Abstract
We report for the first time the use of a selective small-molecule inhibitor of DNA repair to potentiate topoisomerase II (topo II) poisons, identifying DNA-dependent protein kinase (DNA-PK) as a potential target for leukemia therapy. Topo II poisons form cleavable complexes that are processed to DNA double-strand breaks (DSBs). DNA-PK mediates nonhomologous end joining (NHEJ). Inhibition of this DSB repair pathway may sensitize cells to topo II poisons. We investigated the effects of a novel DNA-PK inhibitor, NU7026 (2-(morpholin-4-yl)-benzo[h]chomen-4-one), on the response to topo II poisons using K562 leukemia cells. NU7026 (10 μM) potentiated the growth inhibition of idarubicin, daunorubicin, doxorubicin, etoposide, amsacrine (mAMSA), and mitroxantrone with potentiation factors at 50% growth inhibition ranging from approximately 19 for mAMSA to approximately 2 for idarubicin (potentiation of etoposide was confirmed by clonogenic assay). In contrast, NU7026 did not potentiate camptothecin or cytosine arabinoside (araC). NU7026 did not affect the levels of etoposide-induced topo IIα or β cleavable complexes. NU7026 alone had no effect on cell cycle distribution, but etoposide-induced accumulation in G2/M was increased by NU7026. A concentration-dependent increase in etoposide-induced DSB levels was increased by NU7026. The mechanism of NU7026 potentiation of topo II poisons involves inhibition of NHEJ and a G2/M checkpoint arrest. (Blood. 2004;103:4659-4665)
Introduction
DNA topoisomerase II (topo II) is the target of some of the most important chemotherapeutic drugs used to treat leukemia. For example, induction therapies for adult acute myeloid leukemia (AML) include an anthracycline (eg, idarubicin or daunorubicin) in combination with cytosine arabinoside (araC), and consolidation protocols include other topo II poisons like mitoxantrone or etoposide.1 Elevated expression of P-glycoprotein (P-gp), resulting in decreased cellular retention of topo II poisons, and decreased expression of topo II are important mechanisms of drug resistance in cell culture and in clinical specimens.2-5 Current therapies for AML are limited by associated toxicities (particularly neutropenia) and incomplete responses. Thus, acquisition of resistance to topo II poisons, incomplete responses, and dose-limiting toxicities are major clinical problems, and development of novel therapies remains an important goal.
Topo II is a nuclear enzyme that controls DNA topology. The enzymatic functions include relaxation of supercoiled DNA and decatenation of sister chromatids. There are 2 isoforms of topo II (α, 170 kDa; and β, 180 kDa) encoded by different genes. Although in vitro they perform the same catalytic function, evidence suggests that in vivo they possess distinct functions.6 During the normal catalytic cycle of topo II, the enzyme cleaves the phosphodiester backbone of DNA to generate a transient enzyme-bridged DNA double-strand break (DSB) through which another DNA duplex can pass, and then the DSB is religated. Topo II poisons trap the intermediate in this process, giving rise to a “cleavable complex”.7 Cellular processing converts the protein-DNA cleavable complex into a DSB, which may be repaired by a DSB repair pathway.
Enzyme-mediated repair of DSBs is a major mechanism of resistance to both ionizing radiation (IR) and drugs that cause DSBs as intermediates in repair processes. Two complementary DSB repair pathways in eukaryotes are nonhomologous end joining (NHEJ) and homologous recombination repair (HRR).8 Important components in these repair pathways are the phosphatidylinositol 3-kinase (PI 3-K)-related protein kinase (PIKK) family of enzymes. These DNA damage-activated serine/threonine protein kinases include DNA-dependent protein kinase (DNA-PK), ataxia telangiectasia-mutated kinase (ATM), and ataxia telangiectasia and rad3-related kinase (ATR).9
The DNA-PK holoenzyme comprises a heterodimer of approximately 70- and 80-kDa polypeptides, known as Ku, that binds to DNA strand breaks, recruiting and activating the 470-kDa catalytic subunit, termed DNA-PKcs.10 Numerous studies have shown that cells lacking DNA-PK are hypersensitive to IR and cross-linking agents and defective in DSB repair.10-13 DNA-PK, together with the x-ray cross-complementing (XRCC4)/DNA ligase IV complex and the recently identified cofactor Artemis, is specifically required for NHEJ,14,15 with about 80% of DNA DSBs repaired by this pathway.16
Studies using mutant cell lines and generic inhibitors of the PIKK family have implicated DNA-PK function in the survival of cells treated with topo II poisons. The xrs-1 cell line, which has a deletion mutation in the Ku80 gene rendering DNA-PK inactive, is hypersensitive to a range of topo II poisons and partially defective in the repair of etoposide-induced DSBs.17,18 Etoposide resistance is restored by transfection of Ku80 cDNA into the mutant cell line.19 The first identified inhibitor of the PIKK enzyme family was the fungal metabolite wortmannin. Although primarily a PI 3-K inhibitor, it was also shown to potentiate IR-induced cytotoxicity and inhibit DSB repair at concentrations that inhibited cellular DNA-PK.20-22 Similar results were also obtained with LY294002, another PIKK family inhibitor.22 Potentiation of etoposide cytotoxicity by wortmannin in a DNA-PK-proficient but not a DNA-PK-deficient cell line has also been demonstrated, indicating that this potentiation resulted specifically from DNA-PK inhibition by wortmannin.23 These data support the idea that a specific DNA-PK inhibitor could prove a powerful tool for use in combination with topo II poisons in leukemia therapy.
NU7026 (2-(morpholin-4-yl)-benzo[h]chomen-4-one), a novel and specific inhibitor of DNA-PK, has been evaluated in this study. NU7026 is a competitive and highly selective inhibitor of DNA-PK, with a 60-fold greater potency against this enzyme than PI 3-K, and inactive against both ATM and ATR.24 We have recently demonstrated that NU7026 is a potent radiosensitizer and inhibitor of DSB repair.24 Here we describe the effects of NU7026 on the cytotoxic mechanisms of a range of topo II poisons in a human leukemia cell line. Cell growth and survival, cell cycle phase distribution, cleavable complex formation, and DSB repair were investigated.
Materials and methods
Cell culture
The K562 cell line was derived from a patient with chronic myelogenous leukemia (CML) in terminal blast phase and its metabolism is therefore similar to that of AML blasts. ML1 is an AML cell line. Both lines were maintained as a suspension culture in RPMI 1640 medium supplemented with 10% (vol/vol) fetal bovine serum, penicillin (50 U/mL), and streptomycin (50 μg/mL). Cells were grown at concentrations between 1 × 105 and 1 × 106/mL and were free of mycoplasma contamination. The doubling time of the cells was approximately 24 hours (K562) and 30 hours (ML1). Cell culture reagents were obtained from Invitrogen (Paisley, United Kingdom).
Drugs
Idarubicin, daunorubicin, doxorubicin, mitoxantrone, and araC were made up as 1-mM stocks dissolved in water, stored in aliquots at -20°C, and diluted in water immediately prior to use. NU7026, mitoxantrone and amsacrine (mAMSA), and camptothecin were dissolved in anhydrous dimethyl sulfoxide (DMSO) as 1-mM stocks and stored at -20°C. Etoposide was dissolved in methanol as a 2-mM stock. Drugs were purchased from Sigma (Poole, United Kingdom). Idarubicin was kindly provided by Pharmacia-UpJohn (Cambridge, United Kingdom). NU7026 was synthesized by the Department of Chemistry, University of Newcastle-upon-Tyne, in collaboration with KuDOS Pharmaceuticals (Cambridge, United Kingdom).
XTT Growth inhibition assay
Exponentially growing cells were seeded (2 × 103/well in 100 μL) into 96-well plates. Drug(s) (6 replicates) were added to plates 24 hours later. Following a 5-day incubation, growth inhibition was quantified using a sodium 3′-[1-(phenoaminocarbonyl)-3,4-tetrazolium] bis(4-methoxy-6-nitro)benzene sulfonic acid hydrate (XTT) cell proliferation kit (Roche, Lewes, United Kingdom), per manufacturer's instructions. Plates were read on a Bio-Rad 550 plate reader (Hercules, CA) at 450 nm, and results were expressed as a percentage of controls. NU7026 (10 μM) was found to have a small inhibitory effect on cell growth (∼ 20% after 5 days) and consequently the NU7026 alone samples were used as controls in calculations for drug combinations using a fixed concentration (10 μM) of inhibitor. Potentiation factors (PF50 values) at 50% growth inhibition (IC50) were calculated from the ratios of the concentration of the drug alone that reduced growth by 50% (IC50 value) divided by the IC50 value obtained when the drug was used in combination with NU7026. Data were averaged from at least 3 independent experiments ± SE.
Clonogenic assay
Exponentially growing cells were exposed to increasing concentrations of etoposide in the presence or absence of NU7026 (10 μM) for 16 hours. Alternatively, cells were exposed to increasing concentrations of NU7026 in the presence or absence of a fixed concentration (0.1 μM) of etoposide. After exposure, drugs were removed by centrifugation and cell numbers were estimated by hemocytometer. A known number of cells were seeded in the absence of drug(s) into medium containing 0.125% (wt/vol) SeaKem agarose (Biowhittaker, Wokingham, United Kingdom) and 10% vol/vol fetal bovine serum in 35-mm culture dishes to allow colony formation. Under these conditions, K562 cells formed colonies with a plating efficiency of approximately 50% without the need for a feeder layer. Viable colonies were visualized by staining with MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; 0.5 mg/mL in water). Colonies were counted using an Oxford Optronix colony counter (Oxford, United Kingdom). Survival was calculated as a percentage of control using the relative plating efficiency of control or NU7026-treated cells. Under the conditions used here, NU7026 had no effect on cell survival. PF50 values were calculated as for the XTT assays. Data were averaged from at least 3 independent experiments ± SE.
TARDIS assay
The trapped in agarose DNA immunostaining (TARDIS) assay is used to quantitate cleavable complex formation and has been described in detail previously.25 Briefly, cells were treated with drugs at the concentrations and for the times specified in the Figure legends and embedded in agarose on microscope slides. Proteins that were not covalently bound to the DNA were removed by placing the slides in 1M NaCl plus protease inhibitors followed by lysis buffer (1% sarkosyl; 80 mM phosphate, pH 6.8; 10 mM EDTA[ethylenediaminetetraacetic acid] plus protease inhibitors). Slides were stained with primary antibody (specific for either topo IIα or β) and then a fluorescein isothiocyanate (FITC)-conjugated secondary antibody before being counter-stained with Hoechst 33258. Images were captured using an epi-fluorescence microscope that separately visualizes blue (Hoescht-stained DNA) and green (FITC-stained topo II) fluorescence. Imager 2 software (Astrocam, Cambridge, United Kingdom) was used to analyze and quantify levels of blue and green fluorescence. Data were averaged from at least 3 independent experiments ± SE.
Flow cytometric analysis
Cells were treated with drug(s) for 24 or 48 hours before permeabilization with 70% ethanol. Cells were treated with propidium iodide (400 μg/mL) and RNAse (1 mg/mL) for 30 minutes at 37°C. Flow cytometry was performed on a Becton Dickinson FACScan (Heidelberg, Germany) equipped with an argon ion laser (excitation at 488 nm).
DNA strand break assay
DSB levels were assayed using the neutral filter elution technique.26 The radiolabelling conditions used were exactly as previously described.21 Prelabelled cells were treated with etoposide (in the presence or absence of 10 μM NU7026) for 1 hour before harvesting and preparing for neutral elution. Cells used as internal standards were exposed to IR (100 Gy) and placed immediately on ice. The internal standards were loaded onto the same filters as the experimental samples and eluted at pH 9.6. Data from elution profiles were summarized as relative elution (RE) values using the formula of Fornace and Little.27 This calculates the amount of DNA retained on the filter when 50% of the internal standards have eluted. Data were averaged from 4 independent experiments ± SE.
Results
Growth inhibition
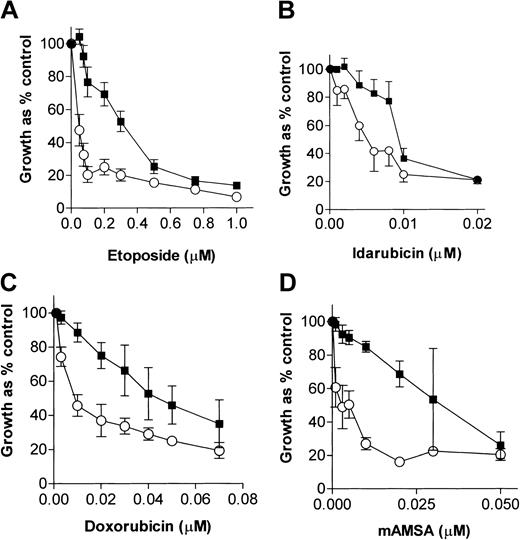
We have previously demonstrated that NU7026 (10 μM) completely inhibits purified human DNA-PK, is not cytotoxic per se in Chinese hamster ovary cells, and maximally potentiates IR-induced cytotoxicity.24 NU7026 was used at 10 μM in all experiments here, because, although it was slightly growth inhibitory (80% control growth), it maximally potentiated etoposide growth inhibition in K562 cells (results not shown) and was not cytotoxic in clonogenic survival assays (see “Cell survival”). Figure 1 shows the concentration-dependent effects of 4 topo II poisons, in the presence or absence of a fixed concentration (10 μM) of NU7026, on cell growth. The IC50 and PF50 values for all the drugs evaluated are summarized in Table 1. NU7026 potentiated the growth inhibitory effects of idarubicin, daunorubicin, doxorubicin, etoposide, mAMSA, and mitoxantrone with PF50 values ranging from approximately 19 for mAMSA to approximately 2 for idarubicin. In contrast, NU7026 did not potentiate the topo I poison, camptothecin, and, although NU7026 appeared to give some protection against the cytotoxicity of the nucleoside analog, araC (PF50 = 0.45 ± 0.29), this was not significant as assessed by the Student t test (P = .1532).
Growth inhibitory effects of topo II poisons in the presence or absence of a fixed concentration of NU7026. K562 cells were treated with increasing concentrations of (A) etoposide, (B) idarubicin, (C) doxorubicin, or (D) mAMSA in the absence (▪) or presence (○) of 10 μM NU7026. After 5 days of exposure, cells were stained with XTT and OD450nm was quantified. Results are means of at least 4 experiments ± SE.
Growth inhibitory effects of topo II poisons in the presence or absence of a fixed concentration of NU7026. K562 cells were treated with increasing concentrations of (A) etoposide, (B) idarubicin, (C) doxorubicin, or (D) mAMSA in the absence (▪) or presence (○) of 10 μM NU7026. After 5 days of exposure, cells were stained with XTT and OD450nm was quantified. Results are means of at least 4 experiments ± SE.
Comparison of IC50 and PF50 values derived from the growth inhibition curves
| Drug . | Drug alone IC50, nM* . | Drug + NU7026 IC50, nM* . | PF50† . |
|---|---|---|---|
| mAMSA | 32 | 1.6 | 19.2 ± 7.5 |
| Mitoxantrone | 3 | 0.18 | 16.6 ± 6.6 |
| Etoposide | 317 | 45 | 8.6 ± 1.0 |
| Doxorubicin | 45 | 10 | 4.0 ± 0.38 |
| Daunorubicin | 205 | 78 | 2.6 ± 0.11 |
| Idarubicin | 9.5 | 5.4 | 1.7 ± 0.17 |
| Camptothecin | 6.8 | 7.3 | 0.88 ± 0.17 |
| araC | 33 | 66 | 0.45 ± 0.29 |
| Drug . | Drug alone IC50, nM* . | Drug + NU7026 IC50, nM* . | PF50† . |
|---|---|---|---|
| mAMSA | 32 | 1.6 | 19.2 ± 7.5 |
| Mitoxantrone | 3 | 0.18 | 16.6 ± 6.6 |
| Etoposide | 317 | 45 | 8.6 ± 1.0 |
| Doxorubicin | 45 | 10 | 4.0 ± 0.38 |
| Daunorubicin | 205 | 78 | 2.6 ± 0.11 |
| Idarubicin | 9.5 | 5.4 | 1.7 ± 0.17 |
| Camptothecin | 6.8 | 7.3 | 0.88 ± 0.17 |
| araC | 33 | 66 | 0.45 ± 0.29 |
IC50 values are the means of at least 3 independent experiments.
All mean PF50 values (± SE) were calculated from the ratio of the individual IC50 values (eg, the concentration of drug that caused 50% growth inhibition divided by the concentration of drug that caused 50% growth inhibition in the presence of NU7026).
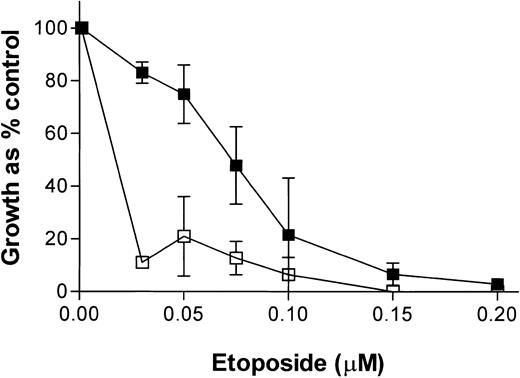
To test whether the effects of NU7026 were cell line specific, we used the AML cell line, ML1. Figure 2 shows that NU7026 also potentiated the growth inhibitory effect of etoposide in this leukemia cell line with a PF50 value of 10.53 ± 4.5. Other data using clonogenic assays have demonstrated that NU7026 also potentiates etoposide- and doxorubicin-induced cytotoxicity in the human epithelial colorectal carcinoma cell lines, LoVo and SW620. Furthermore, NU7026 itself was shown to have little or no effect on the growth of the epithelial lines SW620 and HCT 116 (Yan Zhao, unpublished results, 2003).
Growth inhibitory effect of etoposide ± NU7026 on ML1 cells. Cells were treated for 5 days before XTT staining. ▪ indicates etoposide alone; and □, etoposide + 10 M NU7026. Results are means of 3 experiments ± SE.
Growth inhibitory effect of etoposide ± NU7026 on ML1 cells. Cells were treated for 5 days before XTT staining. ▪ indicates etoposide alone; and □, etoposide + 10 M NU7026. Results are means of 3 experiments ± SE.
Cell survival
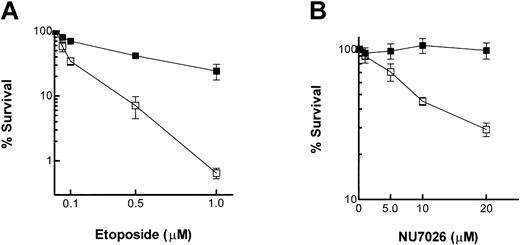
The ability of NU7026 to potentiate cell killing by topo II poisons was evaluated by clonogenic survival assays. Figure 3A shows that treatment of K562 cells with etoposide resulted in a concentration-dependent decrease in survival, and this was potentiated by coincubation with NU7026 (PF50 = 5.4 ± 1.0). It was observed that although 10 μM NU7026 alone was slightly growth inhibitory in a 5-day exposure (see “Growth inhibition”), the 16-hour exposure used here prior to plating for colony formation was completely nontoxic.
Clonogenic survival assay. (A) Cells were treated with increasing concentrations of etoposide in the presence or absence of NU7026 (10 μM) for 16 hours. ▪ indicates etoposide alone; and □, etoposide + NU7026. (B) cells were treated with increasing concentrations of NU7026 in the presence or absence of etoposide (0.1 μM) for 16 hours. ▪ indicates NU7026 alone; and □, NU7026 + etoposide. Cells were then plated in soft agar for colony formation in the absence of drugs. Results are the means of 4 experiments ± SE.
Clonogenic survival assay. (A) Cells were treated with increasing concentrations of etoposide in the presence or absence of NU7026 (10 μM) for 16 hours. ▪ indicates etoposide alone; and □, etoposide + NU7026. (B) cells were treated with increasing concentrations of NU7026 in the presence or absence of etoposide (0.1 μM) for 16 hours. ▪ indicates NU7026 alone; and □, NU7026 + etoposide. Cells were then plated in soft agar for colony formation in the absence of drugs. Results are the means of 4 experiments ± SE.
In order to study the effects of NU7026 alone on cytotoxicity, K562 cells were exposed to increasing concentrations of NU7026 in the presence or absence of 0.1 μM etoposide (a concentration that, used alone, reduced survival to ∼ 65% of controls). Figure 3B demonstrates that NU7026 alone had no effect on cytotoxicity up to a concentration of 20 μM. However, concentrations of 5 to 20 μM all potentiated etoposide-induced cytotoxicity, reducing survival by, for example, 30% (5 μM) and 70% (20 μM).
Cleavable complex levels
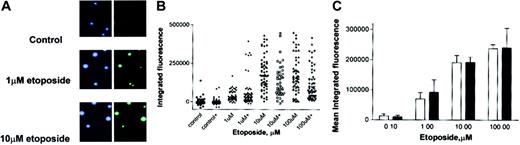
An increase in topo II poison-induced cleavable complex formation by NU7026 is a possible mechanism that would enhance cytotoxicity. Therefore the effect of NU7026 on etoposide-induced cleavable complex levels was investigated using the TARDIS assay. Figure 4A shows the distinctive dose-dependent increase in green immunofluorescence in individual cells after etoposide treatment, corresponding to topo IIα cleavable complex formation.25 The scattergram in Figure 4B is a representative plot from a single experiment to demonstrate the distribution of immunofluorescence in individual cells after treatment with etoposide in the presence or absence of NU7026. Figure 4C (which shows the mean values derived from the scattergram data) demonstrates that coincubation with NU7026 had no significant effect on the levels of topo IIα cleavable complexes. NU7026 also did not alter etoposide-induced topo IIβ cleavable complex levels (data not shown).
Tardis assay of cleavable complex levels. Cells were treated with etoposide in the presence or absence of 10 μM NU7026 for 24 hours. Original magnification × 10. (A) Immunofluorescent micrographs of control cells and cells treated with 1 and 10 μM etoposide. The blue stain indicates DNA; green stain, topo IIα cleavable complexes. (B) Scattergram. (C) Averaged data from scattergram (topo IIα-specific antisera). □ indicates etoposide alone; and ▪, etoposide + NU7026. Results are means of 3 experiments ± SE.
Tardis assay of cleavable complex levels. Cells were treated with etoposide in the presence or absence of 10 μM NU7026 for 24 hours. Original magnification × 10. (A) Immunofluorescent micrographs of control cells and cells treated with 1 and 10 μM etoposide. The blue stain indicates DNA; green stain, topo IIα cleavable complexes. (B) Scattergram. (C) Averaged data from scattergram (topo IIα-specific antisera). □ indicates etoposide alone; and ▪, etoposide + NU7026. Results are means of 3 experiments ± SE.
Cell cycle distribution
Flow cytometric analysis was used to assess the effects of the drugs on cell cycle phase. NU7026 by itself had no effect on cell cycle distribution. Following a 24-hour exposure (equivalent to one cell cycle) to 0.1 μM etoposide, cells accumulated in G2, and this effect was more pronounced after treatment with 0.4 μM etoposide (Figure 5). In the presence of NU7026, the etoposide-induced G2 blockade was enhanced. The percentage of cells in each stage of the cell cycle is summarized in Table 2. For example, at 24 hours the percentage of cells in G2 in NU7026-treated cells was 13% and this was increased to 51% in the presence of etoposide and to 62% when both drugs were present. The differences were even more marked at 48 hours, when cells attempted to progress into a second cell cycle. It was apparent that cells treated with 0.4 μM etoposide alone escaped the 24-hour G2 blockade and progressed into a second cell cycle where they arrested and accumulated in S phase (compare 11% in S phase at 24 h with 40% in S phase at 48 h; Table 2). In contrast, when etoposide and NU7026 were used in conjunction, the majority of cells remained arrested at the initial G2 checkpoint and were unable to progress into the second cell cycle (Figure 5; Table 2). It was also noted that by 48 hours a sub-G1 fraction of cells was accumulating, indicating that cells were undergoing apoptosis (Figure 5).
Flow cytometric analysis of cell cycle distribution. Cells were treated with 0.1 or 0.4 μM etoposide in the presence or absence of NU7026 (10 μM) for 24 or 48 hours before harvesting and preparing for flow cytometric analysis.
Flow cytometric analysis of cell cycle distribution. Cells were treated with 0.1 or 0.4 μM etoposide in the presence or absence of NU7026 (10 μM) for 24 or 48 hours before harvesting and preparing for flow cytometric analysis.
Cell cycle distribution of cells treated with etoposide in the presence or absence of NU7026
. | 24 h . | . | . | 48 h . | . | |||
|---|---|---|---|---|---|---|---|---|
| Cell cycle phase . | NU7026 . | Etoposide . | Etoposide + NU7026 . | Etoposide . | Etoposide + NU7026 . | |||
| G1, % | 71 | 34 | 16 | 32 | 14 | |||
| S, % | 12 | 11 | 10 | 40 | 37 | |||
| G2/M, % | 13 | 51 | 62 | 16 | 44 | |||
. | 24 h . | . | . | 48 h . | . | |||
|---|---|---|---|---|---|---|---|---|
| Cell cycle phase . | NU7026 . | Etoposide . | Etoposide + NU7026 . | Etoposide . | Etoposide + NU7026 . | |||
| G1, % | 71 | 34 | 16 | 32 | 14 | |||
| S, % | 12 | 11 | 10 | 40 | 37 | |||
| G2/M, % | 13 | 51 | 62 | 16 | 44 | |||
Cells were treated with etoposide (0.4 μM) in the presence or absence of NU7026 (10 μM) for 24 or 48 hours, then harvested for flow cytometric analysis. Data are the means of duplicate samples from a representative experiment.
DSB assays
The effect of NU7026 on etoposide-induced DSB levels was assessed by the neutral elution technique. Cells were treated with etoposide for 1 hour in the presence or absence of NU7026. Figure 6A is a representative neutral elution profile showing a concentration-dependent increase in DSBs, represented by decreasing levels of [14C] thymidine (TdR) retained on the filter. In the presence of NU7026, levels of etoposide-induced DSB were higher. For example, (at 50% [3H] retention) 5 μM etoposide resulted in an 82% retention of the [14C], whereas for etoposide + NU7026 this was reduced to 53%. These data were averaged and quantitated by conversion to RE values (Figure 6B). The same trend was observed over all the concentrations of etoposide tested (Figure 6B). For example, the RE value for 5 μM etoposide was 0.056 ± 0.013; in the presence of NU7026 this value was increased to 0.13 ± 0.024, indicating that NU7026 inhibits the rejoining of etoposide-induced DSBs.
DNA DSB assay. (A) Representative neutral elution profile for K562 cells treated with 0 (▪), 5 (▾), 10 (•), or 20 (♦) μM etoposide in the absence (black symbols) or presence (white symbols) of 10 μM NU7026 for 1 hour prior to neutral elution. (B) Mean relative elution values (calculated at 50% [3H] retention). □ indicates etoposide alone; and ▪, etoposide + NU7026. Results are means from 4 independent experiments ± SD. P values were calculated using the 2-tailed unpaired Student t test in order to determine whether DSB levels were significantly different in the presence of NU7026 (P = .01, 5.0 μM; P = .183, 10 μM; P = .06, 20 μM).
DNA DSB assay. (A) Representative neutral elution profile for K562 cells treated with 0 (▪), 5 (▾), 10 (•), or 20 (♦) μM etoposide in the absence (black symbols) or presence (white symbols) of 10 μM NU7026 for 1 hour prior to neutral elution. (B) Mean relative elution values (calculated at 50% [3H] retention). □ indicates etoposide alone; and ▪, etoposide + NU7026. Results are means from 4 independent experiments ± SD. P values were calculated using the 2-tailed unpaired Student t test in order to determine whether DSB levels were significantly different in the presence of NU7026 (P = .01, 5.0 μM; P = .183, 10 μM; P = .06, 20 μM).
Discussion
Topo II poisons are a mainstay of leukemia therapy. Here we demonstrate for the first time that at noncytotoxic concentrations, the DNA-PK inhibitor NU7026 significantly potentiates the growth inhibitory effects of 6 topo II poisons. Furthermore, NU7026 potentiates the cytotoxicity of etoposide in a clonogenic survival assay. NU7026 does not affect the initial lesion of etoposide-induced topo II cleavable complex formation. NU7026 inhibits DSB repair and increases the etoposide-induced G2/M arrest. These data strongly implicate DNA-PK function in the cytotoxic mechanism of topo II poisons downstream of cleavable complex formation. We believe that NU7026 potentiates topo II poisons by inhibiting the DNA-PK-mediated NHEJ of DSBs generated downstream from cleavable complexes. Failure to repair these breaks would lead to the arrest of cells at the DNA damage-induced G2 checkpoint mediated by ATM28 as seen in Figure 5.
PF50 values for these 6 topo II poisons vary from 2 to 19 (Table 1). The wide variation in PF50 is intriguing since the main cytotoxic mechanism of all these agents is poisoning of topo II. There are a number of possibilities to explain this variability. We hypothesize that it is due to the differential targeting of these agents to topo IIα and β. Human topoisomerase II exists in 2 isoforms (α and β), and the agents that give the highest PF50 directly correlate with those that target both topo IIα and topo IIβ. The large PF50 for mAMSA (19.2) and mitoxantrone (16.6) correlates with their known targeting of both isoforms of topo II as part of their cytotoxic mechanism.29 Etoposide shows an intermediate PF50 (8.6) and this agent forms complexes with both isoforms in vivo25 and both isoforms mediate cytotoxicity at high doses of etoposide.29 The 3 topo II poisons that have the lowest PF50 are idarubicin, daunorubicin, and doxorubicin (1.7, 2.55, and 4, respectively) and these are all anthracyclines. We have previously demonstrated that these agents only form cleavable complexes with topo IIα (and not with topo IIβ) in K562 cells and that the sensitivity of mouse embryonic fibroblasts to the anthracyclines is not enhanced by topo IIβ.30 We propose that the dual targeting of both topo IIα and β is the basis for the high levels of potentiation seen with mAMSA, mitoxantrone, and etoposide. However, we cannot exclude the possibility that other factors may play a role in the potentiation by NU7026. These could include the longevity of cleavable complexes30 and their location in the genome,31 both of which are likely to play a role in DNA damage responses. Therefore, for topo II poisons (anthracyclines in particular), it is not always possible to directly correlate levels of DNA strand breaks with cytotoxicity.32,33
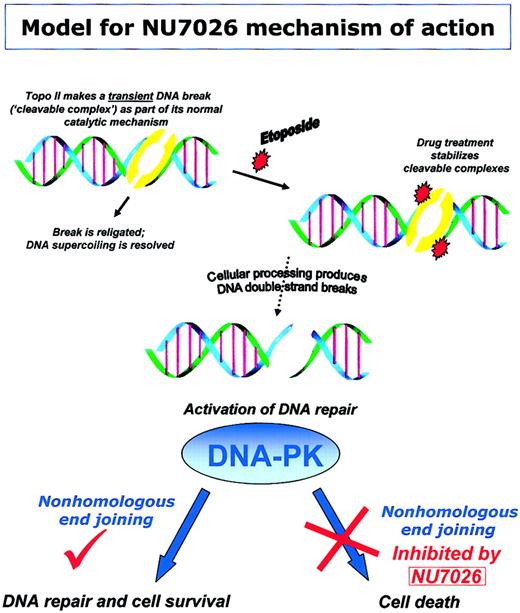
Our model (Figure 7) shows the proposed mechanism by which NU7026 potentiates topo II poison-induced cytotoxicity. Topo II cleavable complexes are transient protein-bridged DNA breaks that are normally religated after the enzyme has passed one DNA duplex through another. Upon drug treatment, these complexes are stabilized and can be processed into DNA DSBs. DNA DSBs can be repaired via recruitment of DNA-PK, and subsequent NHEJ results in cell survival. Alternatively, inhibition of DNA-PK activity by NU7026 results in decreased NHEJ and persisting DNA damage that leads to cell death.
Model for NU7026 mechanism of action. Schematic representing our model for the mechanism by which NU7026 potentiates topo II poison cytotoxicity. The topo II-DNA complex is an intermediate step in the catalytic cycle during which the enzyme passes one DNA duplex through another (via a covalent protein-bridged DNA break) and then religates the cleaved DNA. Treatment with detergent can disrupt or cleave these complexes, hence the term “cleavable complexes.” However, treatment of cells with topo II poisons (eg, etoposide) results in stabilization of the normally transient cleavable complex. After stabilization by drugs, complexes may be processed into DNA DSB, stimulating activation of DNA repair enzymes. Recruitment of DNA-PK to DSB facilitates nonhomologous end joining and therefore repair of DNA. However, in the presence of NU7026, DNA-PK activity is inhibited, NHEJ is impeded, and thus persistence of DNA damage leads to cell death.
Model for NU7026 mechanism of action. Schematic representing our model for the mechanism by which NU7026 potentiates topo II poison cytotoxicity. The topo II-DNA complex is an intermediate step in the catalytic cycle during which the enzyme passes one DNA duplex through another (via a covalent protein-bridged DNA break) and then religates the cleaved DNA. Treatment with detergent can disrupt or cleave these complexes, hence the term “cleavable complexes.” However, treatment of cells with topo II poisons (eg, etoposide) results in stabilization of the normally transient cleavable complex. After stabilization by drugs, complexes may be processed into DNA DSB, stimulating activation of DNA repair enzymes. Recruitment of DNA-PK to DSB facilitates nonhomologous end joining and therefore repair of DNA. However, in the presence of NU7026, DNA-PK activity is inhibited, NHEJ is impeded, and thus persistence of DNA damage leads to cell death.
Our data show that DNA-PK is involved in the repair of DSB following treatment of leukemia cells with topo II poisons. In vitro, DNA-PK is not activated by plasmid DNA bearing an etoposide-stabilized topo II-DNA cleavable complex.34 However, removal of topo II (by proteinase K treatment) revealed a naked DNA break that does stimulate DNA-PK activity. These data are consistent with the requirement for cellular processing to remove topo II from the DNA break before DNA-PK activation can occur. Whether a novel tyrosyl-DNA phosphodiesterase (analogous to the one that acts on DNA topoisomerase I35 ) or a known DNA repair enzyme is able to carry out this function remains to be elucidated.
Two common mechanisms leading to chemoresistance and therapy failure in relapsed leukemias are reduced topo II activity (“atypical drug resistance”) and multidrug resistance (MDR), resulting in reduced intracellular levels of topoisomerase II poisons due to increased drug efflux.36 Increased expression of DNA-PKcs also contributes to chemoresistance and radioresistance by increasing the cellular capacity to carry out NHEJ. Increased DNA-PK activity has been widely demonstrated both in cell culture and in samples from patients with relapsed chronic lymphocytic leukemia (CLL) and correlates with the resistance of tumor cells to IR and bifunctional alkylating agents.37-40 An HL60 cell line selected for resistance to adriamycin was found to have a more than 15-fold increase in DNA-PKcs levels.41 DNA-PK activity and content correlated with sensitivity to doxorubicin and etoposide in clinical samples of cells from patients with CLL.42 Thus, increased DNA-PK activity can contribute to topo II poison-resistance mechanisms. Further research is required to confirm its significance in a clinical setting.
Leukemia-specific alterations in DNA-PK will be important determinants in designing therapeutic regimens for DNA-PK inhibitors. In leukemias where DNA-PK is overexpressed and tumors are radioresistant and chemoresistant, a selective DNA-PK inhibitor would resensitize these resistant cells. In 5 reports, antisense oligonucleotides, small interfering RNAs, and a C-terminal peptide that targets Ku80 and prevents DNA-PKcs binding to Ku have been used to deplete or inhibit DNA-PK function in human cell lines.42-47 Loss of DNA-PK activity correlated with radiosensitization, increased mutation, and inhibition of DNA damage repair.
This is the first report of a very selective small-molecule inhibitor of DNA repair in conjunction with topo II poisons. We believe that potentiation of the cytotoxicity of topo II poisons with DNA-PK inhibitors will be a powerful strategy for sensitizing cells to these agents. NU7026 is a specific DNA-PK inhibitor prototype, ideal for proof-of-principle experiments in cell line models, as reported in this study. The more recent identification of a DNA-PK inhibitor potent enough for pharmacologic evaluation and clinical use is the first step toward being able to test the potential of small-molecule inhibitors of DNA-PK in the clinic. The new compound is now the clinical candidate and is currently undergoing preclinical evaluation in human xenograft studies, cell line models, and leukemia cells derived from patients resistant to topo II poisons in order to test the hypothesis that inhibition of DNA-PK will prove a powerful strategy in overcoming therapeutic resistance.
Prepublished online as Blood First Edition Paper, March 9, 2004; DOI 10.1182/blood-2003-07-2527.
Supported by grant 0171 from the Leukaemia Research Fund, United Kingdom (E.W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


![Figure 6. DNA DSB assay. (A) Representative neutral elution profile for K562 cells treated with 0 (▪), 5 (▾), 10 (•), or 20 (♦) μM etoposide in the absence (black symbols) or presence (white symbols) of 10 μM NU7026 for 1 hour prior to neutral elution. (B) Mean relative elution values (calculated at 50% [3H] retention). □ indicates etoposide alone; and ▪, etoposide + NU7026. Results are means from 4 independent experiments ± SD. P values were calculated using the 2-tailed unpaired Student t test in order to determine whether DSB levels were significantly different in the presence of NU7026 (P = .01, 5.0 μM; P = .183, 10 μM; P = .06, 20 μM).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/12/10.1182_blood-2003-07-2527/5/m_zh80120462640006.jpeg?Expires=1765960489&Signature=mHJJCgQHaUZsLtzWxiK3G9sgpvp4gij0Jx3szM0fMqgm9zI7NSycKNyfW4YcXrwfsIY-GNrNvD2XneZ5gedcCFmArWqYj8pbu7Fcl1N0W26z35vBVQ5AzXsZ0jsFOIGM~~j-Am5dKTaNv1MiBkxjHgtLXSypLO5txjHaYfVNYdFl-rga2bPHwDKHBeGL--AoUfgQZKsBgStwQhNk0Pq-G7u6vy~56j-mNK6emviukKsQbMrrMDxXyS9S0C9bsi4oElYtFLhRarZgr6nsyNBnWxkyjCUNJt74qEgXwGbqOSkwCcoPoRvyVNySBtrSrek9-R52ituZ7watZCnytCwz5g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal