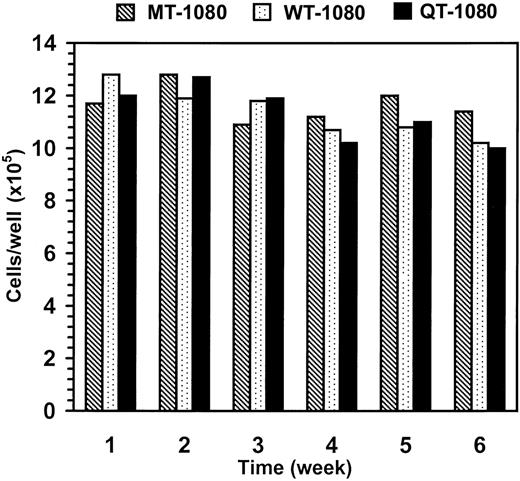
We next evaluated whether the growth rate of TFPI-2-expressing HT-1080 cells in culture were similar to the mock-transfected HT-1080 cells (MT-1080) over a 6-week period as described in “Materials and methods.” Although a slight decline in proliferation rate was noted over time, the proliferative rates of all transfected HT-1080 cells were essentially equivalent, suggesting that TFPI-2 secretion by these cells had no influence on their growth rate in vitro (Figure 2).
Growth rates of MT-1080, WT-1080, and QT-1080 cells in culture. MT-1080 cells, WT-1080 cells, and QT-1080 cells were initially plated at a density of 1 × 105 cells/well in a 6-well plate. Every 7 days, the cells were trypsinized, counted, and replated at the same seeding density.
Growth rates of MT-1080, WT-1080, and QT-1080 cells in culture. MT-1080 cells, WT-1080 cells, and QT-1080 cells were initially plated at a density of 1 × 105 cells/well in a 6-well plate. Every 7 days, the cells were trypsinized, counted, and replated at the same seeding density.
In separate experiments, transfected HT-1080 cells cultured in the absence of G418 also exhibited growth rates virtually identical to transfected cells grown in the presence of G418 (data not shown), providing evidence that G418 was also not affecting the proliferative rate of these cells. Moreover, transfection of these cells had no effect on their proliferative rates as transfected tumor cells, cultured in the absence of G418, grew at a rate indistinguishable from the parental HT-1080 cell line (data not shown).
Growth of transfected HT-1080 tumors in athymic mice
Athymic male Balb/c mice, grouped randomly, were inoculated subcutaneously with 5 × 106 transfected HT-1080 cells. Subcutaneous tumor growth of MT-1080 cells was linear with time and achieved an average volume of 837 ± 104 mm3 5 weeks after inoculation (Table 1). Subcutaneous tumor growth of WT-1080 cells was also linear, but in sharp contrast to MT-1080 subcutaneous tumors, these tumors were on average 28% to 60% smaller than the MT-1080 tumors at comparable times after inoculation (Table 1). QT-1080 subcutaneous tumors exhibited essentially the same tumor growth rate as the MT-1080 tumors (Table 1), suggesting that secretion of wild-type TFPI-2 by WT-1080 cells was associated with reduced tumor size.
Effect of TFPI-2 expression on the growth of HT-1080 tumors
. | Tumor volume, mm3* . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Tumor type . | Week 1 . | Week 2 . | Week 3 . | Week 4 . | Week 5 . | ||||
| MT-1080 | 24.1 ± 4.7 | 165.9 ± 17.5 | 336.3 ± 38.2 | 570 ± 59.1 | 837 ± 103.8 | ||||
| WT-1080 | 5.5 ± 0.8 | 66.9 ± 14.4 | 161.0 ± 30.6 | 327 ± 57.2 | 462 ± 70.5 | ||||
| OT-1080 | 20.5 ± 4.0 | 155.6 ± 18.9 | 302.4 ± 36.2 | 515 ± 57.4 | 731 ± 78.1 | ||||
. | Tumor volume, mm3* . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Tumor type . | Week 1 . | Week 2 . | Week 3 . | Week 4 . | Week 5 . | ||||
| MT-1080 | 24.1 ± 4.7 | 165.9 ± 17.5 | 336.3 ± 38.2 | 570 ± 59.1 | 837 ± 103.8 | ||||
| WT-1080 | 5.5 ± 0.8 | 66.9 ± 14.4 | 161.0 ± 30.6 | 327 ± 57.2 | 462 ± 70.5 | ||||
| OT-1080 | 20.5 ± 4.0 | 155.6 ± 18.9 | 302.4 ± 36.2 | 515 ± 57.4 | 731 ± 78.1 | ||||
Values are expressed as the mean ± SEM. Differences between all mean values at a given time point were tested pairwise by one-way analysis of variance (ANOVA), and statistical significance was accepted at P ≤ .05.
Mice were killed 5 weeks after inoculation. At sacrifice, the average weight of the resected MT-1080 and QT-1080 tumors was 2.65 ± 0.56 g, whereas the average weight of the resected WT-1080 tumors was 1.47 ± 0.34 g. Metastasis of MT-1080, WT-1080, and QT-1080 tumor cells in the SCID mice from the primary tumor location was confined exclusively to the lungs. Of the 12 SCID mice inoculated with MT-1080 cells, 9 mice (75%) developed metastatic lesions/nodules in the lungs compared with a metastatic incidence of 42% (5/12) in mice inoculated with WT-1080 cells (P < .001). The metastatic incidence of SCID mice inoculated with QT-1080 cells was identical to that observed for mice inoculated with MT-1080 cells. In order to assess the degree of metastasis in each experimental group, 4 randomly selected paraffin-embedded lungs from each group were sectioned in their entirety, and all sections (except every fourth section) were H&E stained. Examination of these sections revealed as many as 5 to 8 metastatic sites per tumor-positive lung in mice injected with MT-1080 cells, whereas only 1 to 2 metastatic sites were observed in tumor-positive lungs of mice inoculated with WT-1080 or QT-1080. The metastatic tumor size, evaluated from the number of sections they spanned, varied from 30 to 300 μ. As with the growth of subcutaneous tumors in athymic mice, these findings provide evidence that metastasis of HT-1080 tumors was markedly inhibited through their ability to secrete inhibitory TFPI-2.
Histologic and immunohistochemical analyses of primary and metastatic tumors
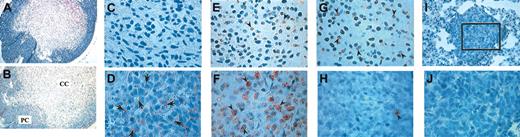
The histology of all primary subcutaneous tumors (Figure 3A) exhibited a peculiar morphology with loosely distributed cells in a core region (referred to as core cells, or CCs) encompassed by rather tightly packed peripheral cells (PCs). Initial immunohistochemical analyses of MT-1080, WT-1080, and QT-1080 cell cytospins revealed that MT-1080 cells were negative for TFPI-2 antigen, consistent with the absence of detectable TFPI-2 in parental HT-1080 cell-conditioned media by ELISA. On the other hand, WT-1080 and QT-1080 cells stained strongly positive for TFPI-2 antigen in cell cytospins (data not shown). In agreement with the cell cytospin analyses, primary MT-1080 tumors exhibited negative immunoreactivity for TFPI-2 antigen both in the PC and CC regions. In contrast, WT-1080 and QT-1080 primary tumors stained positive for TFPI-2 antigen in the PC region (Figure 3D) but failed to stain in the CC region (Figure 3C). Interestingly, metastatic tumors from all groups of mice, when subjected to immunohistochemical analyses, failed to show any detectable TFPI-2 antigen (Figure 3I-J).
Histologic and immunohistochemical analyses of paraffin-embedded subcutaneous and lung tumors. Tumor sections (5 μ) were either stained with hematoxylin and eosin (H&E) or treated with antibody as described in “Materials and methods.” (A) H&E-stained subcutaneous tumor; (B-D) immunohistochemical detection of TFPI-2 (arrows) in subcutaneous tumors; (E-F) immunohistochemical detection of BrdU-positive cells (arrowheads) in subcutaneous tumors; (G-H) TUNEL staining for apoptotic cells (arrowheads) in subcutaneous tumors; (I-J) anti-TFPI-2 IgG immunohistochemistry of metastatic lung tumors. Boxed area in I is magnified an additional 2.5-fold in panel J. PC indicates peripheral cells (D,F,H). CC indicates core cells (C,E,G). Original magnifications: × 250 (A-B); × 1000 (I); × 2500 (C-H,J).
Histologic and immunohistochemical analyses of paraffin-embedded subcutaneous and lung tumors. Tumor sections (5 μ) were either stained with hematoxylin and eosin (H&E) or treated with antibody as described in “Materials and methods.” (A) H&E-stained subcutaneous tumor; (B-D) immunohistochemical detection of TFPI-2 (arrows) in subcutaneous tumors; (E-F) immunohistochemical detection of BrdU-positive cells (arrowheads) in subcutaneous tumors; (G-H) TUNEL staining for apoptotic cells (arrowheads) in subcutaneous tumors; (I-J) anti-TFPI-2 IgG immunohistochemistry of metastatic lung tumors. Boxed area in I is magnified an additional 2.5-fold in panel J. PC indicates peripheral cells (D,F,H). CC indicates core cells (C,E,G). Original magnifications: × 250 (A-B); × 1000 (I); × 2500 (C-H,J).
Differential cellular proliferation and apoptosis
The relative distribution of proliferating (BrdU positive) cells was also assessed in sections of primary tumors from each experimental group. More than 90% of the PCs in all 3 tumor types was proliferating (Figure 3F), whereas only 8% to 10% of CCs were proliferating in MT-1080 or QT-1080 primary tumors. By comparison, more than 20% of CC region cells in WT-1080 tumors were positive for proliferation (Figure 3E). When examined for apoptosis by TUNEL assay, very few cells in the PC or CC regions in MT-1080 and QT-1080 primary tumors stained positive. In the case of the WT-1080 primary tumors, negligible numbers of cells were undergoing apoptosis in the PC region, whereas more than 40% of cells in the CC region stained positive (Figure 3G-H).
Tumor cell DNA analyses
To address the possibility that the core cells were of murine origin recruited into the growing tumor mass, a PCR-based qualitative analysis was performed on each cell type (PC and CC) found in the 3 primary tumors, as well as cells of metastatic tumors. Cellular DNA was prepared from microdissected cells as described in “Materials and methods.” By qualitative PCR amplification using human mitochondrial DNA (mtDNA)-specific primers, both PC and CC cell types were positive for human mitochondrial DNA (Figure 4, top), providing evidence that these regions contain human cells. PCR amplification of the recombinant vector/construct DNA (rvcDNA)-specific region revealed that only cellular DNA from the PC regions of WT-1080 and QT-1080 tumors was positive (Figure 4, bottom). No rvcDNA amplification was observed in MT-1080 tumor cells (Figure 4, bottom). The metastatic lung tumors also demonstrated the presence of human mitochondrial DNA, attesting to their human origin (Figure 4, top). Somewhat surprisingly, cellular DNA derived from WT-1080 and QT-1080 lung tumors tested positive for the intact rvcDNA region following PCR amplification (Figure 4, bottom), in spite of undetectable TFPI-2 antigen in these cells. The reason for this discrepancy is not known but most probably relates to the relative sensitivities between immunohistochemistry and PCR amplification techniques. Alternatively, TFPI-2 synthesis and expression may be down-regulated by lung-specific signaling molecules from the metastatic tumor microenvironment.
Qualitative PCR analyses of cellular DNA obtained from various tissues by microdissection. Cellular DNA used as a template in these PCR reactions was obtained from microdissected cells as described in “Materials and methods,” and PCR products resolved electrophoretically in 1.2% agarose gels. Cellular DNA from mouse gastric and lung cells were processed in an identical manner to exclude the possibility that the human-specific primer pair cross-reacted with mouse DNA.
Qualitative PCR analyses of cellular DNA obtained from various tissues by microdissection. Cellular DNA used as a template in these PCR reactions was obtained from microdissected cells as described in “Materials and methods,” and PCR products resolved electrophoretically in 1.2% agarose gels. Cellular DNA from mouse gastric and lung cells were processed in an identical manner to exclude the possibility that the human-specific primer pair cross-reacted with mouse DNA.
VEGF expression in tumors
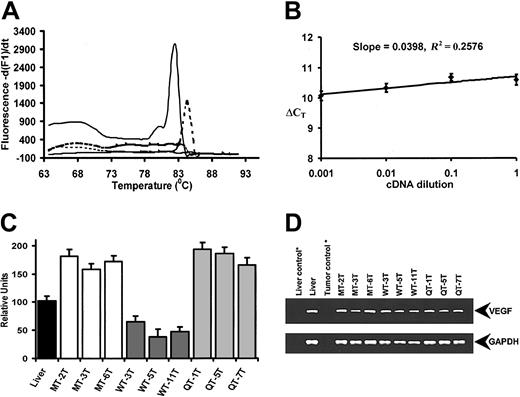
As VEGF expression is critical for tumor microvasculature formation,19 we performed a real-time quantitative RT-PCR analysis to assess murine VEGF gene expression levels in 3 tumor samples randomly selected from each tumor group. Melting curve analyses of the amplified PCR products revealed predominately a single product with distinct melting temperature (Tm) values (Tm = 82.6°C for GAPDH; Tm = 84.4°C for VEGF; Figure 5A). The efficiencies for the VEGF and GAPDH amplification were similar as the slope obtained from a plot of log cDNA dilution versus ΔCT was less than 0.1 (Figure 5B), thus validating the primer sets. The relative VEGF levels (mean ± SEM) for mouse liver and 3 representative samples of each tumor type were then plotted as shown in Figure 5C. Tumors arising from MT-1080 cells showed a 3- to 6-fold higher expression of VEGF mRNA than tumors derived from WT-1080 cells, whereas VEGF mRNA expression in QT-1080 cells was essentially identical to VEGF mRNA levels found in MT-1080 cells. The final PCR reaction products revealed amplification of a single, specific band for VEGF (Figure 5D top) and GAPDH (Figure 5D bottom) on agarose gel electrophoresis.
Real-time quantitative RT-PCR analysis of murine VEGF gene expression in tumors. (A) Melting curve analysis of the VEGF and GAPDH amplicons. Distinct melting curves of VEGF (dotted line) and GAPDH (solid line) are shown together with controls. (B) Relative efficiency plot of VEGF and GAPDH. The ΔCT (difference in CT values of VEGF and GAPDH) were calculated for each cDNA dilution. (C) Murine VEGF gene expression levels in MT-1080, QT-1080, and WT-1080 tumor samples. Mouse liver RNA (black bar) was used as a positive control. Each column represents the average of 3 amplification reactions (error bars represent standard deviation) performed on a single cDNA sample reverse-transcribed from RNA derived from each tumor sample. Samples MT-2T, MT-3T, and MT-6T are representative MT-1080 tumors (white bars). Samples WT-3T, WT-5T, and WT-11T are representative WT-1080 tumors (dark grey bars), while samples QT-1T, QT-5T, and QT-7T are representative QT-1080 tumors (light gray bars). (D) Agarose gel analyses of PCR products obtained following specific amplification of murine VEGF (top) and murine GAPDH (bottom) amplicons. * indicates negative controls lacking reverse transcriptase in first-strand cDNA synthesis.
Real-time quantitative RT-PCR analysis of murine VEGF gene expression in tumors. (A) Melting curve analysis of the VEGF and GAPDH amplicons. Distinct melting curves of VEGF (dotted line) and GAPDH (solid line) are shown together with controls. (B) Relative efficiency plot of VEGF and GAPDH. The ΔCT (difference in CT values of VEGF and GAPDH) were calculated for each cDNA dilution. (C) Murine VEGF gene expression levels in MT-1080, QT-1080, and WT-1080 tumor samples. Mouse liver RNA (black bar) was used as a positive control. Each column represents the average of 3 amplification reactions (error bars represent standard deviation) performed on a single cDNA sample reverse-transcribed from RNA derived from each tumor sample. Samples MT-2T, MT-3T, and MT-6T are representative MT-1080 tumors (white bars). Samples WT-3T, WT-5T, and WT-11T are representative WT-1080 tumors (dark grey bars), while samples QT-1T, QT-5T, and QT-7T are representative QT-1080 tumors (light gray bars). (D) Agarose gel analyses of PCR products obtained following specific amplification of murine VEGF (top) and murine GAPDH (bottom) amplicons. * indicates negative controls lacking reverse transcriptase in first-strand cDNA synthesis.
Genes regulated by TFPI-2 expression
Using 4 independent tumor samples in the Affymetrix GeneChip microarray system, a relative gene expression profile was obtained. Comparative differential gene expression analysis revealed that 80 genes had significantly altered levels of expression directly or indirectly regulated by TFPI-2 expression in these tumors. Among these, 43 genes were up-regulated and 37 genes were down-regulated. Further analysis revealed that 15 mRNA species were induced by more than 4-fold and 10 mRNA species were repressed by 4-fold or more. In Table 2, the proteins encoded by these genes are grouped according to their functions. The analysis of the genes according to a gene ontology system showed that TFPI-2 expression regulated genes in almost every category including those implicated in transcription, signal transduction, cell growth and proliferation, extracellular matrix, oncogenesis, invasion, metastasis, apoptosis, and angiogenesis.
Genes regulated by TFPI-2 expression in fibrosarcoma xenografts obtained from SCID mice
GeneBank accession no. . | Fold change* . | Description . |
|---|---|---|
| Transcription factors | ||
| NM_001186 | −2.11 | Helicase, basic leucine zipper transcription factor-1 |
| NM_006963, AA744771 | 29.27 | Zinc finger protein 22 (KOX 15) |
| NM_006291 | 5.05 | Zinc finger protein 185 (LIM domain) |
| NM_014368 | 2.91 | LIM homeobox protein 6 |
| NM_001290 | −5.20 | LIM domain binding 2 |
| NM_002586 | −2.75 | Pre-B-cell leukemia transcription factor-2 (PBX-2) |
| X16155 | −2.79 | COUP-transcription factor |
| Signal transduction | ||
| NM_004445 | −2.38 | Erythropoietin-producing hepatocyte kinase (EphB6) |
| NM_002547 | 2.95 | Oligophrenin 1 |
| NM_002821 | 2.33 | Protein tyrosine kinase 7 |
| U71075 | −2.48 | Protein tyrosine phosphatase, receptor type, U |
| D30751 | 2.79 | Bone morphogenetic protein 4 |
| NM_022159 | −2.92 | EGF-TM-7-latrophilin-related protein |
| Cell growth, proliferation, and maintenance | ||
| NM_013975 | 2.04 | Ligase III, DNA |
| NM_006567 | 2.11 | Phenylalanine-tRNA synthetase |
| AU118882, NM_001957 | −2.90 | Endothelin receptor type A |
| NM_002349 | 14.32 | Lymphocyte antigen 75, gp200-MR6 |
| NM_005330 | −7.63 | Hemoglobin, epsilon 1, oxygen transport |
| NM_000385 | 2.30 | Aquaporin 1 |
| AF052169 | 2.48 | Voltage-gated potassium channel activity |
| BE742268 | −4.21 | Sortilin 1 (SORT 1) |
| AW206786 | −2.32 | Enigma (LIM domain protein) |
| Invasion and metastasis | ||
| M34064 | −2.13 | N-cadherin |
| NM_014751 | 2.06 | Metastasis suppressor gene |
| NM_002961 | 2.79 | S100 calcium-binding protein A4 |
| NM_021111 | 2.64 | Reversion-inducing cysteine-rich protein with kazal motif (RECK) |
| AF348491, AJ224869 | −4.21, −27.40 | Chemokine (C-X-C motif), receptor 4 (fusin) |
| NM_022842 | −18.70 | CUB domain-containing protein 1 (CDCP1) |
| Oncogenes/tumor suppressor genes | ||
| NM_021991 | −2.21 | Junction plakoglobin |
| NM_003287 | 2.10 | Tumor protein D52-like 1 |
| NM_001958 | 6.85 | Eukaryotic translation elongation factor 1 alpha 2 (EEF 1A2) |
| Apoptosis | ||
| NM_005892 | −2.14 | Formin-like (FRL) |
| NM_020371 | −2.11 | Cell death regulator Aven |
| Angiogenesis | ||
| NM_002019 | −2.24 | FLT, VEGFR1, Fms-related tyrosine kinase receptor |
| NM_000584 | −6.99 | Interleukin-8 (IL-8, C-X-CL8) |
| A1812030 | −3.39 | Thrombospondin 1 precursor |
| L01639 | −8.03 | Neuropeptide Y receptor |
| U58111 | −2.41 | Vascular endothelial growth factor C |
| NM_006291 | 2.98 | Tumor necrosis factor, alpha-induced protein 2 |
| Extracellular matrix | ||
| NM_002607 | 2.53 | Platelet-derived growth factor alpha polypeptide |
| NM_021599 | −4.62 | ADAM-TS2 |
| BC002416 | −2.29 | Biglycan |
| NM_000088 | 3.51 | Collagen, type I, alpha 1 |
| AI264196 | −2.48 | Fibrillin 1 precursor |
| D32039 | −2.13 | Chondroitin sulfate proteoglycan 2 (versican) |
| AI146848 | 2.87 | Dermatopontin precursor |
| AJ276395 | −3.80 | Fibronectin 1 |
| AA669336 | 7.26 | Alpha 1 chain of type XII collagen |
| Others | ||
| NM_002759 | 2.51 | EIF2Ak1, protein kinase, PKR |
| NM_004988 | −2.95 | Melanoma antigen family A1 (MAGE-A1) |
| NM_016931 | −2.46 | NADPH oxidase 4 (Nox 4) |
| NM_021822 | 2.80 | Phorbolin-like protein MDS019 |
| NM_001785 | 35.30 | Cytidine deaminase |
GeneBank accession no. . | Fold change* . | Description . |
|---|---|---|
| Transcription factors | ||
| NM_001186 | −2.11 | Helicase, basic leucine zipper transcription factor-1 |
| NM_006963, AA744771 | 29.27 | Zinc finger protein 22 (KOX 15) |
| NM_006291 | 5.05 | Zinc finger protein 185 (LIM domain) |
| NM_014368 | 2.91 | LIM homeobox protein 6 |
| NM_001290 | −5.20 | LIM domain binding 2 |
| NM_002586 | −2.75 | Pre-B-cell leukemia transcription factor-2 (PBX-2) |
| X16155 | −2.79 | COUP-transcription factor |
| Signal transduction | ||
| NM_004445 | −2.38 | Erythropoietin-producing hepatocyte kinase (EphB6) |
| NM_002547 | 2.95 | Oligophrenin 1 |
| NM_002821 | 2.33 | Protein tyrosine kinase 7 |
| U71075 | −2.48 | Protein tyrosine phosphatase, receptor type, U |
| D30751 | 2.79 | Bone morphogenetic protein 4 |
| NM_022159 | −2.92 | EGF-TM-7-latrophilin-related protein |
| Cell growth, proliferation, and maintenance | ||
| NM_013975 | 2.04 | Ligase III, DNA |
| NM_006567 | 2.11 | Phenylalanine-tRNA synthetase |
| AU118882, NM_001957 | −2.90 | Endothelin receptor type A |
| NM_002349 | 14.32 | Lymphocyte antigen 75, gp200-MR6 |
| NM_005330 | −7.63 | Hemoglobin, epsilon 1, oxygen transport |
| NM_000385 | 2.30 | Aquaporin 1 |
| AF052169 | 2.48 | Voltage-gated potassium channel activity |
| BE742268 | −4.21 | Sortilin 1 (SORT 1) |
| AW206786 | −2.32 | Enigma (LIM domain protein) |
| Invasion and metastasis | ||
| M34064 | −2.13 | N-cadherin |
| NM_014751 | 2.06 | Metastasis suppressor gene |
| NM_002961 | 2.79 | S100 calcium-binding protein A4 |
| NM_021111 | 2.64 | Reversion-inducing cysteine-rich protein with kazal motif (RECK) |
| AF348491, AJ224869 | −4.21, −27.40 | Chemokine (C-X-C motif), receptor 4 (fusin) |
| NM_022842 | −18.70 | CUB domain-containing protein 1 (CDCP1) |
| Oncogenes/tumor suppressor genes | ||
| NM_021991 | −2.21 | Junction plakoglobin |
| NM_003287 | 2.10 | Tumor protein D52-like 1 |
| NM_001958 | 6.85 | Eukaryotic translation elongation factor 1 alpha 2 (EEF 1A2) |
| Apoptosis | ||
| NM_005892 | −2.14 | Formin-like (FRL) |
| NM_020371 | −2.11 | Cell death regulator Aven |
| Angiogenesis | ||
| NM_002019 | −2.24 | FLT, VEGFR1, Fms-related tyrosine kinase receptor |
| NM_000584 | −6.99 | Interleukin-8 (IL-8, C-X-CL8) |
| A1812030 | −3.39 | Thrombospondin 1 precursor |
| L01639 | −8.03 | Neuropeptide Y receptor |
| U58111 | −2.41 | Vascular endothelial growth factor C |
| NM_006291 | 2.98 | Tumor necrosis factor, alpha-induced protein 2 |
| Extracellular matrix | ||
| NM_002607 | 2.53 | Platelet-derived growth factor alpha polypeptide |
| NM_021599 | −4.62 | ADAM-TS2 |
| BC002416 | −2.29 | Biglycan |
| NM_000088 | 3.51 | Collagen, type I, alpha 1 |
| AI264196 | −2.48 | Fibrillin 1 precursor |
| D32039 | −2.13 | Chondroitin sulfate proteoglycan 2 (versican) |
| AI146848 | 2.87 | Dermatopontin precursor |
| AJ276395 | −3.80 | Fibronectin 1 |
| AA669336 | 7.26 | Alpha 1 chain of type XII collagen |
| Others | ||
| NM_002759 | 2.51 | EIF2Ak1, protein kinase, PKR |
| NM_004988 | −2.95 | Melanoma antigen family A1 (MAGE-A1) |
| NM_016931 | −2.46 | NADPH oxidase 4 (Nox 4) |
| NM_021822 | 2.80 | Phorbolin-like protein MDS019 |
| NM_001785 | 35.30 | Cytidine deaminase |
+ and − indicate increased and decreased mRNA levels, respectively; LIM, the Lin-1, IsI-1, and Mec-3 protein domain; COUP, chicken ovalbumin upstream promoter-transcription factors; TM7, transmembrane domain 7; gp200-MR6, a 200 000 Mr antigen detected by monoclonal antibody MR6; Fms, a protooncogene from the feline McDonough strain of sarcoma virus; ADAM-TS2, A disintegrin-like and metalloproteinase with thrombospondin type 1 motifs, 2; EIF2Ak1, eukaryotic translation initiation factor 2 α kinase 1; and PKR, double-stranded RNA-dependent protein kinase.
The number indicates the fold change in mRNA abundance in TFPI-2-expressing tumors over mock-transfected tumors determined by microarray data analysis.
Discussion
In the present study, we have prepared stably transfected human HT-1080 fibrosarcoma cell lines expressing either wild-type human TFPI-2 or an inactive mutant TFPI-2 (R24Q TFPI-2) and assessed their ability to grow and metastasize in athymic Balb/c mice in relation to a mock-transfected HT-1080 cell line. We observed that stably transfected WT-1080 cell tumors grew at a substantially lower (approximately 28%-60%) rate than MT-1080 solid tumors. QT-1080 produced subcutaneous tumor masses essentially identical in volume to that observed for the MT-1080, providing suggestive evidence that the expression of inhibitory TFPI-2 was associated with restricted tumor growth. In addition to the substantially decreased growth rate of WT-1080 tumors, the metastatic rate of WT-1080 cells (42%) was also markedly lower than that observed for QT-1080 or MT-1080 cells (75%). The decreased metastatic rate of WT-1080 cells in all likelihood relates to a smaller primary tumor mass burden rather than TFPI-2 expression, as immunohistochemical analyses revealed that the WT-1080 metastatic tumors paradoxically failed to stain for immunoreactive TFPI-2 but retained the vector/construct as shown by PCR. The reason(s) for this discrepancy is not known but may be related to different sensitivities between these 2 techniques and/or down-regulation of TFPI-2 expression in the metastatic tumor microenvironment.
Our in vivo findings are clearly consistent with and extend previous in vitro results demonstrating a dose-dependent inhibition of HT-1080 invasiveness in Matrigel and ECM degradation by exogenous TFPI-2.16 Our results are also consistent with a recent report by Konduri and colleagues20 demonstrating that high-grade SNB19 glioma cells stably transfected with the human TFPI-2 expression vector formed smaller intracerebral tumors in contrast to its mock-transfected counterpart. Finally, our data agree, in part, with that very recently published by Jin and coworkers21 who demonstrated that TFPI-2-expressing human choriocarcinoma cells (JAR) exhibited decreased invasive properties in Matrigel relative to mock-transfected JAR tumor cells as well as decreased invasiveness in vivo in nude mice following subcutaneous transplantation. However, in contrast to our findings using HT-1080 fibrosarcoma cells, mock-transfected and TFPI-2-expressing human choriocarcinoma tumors were essentially identical in mass and failed to metastasize.21
Histologic analyses on primary and metastatic tumors were performed to evaluate the effects of TFPI-2 on tumor growth and metastasis. HT-1080 tumors consisted of 2 distinct regions: a homogeneous core of cells with condensed nuclei and peripheral cells that appeared morphologically similar to cultured HT-1080 cells. Although both regions demonstrated the presence of human mtDNA, only cells occupying the peripheral regions were positive for TFPI-2 antigen and demonstrated presence of the vector construct. BrdU and TUNEL staining confirmed that cells present in the peripheral region were still proliferating, while most of the cells occupying the core region were undergoing apoptosis. Since tumor volumes were significantly smaller in WT-1080-treated mice, TFPI-2 most likely affects those processes involved in tumor mass formation in vivo, such as neovascularization, rather than inhibiting the proliferative rate of individual tumor cells.
The precise mechanism(s) whereby genetically engineered expression of functional TFPI-2 by a TFPI-2 null cell reduces tumor size and its aggressive phenotype in vivo is unclear. During tumor growth, malignant cells invade normal adjacent tissues, and regulation of plasmin activity on the surface of tumor cells has been shown to influence the invasive and metastatic behavior of tumor cells.22-24 However, plasmin associated with the ECM or the membranes of cultured cells is resistant to inhibition by known physiologically relevant proteinase inhibitors,25-27 and it has been suggested that metastatic tumor cells generate “unregulated” plasmin activity that potentiates metastatic behavior.28,29 Many tumor cells, including the HT-1080 fibrosarcoma cell line used in this study, employ the uPA-urinary plasminogen activator receptor (uPAR) system to activate plasminogen, resulting in plasmin-mediated ECM degradation and invasion, as well as proMMP-1 and proMMP-3 activation that further enhances tumor invasion and metastasis.13 In addition, tumor growth is highly dependent on an adequate blood supply, and plasmin presumably plays an important role in tumor angiogenesis.30 In this regard, Soff and coworkers31 have reported that expression of plasminogen activator inhibitor 1 (PAI-1) by a stably transfected human prostate carcinoma cell line (PC-3) markedly reduced the growth rate of these primary tumors in an athymic mouse model in relation to the parental PC-3 cell line, providing clear evidence that regulation of plasmin formation reduced the aggressive phenotype of these cells. In view of its ability to strongly inhibit plasmin in vitro, it is not unreasonable to speculate that ECM-associated TFPI-2 generated by WT-1080 tumors inhibits ECM turnover mediated by plasmin and plasmin-activated matrix metalloproteinases, thereby inhibiting tumor invasiveness and metastases in vivo. In this connection, preliminary studies have shown that wild-type human TFPI-2 exhibited a potent and dose-dependent antiangiogenic effect in both the VEGF-induced chorioallantoic membrane assay and the fibroblast growth factor 2 (FGF-2)-induced cornea pocket assay (L. Arispe and W.K., unpublished data, July 2000). Accordingly, the ability of secreted TFPI-2 to reduce tumor size may be dependent, in part, on its antiangiogenic properties.
To establish the role of TFPI-2, if any, in neovascularization essential for tumor growth and metastasis, host VEGF gene expression was quantitated in these tumors by real-time quantitative RT-PCR. MT- and QT-1080 tumors expressed essentially the same levels of VEGF mRNA, whereas WT-1080 tumor VEGF mRNA levels were reduced 3- to 6-fold in relation to MT- and QT-1080 tumors. Accordingly, a clear quantitative correlation was observed between murine VEGF expression levels and tumor size, suggesting that active TFPI-2 plays a suppressive role on host-derived VEGF gene expression and, by extension, on VEGF-mediated angiogenesis. The cellular origin of murine VEGF mRNA isolated from these tumors is not known, although host stromal cells may be responsible for VEGF synthesis. Clearly, the higher apoptotic rate of WT-1080 tumor core cells strongly suggests decreased tumor angiogenesis in this portion of the tumor architecture that may be related to either lower host VEGF mRNA expression in these tumors or elevated levels of angiostatin, or both. Although wild-type HT-1080 cells constitutively secrete human VEGF32 that presumably contributes greatly to neovascularization in these tumors, recent studies have shown that complete inhibition of rhabdomyosarcoma xenograft growth and neovascularization in nude mice required inhibition of both tumor- and host-derived VEGF.33
The relative assessment of genes in the snap-frozen tumor xenograft by oligonucleotide-based microarray analysis revealed no significant change in the human (tumor) VEGF gene levels, although a 2-fold or greater decrease in FLT-1 (VEGFR1) and VEGF-C (the C-isoform of VEGF) mRNA levels was observed. VEGF-C and VEGFR1, a VEGF receptor, have been implicated in tumor-related angiogenesis.34-36 Thus, induced TFPI-2 expression does not regulate tumor VEGF mRNA levels but rather appears to suppress its angiogenic effect by down-regulating receptor (VEGFR1) levels. Among other angiogenic regulators, interleukin-8 (IL-8), thrombospondin 1 (THBS-1), and neuropeptide-Y receptor gene levels were also down-regulated, whereas the tumor necrosis factor alpha-induced protein 2 (TNF-AIP2) levels were up-regulated. Interleukin-8 is not expressed constitutively but on TNF-α induction it inhibits apoptosis via nuclear factor-kappaB (NF-kappaB) and Akt signaling pathways.37 IL-8 also exhibits potent angiogenic activity and thus may play a role in tumor progression. Thrombospondin (THBS-1) suppresses tumor growth, inhibits activation of MMP-9, and inhibits VEGF binding to receptor suppressing capillary morphogenesis.38 Surprisingly, its expression is down-regulated by TFPI-2, partially reducing its anti-tumor growth function. However, another antiangiogenic gene, neuropeptide Y receptor-2, regulates angiogenesis-dependent tumor repair39 and is down-regulated. Thus, at the transcriptional level, TFPI-2 expression regulates both proangiogenic and antiangiogenic regulators, which, in concert, could affect tumor angiogenesis.
The proinvasive and prometastatic genes such as N-cadherin, CDCP1, and chemokine receptor 4 are suppressed by TFPI-2 induction in these tumor cells. N-cadherin, a cell adhesion molecule, makes heterotypic contacts with catenin (α, β, γ)-p120ctn promoting matrix invasion and transendothelial migration by convergence of transforming growth factor β (TGF-β) signaling.40 The junction plakoglobin (γ-catenin)41 is also suppressed by TFPI-2. Protein tyrosine phosphatase receptor μ (PTPRmu),42 another component of this cadherin-catenin complex that dephosphorylates p120ctn, is also down-regulated. Thus, most of the components of the cadherin-catenin complex are suppressed, possibly leading to an imbalance between levels of activated N-cadherin and E-cadherin necessary for cell motility and tumor invasion.40 Furthermore, the ectodomain of E-cadherin (sE-CAD) is shed by plasmin, stromelysin-1, and matrilysin (MMP7) cleavage thereby stimulating tumor invasion, in part, by up-regulation of MMP-2, MMP-9, and membrane type 1-matrix metalloproteinase (MT1-MMP).40,43,44 Since it is thought that metastatic tumor cells generate “unregulated” plasmin activity,28,29 the tumor growth suppression could also be affected by the plasmin-inhibitory activity of TFPI-2 suppressing sE-CAD production.
Tumor invasion metastasis suppressor genes, MIM45 and RECK, are induced more than 2-fold in TFPI-2-overexpressing tumors. Overexpression of RECK has been shown to form HT-1080 tumors defective in vasculature due to inhibition of angiogenic sprouting through excessive degradation of the ECM.46 Moreover, RECK negatively regulates matrix-metalloproteinases MMP-2, MMP-9, and MT1-MMP, thereby inhibiting tumor invasion, metastasis, and angiogenesis.47,48 However, the prometastatic gene, S100A4/MTS1/metastasin, up-regulated in medulloblastoma, brain cancer cells, and murine melanoma,49 was also up-regulated in these tumors.
Two of the proapoptotic genes, FRL50 and chemokine receptor 4 (CXC-R4),51 are down-regulated, suggesting the induction of genes that support tumor cell growth and survival. In contrast, the antiapoptotic gene, Aven,52 is down-regulated. Genes encoding for extracellular matrix constituents like ADAM-TS2, biglycan, fibronectin 1, versican, and fibrillin 1 precursors are suppressed, whereas dermatopontin and collagen I alpha 1 genes are up-regulated, suggesting regulation of ECM remodeling at the transcriptional level. In addition, a large number of genes found to be regulated by TFPI-2 in this model are implicated in general cellular functions including signal transduction, cell growth and proliferation, and the synthesis of some transcription factors, which in turn regulate other genes that affect other cellular processes.
While the genomic response to TFPI-2 overexpression appears complex, most of the genes regulated by TFPI-2 would result in an overall decrease in tumor growth and metastatic potential. The molecular mechanism whereby TFPI-2 expression and function affects tumor cell gene expression is not known but presumably involves its ability to regulate proteinases such as trypsin, plasmin, or the factor VIIa-tissue factor complex either on the tumor cell or in the tumor microenvironment that, in turn, affect a variety of tumor cell signaling processes involved in growth and angiogenesis. Future studies focused on the regulation and functional significance of the target genes reported here are likely to increase our understanding of the role of TFPI-2 in the regulation of pericellular ECM remodeling in normal and tumor cells.
Prepublished online as Blood First Edition Paper, October 2, 2003; DOI 10.1182/blood-2003-06-1930.
Supported in part by Research Grant HL64119 from the National Institutes of Health (W.K.) and Contract no. N00178-01-C-3069 from the US Department of Defense (H.D.I.). S.K. was supported by a grant from the Chemo-Sero-Therapeutic Research Institute (KAKETSUKEN), Kumamoto, Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors gratefully acknowledge the helpful discussions of Drs Rick Lyons, Gow Arepally, and Barbara Griffith during these studies. The authors would also like to acknowledge the technical assistance of Gavin Pickett and Marilee Morgan of the Keck-UNM Genomics Shared Resource, which is supported by the UNM Cancer Center, the WM Keck Foundation, and the State of New Mexico. Finally, the authors are indebted to Keiko Kamei and Joseph Prescott for technical assistance in the preparation of the monoclonal antibodies and standardizing the real-time RT-PCR, respectively.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal