Abstract
Severe deficiency of the von Willebrand Factor (VWF)–cleaving proteinase, ADAMTS13, is associated with the development of thrombotic thrombocytopenic purpura (TTP). Several mutations spread across the ADAMTS13 gene have been identified in association with a deficiency of VWF-cleaving proteinase activity in patients with congenital TTP. The spread of these dysfunctional mutations and the domain structure of ADAMTS13 are suggestive of a complex interaction between the enzyme and its substrate. We have studied a patient with congenital TTP who is a compound heterozygote for the Thr196Ile mutation in the metalloproteinase domain and a frameshift mutation (4143-4144insA) in the second CUB domain that results in loss of the last 49 amino acids of the protein. The VWF-cleaving proteinase activity of the truncated enzyme was comparable to that of the wild-type enzyme but its secretion from transfected COS-7 cells was about 14% of the wild type.
Introduction
Thrombotic thrombocytopenic purpura (TTP) is a thrombotic microangiopathy characterized by a schistocytic hemolytic anemia and consumptive thrombocytopenia with varying degrees of neurologic and renal impairment. The presence of ultralarge von Willebrand factor (VWF) multimers in the plasmas of patients with chronic relapsing TTP during remission that disappear during an attack led to the implication of these multimers in the pathogenesis of the platelet-rich, fibrin-poor thrombi that occlude arterioles and are the hallmark of this disorder.1 The VWF-cleaving metalloproteinase, ADAMTS13, hydrolyzes the ultralarge VWF multimers as they emerge from endothelial cells and undergo conformational strain due to shear stress in arterioles and capillaries.2 A severe deficiency of VWF-cleaving proteinase activity is associated with the pathogenesis of congenital TTP (the Schulman-Upshaw syndrome).3,4
A number of mutations, spread throughout the ADAMTS13 gene, have been reported in association with congenital TTP.3,5-9 The CUB domains at the C-terminus of ADAMTS13 are of uncertain physiologic relevance. The findings that the CUB domains are not required for VWF-cleaving proteinase activity measured under static conditions10,11 and that certain strains of mice have a variant form of murineADAMTS13 that lacks the CUB domains (together with the seventh and eighth thrombospondin type 1 (TSP-1) repeats) support the premise that these domains are dispensable in vivo.12 However, a report that peptides from the CUB domains inhibit VWF-cleaving proteinase activity under flow, but not static, conditions suggests a functional role for these domains.13
We studied a patient with congenital TTP who is a compound heterozygote for the Thr196Ile mutation in the metalloproteinase domain and the insertion of a nucleotide (A) at 4143-4144. This insertion results in a frameshift in the second CUB domain and loss of the last 49 amino acids of the protein. The CUB mutation had little effect on the specific activity of the enzyme but its secretion from COS-7 cells was impaired.
Study design
Case history
We investigated an 8-year-old white girl with congenital TTP. Diagnosis was made when she was 18 months of age; she receives a plasma infusion every 2 to 3 months when her platelet count falls below 30 × 109/L.
Assay of plasma VWF-cleaving proteinase activity
An assay based on the collagen-binding activity of digested VWF14 was used to measure the VWF-cleaving proteinase activity in the plasma of the patient (during remission) and her family.
ADAMTS13 sequence analysis
DNA was extracted from peripheral blood leukocytes and all the exons and intron-exon boundaries of the ADAMTS13 gene were amplified by polymerase chain reaction (PCR) and sequenced.5 The study was conducted with parental consent and institutional ethics approval.
Transient expression of the ADAMTS13 mutant
The 4143insA mutant of the ADAMTS13 cDNA15 was created using PCR-based mutagenesis. The wild-type and mutant constructs were subcloned into the mammalian expression vector pCXN and transiently expressed in COS-7 cells.10 Serum-free medium was applied to equivalent numbers of confluent cells at 24 hours and the conditioned medium collected at 48 hours was clarified by centrifugation and concentrated 50-fold. Equivalent numbers of cells were lysed with 0.5% Triton X-100, 5 mM EDTA (ethylenediaminetetraacetic acid), and 1 mM AEBSF ([4-(2-aminoethyl)benzenesulfonylfluoride HCl]). The samples were resolved on 8% to 16% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions and visualized by Western blot using the anti-ADAMTS13 monoclonal antibody, 242/H2.16
Assay of ADAMTS13 activity
VWF-cleaving proteinase activity was determined as previously described.16,17 Dilutions of purified recombinant ADAMTS13 of known concentration were used for assay calibration. Proteolysis of VWF was measured by resolving reactions on 1% agarose gel electrophoresis or 8% SDS-PAGE under reducing conditions and blotting the VWF and fragments with iodinated or peroxidase-conjugated anti-VWF polyclonal antibodies (Dako, Carpinteria, CA), respectively.18
Results and discussion
Three mutations (587C>T, 1342G>C, and 4143-4144insA) were identified in this patient (Figure 1). The nucleotides are numbered from the A of the initiation Met codon. The 587C>T (Thr196Ile) missense mutation has been previously identified in patients with familial TTP and the recombinant mutant protein is reported to have less than 25% VWF-cleaving proteinase activity of the wild type.3,19 The 1342G>C (Gln448Glu) missense mutation on the other hand is not associated with reduced VWF-cleaving proteinase activity and is a single nucleotide polymorphism (SNP) with a reported heterozygosity of 31.2% to 42.5%.5,6 The insertion of a nucleotide (A) at 4143-4144 results in a frameshift in the second CUB domain after Ser1381 followed by a predicted 5–amino acid extension (Arg-Glu-Gln-Pro-Gly) and premature termination of the protein with the loss of 49 amino acids.
VWF-cleaving proteinase activity and ADAMTS13 mutations in a patient with congenital TTP. (A) The pedigree with the ADAMTS13 activity levels of the father (I-1), the mother (I-2), the patient (II-1), and the patient's brother (II-2). The values are expressed as a percentage of VWF-cleaving proteinase activity in normal pooled human plasma. (B) The 1427–amino acid precursor protein of ADAMTS13 contains a signal peptide (SP), a propeptide (Pro), a metalloprotease domain (MP), a disintegrin-like domain (Dis), a type-1 TSP module (T), a cysteine-rich (Cys) and spacer domain, 7 additional type-1 TSP modules, and 2 CUB (complement components C1r/C1s, sea urchin epidermal growth factor, and bone morphogenetic protein) domains. The mutation at Thr196Ile on exon 6 is positioned within the metalloprotease domain and the mutation after Ser1381 truncates the second CUB domain of the protein.
VWF-cleaving proteinase activity and ADAMTS13 mutations in a patient with congenital TTP. (A) The pedigree with the ADAMTS13 activity levels of the father (I-1), the mother (I-2), the patient (II-1), and the patient's brother (II-2). The values are expressed as a percentage of VWF-cleaving proteinase activity in normal pooled human plasma. (B) The 1427–amino acid precursor protein of ADAMTS13 contains a signal peptide (SP), a propeptide (Pro), a metalloprotease domain (MP), a disintegrin-like domain (Dis), a type-1 TSP module (T), a cysteine-rich (Cys) and spacer domain, 7 additional type-1 TSP modules, and 2 CUB (complement components C1r/C1s, sea urchin epidermal growth factor, and bone morphogenetic protein) domains. The mutation at Thr196Ile on exon 6 is positioned within the metalloprotease domain and the mutation after Ser1381 truncates the second CUB domain of the protein.
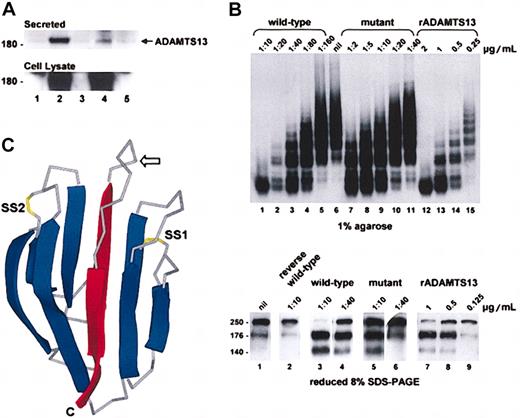
The CUB domain mutation impaired secretion of the enzyme from transfected COS-7 cells but had little effect on VWF-cleaving proteinase activity. The secretion (Figure 2A) and the specific activity (Figure 2B) of the mutant were about 14% and about 85%, respectively, of the wild-type enzyme. Whether this in vitro activity translates into full functional activity in vivo is unknown. The reported VWF-cleaving proteinase activities in patients with hereditary TTP and ADAMTS13 mutations are all less than 10%. The plasma VWF-cleaving proteinase activity in our patient was below the detection threshold of 10% and has been noted to be less than 2% to 5% in other compound heterozygotes with this mutation.7 Asymptomatic heterozygotes, including the mother of our patient, have VWF-cleaving proteinase levels ranging from 21% to 55%.7 It is possible that the plasma level of this mutant is further compromised after secretion due to increased clearance. Complete absence of enzyme activity is thought to be lethal in utero.3 A recent report that 2 brothers with congenital TTP are homozygous for this mutation8 supports the notion that the mutant retains at least some of its biologic activity in vivo.
Expression and activity of the ADAMTS13 mutant. (A) COS-7 cells were transfected with plasmids encoding wild-type or mutant ADAMTS13. The ADAMTS13 in the conditioned media (top panel) and cell lysates (bottom panel) were analyzed by Western blotting. Controls were cells transfected with empty vector (lane 1), wild-type cDNA (lane 3), or mutant cDNA (lane 5) inserted in the reverse orientation. The band density of the secreted mutant (lane 4) was about 14% of the wild-type protein (lane 2). (B) ADAMTS13 activity in the conditioned medium of the transfected COS-7 cells was assayed using purified VWF. Reactions were resolved on 1% agarose and visualized by autoradiography (top panel) and 8% SDS-PAGE under reducing conditions and visualized by Western blotting (bottom panel). Purified rADAMTS13 (lanes 12-15 top panel; 7-9 bottom panel) was used to calibrate the assay. Wild-type conditioned medium (lanes 1-5 top panel; 3 and 4 bottom panel) had about 0.36 μg/mL VWF-cleaving proteinase activity, whereas mutant conditioned medium (lanes 7-11 top panel; 5 and 6 bottom panel) had about 0.04 μg/mL activity. Conditioned medium of cells transfected with wild-type ADAMTS13 cDNA in the reverse orientation had no activity (lane 2 bottom panel). Based on a secretion efficiency of 14% (panel A), the VWF-cleaving proteinase activity of the mutant was about 85% of the wild-type protein. (C) A 3-dimensional representation of the structure of acidic seminal fluid protein illustrating the CUB domain architecture. The β strands are shown as ribbons (blue) and the connecting loops as Cα traces (gray). The expected position of the free cysteine in the first CUB domain of ADAMTS13 is indicated with an arrow. The side chains of disulfides (SS1 and SS2) are shown in yellow. Note that SS2 can exist in the reduced dithiol form in the acidic seminal fluid protein and is absent from the second CUB domain in ADAMTS13. The 2 central β strands that are predicted to be deleted in the first or second CUB domain of ADAMTS13 as a result of the 3769-3770insT or 4143-4144insA mutation, respectively, are shown in red. C indicates C-terminus.
Expression and activity of the ADAMTS13 mutant. (A) COS-7 cells were transfected with plasmids encoding wild-type or mutant ADAMTS13. The ADAMTS13 in the conditioned media (top panel) and cell lysates (bottom panel) were analyzed by Western blotting. Controls were cells transfected with empty vector (lane 1), wild-type cDNA (lane 3), or mutant cDNA (lane 5) inserted in the reverse orientation. The band density of the secreted mutant (lane 4) was about 14% of the wild-type protein (lane 2). (B) ADAMTS13 activity in the conditioned medium of the transfected COS-7 cells was assayed using purified VWF. Reactions were resolved on 1% agarose and visualized by autoradiography (top panel) and 8% SDS-PAGE under reducing conditions and visualized by Western blotting (bottom panel). Purified rADAMTS13 (lanes 12-15 top panel; 7-9 bottom panel) was used to calibrate the assay. Wild-type conditioned medium (lanes 1-5 top panel; 3 and 4 bottom panel) had about 0.36 μg/mL VWF-cleaving proteinase activity, whereas mutant conditioned medium (lanes 7-11 top panel; 5 and 6 bottom panel) had about 0.04 μg/mL activity. Conditioned medium of cells transfected with wild-type ADAMTS13 cDNA in the reverse orientation had no activity (lane 2 bottom panel). Based on a secretion efficiency of 14% (panel A), the VWF-cleaving proteinase activity of the mutant was about 85% of the wild-type protein. (C) A 3-dimensional representation of the structure of acidic seminal fluid protein illustrating the CUB domain architecture. The β strands are shown as ribbons (blue) and the connecting loops as Cα traces (gray). The expected position of the free cysteine in the first CUB domain of ADAMTS13 is indicated with an arrow. The side chains of disulfides (SS1 and SS2) are shown in yellow. Note that SS2 can exist in the reduced dithiol form in the acidic seminal fluid protein and is absent from the second CUB domain in ADAMTS13. The 2 central β strands that are predicted to be deleted in the first or second CUB domain of ADAMTS13 as a result of the 3769-3770insT or 4143-4144insA mutation, respectively, are shown in red. C indicates C-terminus.
In addition to the 4143-4144insA mutation in the second CUB domain, another frameshift mutation 3769-3770insT that leads to truncation at Leu1257 in the first CUB domain and a predicted extension of 34 amino acids has been described in association with congenital TTP.3 The structural consequences of the truncations in the CUB domains were evaluated by searching the Superfamily 1.61 database (http://supfam.org) of all known 3-dimensional protein structures20 using either residues 1131-1298 or residues 1287-1427 as templates. These stretches correspond to the first and the second CUB domains and their flanking regions, respectively. This analysis suggested that residues 1192-1286 and 1299-1407 have structural similarity to the CUB domains of the spermadhesins, porcine major seminal plasma glycoprotein Psp-I and bovine acidic seminal fluid protein, respectively. The CUB domain is a sandwich of two 5-stranded β sheets with one or 2 disulfides.21 The mutations 3769-3770insT and 4143-4144insA are predicted to remove the central β strands from both sheets in the affected CUB domain thereby destroying its architecture.
The first CUB domain of ADAMTS13 contains 5 cysteines of which one (Cys1275) is predicted to be unpaired and surface exposed in the loop connecting the 2 central β strands in this domain (Figure 2C). Furthermore, the second disulfide in the acidic seminal fluid protein CUB domain can exist in the cleaved dithiol form.21 The first ADAMTS13 CUB domain, therefore, may contain one to 3 free thiols. This is noteworthy, considering that the Cys974 thiol of TSP-1 can reduce VWF multimer size by facilitating reduction of inter-subunit disulfide bonds.22 It is possible, therefore, that the first CUB domain has a similar activity. CUB domains in general are involved in protein-protein and protein-carbohydrate interactions,23-25 but the specific ligands for the CUB domains of ADAMTS13 are not known. The sequences of the CUB domains of ADAMTS13 did not align sufficiently with those of other human proteins to predict the identity of these ligands.
Prepublished online as Blood First Edition Paper, September 25, 2003; DOI 10.1182/blood-2003-04-1346.
Supported by the National Health and Medical Research Council of Australia, the National Heart Foundation of Australia, the New South Wales Health Department, and the Danish Natural Science Research Council.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Dr J. Evan Sadler for the ADAMTS13 cDNA, Dr Freidrich Scheiflinger for the 242/H2 antibody, Dr Robert Andrews for the purified VWF, Mr Tim Ganderton for technical assistance, and Mr Peter Taylor for helpful comments.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal