Abstract
The t(12;21)(p13;q22) translocation is the most common chromosomal abnormality yet identified in any pediatric leukemia and gives rise to the TEL-AML1 fusion product. To investigate the effects of TEL-AML1 on hematopoiesis, fetal liver hematopoietic progenitor cells (HPCs) were transduced with retroviral vectors expressing this fusion protein. We show that TEL-AML1 dramatically alters differentiation of HPCs in vitro, preferentially promoting B-lymphocyte development, enhancing self-renewal of B-cell precursors, and leading to the establishment of long-term growth factor–dependent pre–B-cell lines. However, it had no effect on myeloid development in vitro. Further experiments were performed to determine whether TEL-AML1 also demonstrates lineage-specific activity in vivo. TEL-AML1–expressing HPCs displayed a competitive advantage in reconstituting both B-cell and myeloid lineages in vivo but had no effect on reconstitution of the T-cell lineage. Despite promoting these alterations in hematopoiesis, TEL-AML1 did not induce leukemia in transplanted mice. Our study provides a unique insight into the role of TEL-AML1 in leukemia predisposition and a potential model to study the mechanism of leukemogenesis associated with this fusion.
Introduction
The t(12;21)(p13;q22) translocation is present in up to 25% of children with pre–B-cell acute lymphoblastic leukemia (ALL).1,2 This translocation results in the fusion of the AML1 (acute myeloid leukemia-1) gene with the TEL (translocation-Ets-leukemia) gene and generates a TEL-AML1 fusion transcription factor.1,3 Both the AML1 and TEL genes are frequently rearranged in human myeloid and lymphoid leukemias, suggesting that they may be key regulators of hematopoiesis. Indeed, inactivation of these genes by homologous recombination established that AML1 is necessary for definitive hematopoiesis of all lineages4,5 and that TEL is required for hematopoiesis of all lineages in the bone marrow.6,7
Two recent studies have examined whether TEL-AML1 can induce leukemia. Transgenic mice in which expression of TEL-AML1 was driven by the immunoglobulin heavy gene enhancer/promoter failed to develop leukemia or any signs of hematologic disorder.8 However, transfer of adult bone marrow infected with TEL-AML1–expressing retrovirus was found to induce ALL in 2 of 9 irradiated syngeneic mice after a long latency.9 Furthermore, loss of the p16INK4ap19ARF genes, which encode cyclin-dependent kinase inhibitors, was found to cooperate with TEL-AML1 in this model of leukemogenesis. Taken together with the relatively low incidence and long latency of leukemia induced by TEL-AML1 alone, this suggests that secondary tumorigenic mutations are necessary for leukemia associated with TEL-AML1. This conclusion also can be drawn from studies on the occurrence of the TEL-AML1 translocation in the human population. Screening of unselected human cord blood samples established that approximately 1% have a leukemic TEL-AML1 gene fusion.10 This frequency is 100 times the incidence rate of ALL with TEL-AML1, suggesting that generation of a functional TEL-AML1 fusion is insufficient to generate overt ALL and that some additional secondary mutation is necessary. In support of this hypothesis, the onset of leukemia in identical twin children with concordant ALL has a variable and protracted latency, despite evidence that leukemic cells from both twins in a pair probably arose from a single hematopoietic progenitor cell.11,12
These studies have led to the hypothesis that the TEL-AML1 gene fusion generates a covert preleukemic clone that can persist for many years before acquiring secondary mutations and producing overt leukemia.13,14 However, to date there is no direct evidence for any preleukemic activity of TEL-AML1 or indeed any effects of this fusion on hematopoiesis in vivo. Also, apart from inhibiting the myeloid differentiation of 32Dcl3 cells,15 TEL-AML1 has not been found to alter the growth properties of cell lines in vitro.8
To determine whether TEL-AML1 affects hematopoiesis, we transduced mouse fetal liver hematopoietic progenitor cells with a retroviral vector expressing TEL-AML1 and analyzed their differentiation in vitro and in vivo. In children, the t(12;21) translocation is thought to occur before birth, in utero.16 For this reason, we used hematopoietic progenitor cells from mouse fetal liver to model more accurately the effects of TEL-AML1 in human progenitor cells.
Materials and methods
Mice
All mice were maintained in the animal facilities of the National Institute for Medical Research, and experiments were performed according to United Kingdom institutional guidelines and National Institute for Medical Research Home Office regulations.
Retroviral constructs and infection of hematopoietic progenitor cells
The pMSCV-IRES-EGFP (MSCV-EGFP) vector was derived from pMSCV-neo (BD Clontech, Palo Alto, CA) by replacement of phosphoglycerate kinase promoter (PGK) and neor with an internal ribosome entry site (IRES)–enhanced green fluorescent protein (EGFP) cassette. The hemagglutinin (HA)–tagged TEL-AML1 cDNA was obtained by ligating a 5′ TEL cDNA fragment (aa 1-336) (kindly provided by G. Nucifora [Loyola University]17 ) to the 3′ AML1 cDNA fragment (aa 21-453) obtained from Jurkat cell RNA by reverse transcriptase–polymerase chain reaction (PCR) and modified to include a 3′ HA tag. The fusion cDNA was sequenced to confirm that there were no PCR-introduced errors and was cloned into the pMSCV-IRES-EGFP upstream of the IRES-EGFP cassette to generate the MSCV-T/A vector.
The LinXE ecotropic retrovirus packaging cell line18 was transfected with retroviral vectors and supernatant was harvested after 48 hours. Retroviral supernatant, cleared of cell debris, was concentrated 10-fold in some experiments by centrifugation for 1 hour at 16 000g. Viral titers, as determined by EGFP expression by infected 3T3 cells, were routinely in excess of 5 × 106 infectious particles per milliliter for concentrated supernatant. c-Kit+Ter-119– hematopoietic progenitor cells (HPCs) were purified from embryonic day–12 (E12) fetal liver of C57BL/10 mice by fluorescence-activated cell sorting (FACS) using a MoFlo sorter (DakoCy-tomation, Glostrup, Denmark) and mAb specific to c-Kit (2B8) and Ter-119 (Ter-119) (BD Pharmingen, San Diego, CA). HPCs were cultured in recombinant growth factors for 24 hours, infected with retrovirus by spinoculation (centrifugation at 700g, 25° C, 45 minutes) in the presence of 5 μg/mL polybrene (Sigma-Aldrich, Poole, United Kingdom), and returned to culture with the same growth factors for 48 hours. Infection efficiency was determined by analyzing EGFP expression. HPCs were then used for colony-forming assays or in vivo reconstitution assays. For B-cell colony assays and in vivo reconstitution, HPCs were infected in the presence of 10 ng/mL fms-like tyrosine kinase-3 ligand (Flt-3L), 20 ng/mL interleukin-7 (IL-7), and 100 ng/mL stem cell factor (SCF), and for myeloid assays, 10 ng/mL IL-3, 10 ng/mL IL-6, and 100 ng/mL SCF. All recombinant growth factors were of mouse origin (Peprotech EC, London, United Kingdom).
Transplantation assays
Transplantation assays using both lethally and sublethally irradiated mice were used. The former were used to examine the leukemic potential of TEL-AML1–expressing HPCs, whereas the latter were used to establish whether TEL-AML1 conferred any advantage to hematopoietic reconstitution by transduced HPCs, in competition with untransduced donor HPCs and recipient cells. Lethally γ-irradiated (9 Gy) recipient C57BL/6-Ly5.1 mice were injected intravenously with 2 × 105 c-Kit+Ter-119– HPCs 48 hours after infection with unconcentrated MSCV-T/A or MSCV-EGFP retroviral supernatant or with uninfected HPCs. Reconstitution was determined 3 weeks later by flow cytometric analysis of peripheral blood. Donor C57BL/10 cells were distinguished from recipient C57BL/6-Ly5.1 cells by virtue of allotypic markers Ly5.2 and Ly5.1, respectively. Peripheral blood of recipient mice was regularly analyzed for signs of leukemia throughout the experiment. In some experiments, sublethally γ-irradiated (6 Gy) C57BL/6-Ly5.1 mice were used as recipients. Reconstitution of hematopoietic lineages was determined by analysis of peripheral blood 3 and 6 weeks after transplantation. Such analysis gave similar results in 3 independent experiments. Data from a representative experiment is presented in Figures 5 and 6. Mice from this experiment were killed 6 weeks after transplantation, and the contribution of transplanted HPCs to different hematopoietic lineages in spleen and bone marrow was assessed. Statistical significance of differences between data obtained from mice transplanted with MSCV-T/A–transduced HPCs compared to mice transplanted with uninfected HPCs or MSCV-EGFP–transduced HPCs was determined using a 2-tailed Student t test. To analyze the tumorigenicity of TEL-AML1–expressing pre–B-cell lines in vivo, sublethally γ-irradiated (6 Gy) C57BL/6-Ly5.1 mice were injected intravenously with 1 × 106 MSCV-T/A pre–B cells, derived from liquid culture of tertiary MSCV-T/A methylcellulose cultures (see next section).
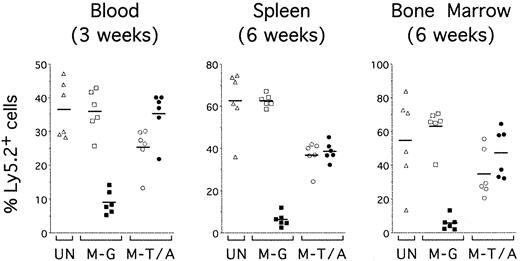
TEL-AML1 confers an advantage on reconstitution by HPCs in vivo. Sublethally irradiated mice were reconstituted with uninfected HPCs (UN; triangles) or HPCs transduced with MSCV-EGFP (M-G; squares) or MSCV-T/A (M-T/A; circles) (n = 6 for each group). Flow cytometry was performed on peripheral blood 3 weeks after transplantation (left panel) and on splenocytes (middle panel) and bone marrow cells (right panel) from the same mice 6 weeks after transplantation. Ly5.2+Ly5.1– cells were gated and EGFP expression was analyzed. Plots show the percentage of EGFP–Ly5.2+ (open symbols) and EGFP+Ly5.2+ (filled symbols) donor-derived cells in the mice that received transplantations. Each symbol represents data from an individual mouse, and bars represent the means within each group.
TEL-AML1 confers an advantage on reconstitution by HPCs in vivo. Sublethally irradiated mice were reconstituted with uninfected HPCs (UN; triangles) or HPCs transduced with MSCV-EGFP (M-G; squares) or MSCV-T/A (M-T/A; circles) (n = 6 for each group). Flow cytometry was performed on peripheral blood 3 weeks after transplantation (left panel) and on splenocytes (middle panel) and bone marrow cells (right panel) from the same mice 6 weeks after transplantation. Ly5.2+Ly5.1– cells were gated and EGFP expression was analyzed. Plots show the percentage of EGFP–Ly5.2+ (open symbols) and EGFP+Ly5.2+ (filled symbols) donor-derived cells in the mice that received transplantations. Each symbol represents data from an individual mouse, and bars represent the means within each group.
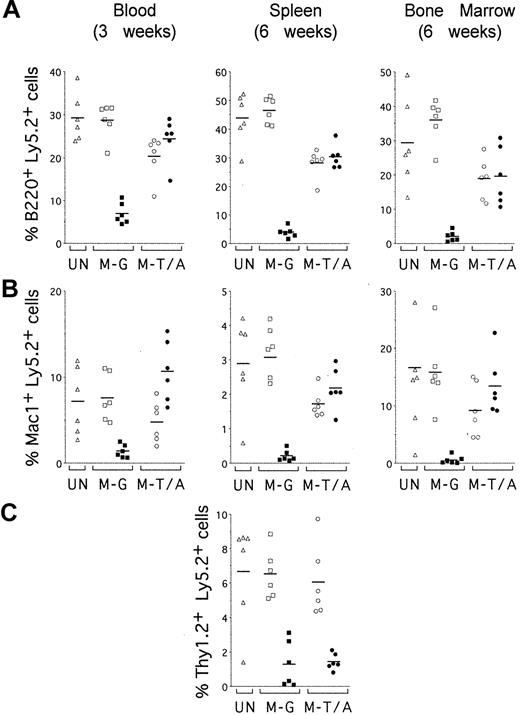
TEL-AML1 promotes development of B and myeloid cells but not T cells in vivo. Sublethally irradiated mice were reconstituted with uninfected HPCs (UN; triangles) or HPCs transduced with MSCV-EGFP (M-G; squares) or MSCV-T/A (M-T/A; circles) (n = 6 for each group). Flow cytometry was performed on peripheral blood 3 weeks after transplantation (left panels) and on splenocytes (middle panels) and bone marrow cells (right panels) from the same mice 6 weeks after transplantation. Ly5.2+Ly5.1– cells were gated and analyzed for the expression of EGFP and lineage-specific cell surface markers. Plots show the percentage of EGFP–Ly5.2+ (open symbols) and EGFP+Ly5.2+ (filled symbols) donor-derived cells that also expressed (A) B220, (B) Mac1, and (C) Thy1.2. Each symbol represents data from an individual mouse, and bars represent the means within each group.
TEL-AML1 promotes development of B and myeloid cells but not T cells in vivo. Sublethally irradiated mice were reconstituted with uninfected HPCs (UN; triangles) or HPCs transduced with MSCV-EGFP (M-G; squares) or MSCV-T/A (M-T/A; circles) (n = 6 for each group). Flow cytometry was performed on peripheral blood 3 weeks after transplantation (left panels) and on splenocytes (middle panels) and bone marrow cells (right panels) from the same mice 6 weeks after transplantation. Ly5.2+Ly5.1– cells were gated and analyzed for the expression of EGFP and lineage-specific cell surface markers. Plots show the percentage of EGFP–Ly5.2+ (open symbols) and EGFP+Ly5.2+ (filled symbols) donor-derived cells that also expressed (A) B220, (B) Mac1, and (C) Thy1.2. Each symbol represents data from an individual mouse, and bars represent the means within each group.
Colony-forming assays and generation of pre–B-cell lines
Uninfected HPCs or HPCs transduced with MSCV-T/A or MSCV-EGFP retroviral constructs were cultured in 1.1-mL duplicate methylcellulose cultures in 35-mm plates (1 × 104 cells/plate) 48 hours after infection with concentrated retroviral supernatant. For B-cell assays, cells were plated in Methocult M3231 (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with IL-7, Flt3L, and SCF. For myeloid assays, cells were plated in Methocult M3534 (Stem Cell Technologies) containing IL-3, IL-6, and SCF and supplemented with 10 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF). After 6 to 10 days, colonies containing 50 cells or more were scored, pooled, and cells counted. Secondary assays and subsequent rounds were performed by replating 1 × 104 cells/mL under identical conditions. Colonies were stained with 1 mg/mL p-iodonitrotetrazolium (INT) in phosphate-buffered saline (PBS). To generate pre–B-cell lines in liquid culture, cells were pooled from tertiary assays and cultured at 1 × 105 cells/mL in Iscoves modified Dulbecco medium (IMDM) (Invitrogen, Carlsbad, CA) with 10% fetal calf serum (FCS), l-glutamine, and 50 μM 2-ME (basic medium) in the presence of IL-7, Flt-3L, and SCF.
Flow cytometry
Cells were stained with phycoerythrin (PE)–, PerCP-, allophycocyanin (APC)–, or biotin-conjugated mAb specific for c-Kit (2B8), B220 (RA3-6B2), CD19 (1D3), BP-1 (6C3), Mac-1 (M1/70), Gr-1 (RB6-8CS), Ly5.2 (104), Ly5.1 (A20), TER-119, and isotope control antibodies (all from BD Pharmingen, San Diego, CA). Cells were resuspended in PBS, 0.5% bovine serum albumin (BSA), and 0.05% sodium azide, and preincubated with unlabeled anti–Fcγ III/II receptor mAb2.4G2 prior to staining with primary antibody. Biotin-conjugated mAbs were visualized using Streptavidin-Tricolour (Caltag, Burlingame, CA) or Streptavidin-PerCP (BD Pharmingen). Four-color cytometry was performed on a FACSCalibur (BD Biosciences, San Jose, CA) and data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Western blot analysis of protein expression
Cell extracts were prepared using radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl; 1% Triton X-100; 0.5% sodium deoxycholate; 0.1% sodium dodecyl sulfate [SDS]; 50 mM Tris [tris(hydroxymethyl)aminomethane] [pH8.0]; 5 mM ethylenediaminetetraacetic acid [EDTA]; 10 μg/mL aprotinin; 250 μg/mL phenylmethylsulfonyl fluoride [PMSF]; 10 μg/mL leupeptin; 10 μg/mL pepstatin A). Protein samples were resolved on a 10% SDS-polyacrylamide gel and transferred to a polyvinylidenefluoride (PVDF) membrane (Immobilon-P; Millipore, Billerica, MA). Membranes were blocked in PBS with 5% nonfat milk and 0.2% Tween-20, and then stained with anti-TEL (H-214; Santa Cruz Biotechnology, Santa Cruz, CA), anti-AML1 (Zymed Laboratories, San Francisco, CA), anti-HA (clone 3F10; Roche Applied Science, Indianapolis, IN), or anti–alpha tubulin (clone YL1/2; Serotec, Oxford, United Kingdom). Proteins were detected by the appropriate secondary horseradish peroxidase (HRP)–conjugated antibodies and a chemiluminescent Reagent (ECL; Amersham Biosciences, Arlington Heights, IL), according to the manufacturers instructions.
Southern blot analysis of DNA
Genomic DNA was digested with EcoRI and NotI. Southern blotting was performed according to standard protocols. The hybridization probe was a 1-kb EcoRI/XbaI cDNA fragment from MSCV-T/A (Figure 1A) labeled with [32P] α-dCTP using random primer labeling according to standard protocols and visualized using a Typhoon phosphorimager (Amersham Biosciences).
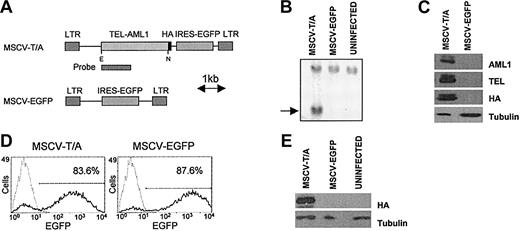
Expression of TEL-AML1 in murine hematopoietic progenitor cells. (A) Schematic diagram of the MSCV constructs used to express TEL-AML1(HA) and EGFP in hematopoietic progenitor cells. E indicates EcoRI; N, NotI. (B) Southern blot analysis of genomic DNA from retrovirally transduced 3T3 cells demonstrating integration of the MSCV-T/A provirus. The indicated cDNA fragment (A) was used as a probe to detect the 2.4-kilobase proviral band (indicated by an arrow) following EcoRI/Not I digestion of genomic DNA. (C) Western blot analysis of protein extracts from 3T3 cells transduced with MSCV-T/A or MSCV-EGFP. Anti-TEL, anti-AML1, or anti-HA antibodies detect TEL-AML1(HA) as a doublet of 95 kDa, due to alternative TEL translational start sites.33 (D) Flow cytometric analysis of HPCs 48 hours following infection with concentrated viral supernatants. Gray lines represent uninfected cells, black lines depict transduced cells, and numbers refer to the percentage of cells transduced. (E) Detection of TEL-AML1(HA) protein in HPCs 48 hours after infection, with anti-HA antibody by Western blot analysis.
Expression of TEL-AML1 in murine hematopoietic progenitor cells. (A) Schematic diagram of the MSCV constructs used to express TEL-AML1(HA) and EGFP in hematopoietic progenitor cells. E indicates EcoRI; N, NotI. (B) Southern blot analysis of genomic DNA from retrovirally transduced 3T3 cells demonstrating integration of the MSCV-T/A provirus. The indicated cDNA fragment (A) was used as a probe to detect the 2.4-kilobase proviral band (indicated by an arrow) following EcoRI/Not I digestion of genomic DNA. (C) Western blot analysis of protein extracts from 3T3 cells transduced with MSCV-T/A or MSCV-EGFP. Anti-TEL, anti-AML1, or anti-HA antibodies detect TEL-AML1(HA) as a doublet of 95 kDa, due to alternative TEL translational start sites.33 (D) Flow cytometric analysis of HPCs 48 hours following infection with concentrated viral supernatants. Gray lines represent uninfected cells, black lines depict transduced cells, and numbers refer to the percentage of cells transduced. (E) Detection of TEL-AML1(HA) protein in HPCs 48 hours after infection, with anti-HA antibody by Western blot analysis.
Results
Expression of the TEL-AML1 fusion in primary hematopoietic progenitor cells using retroviral vectors
The MSCV-T/A vector was made by subcloning the TEL-AML1(HA) cDNA into the murine stem cell virus (MSCV)–based retroviral vector pMSCV-IRES-EGFP, 5′ of the internal ribosome entry site (IRES) and a cDNA encoding the enhanced green fluorescent protein (EGFP) (Figure 1A). The empty vector (MSCV-EGFP) was used as a control (Figure 1A). Integration of the TEL-AML1 provirus into 3T3 cells was confirmed by Southern blot analysis (Figure 1B), and expression of full-length TEL-AML1(HA) protein in cells transduced with MSCV-T/A was confirmed by detection of a 95-kDa band in Western blot analysis using anti-HA, anti-TEL, or anti-AML1 antibodies (Figure 1C). Primary mouse c-Kit+TER-119– HPCs, purified from embryonic day-12 (E12) C57BL/10 fetal liver, were then transduced with MSCV-T/A or MSCV-EGFP vectors. Routinely, in excess of 80% HPCs were infected with concentrated retroviral supernatant in these experiments, based on EGFP expression (Figure 1D). The level of EGFP expressed was consistently lower in MSCV-T/A–infected cells than cells infected with MSCV-EGFP. Western blot analysis using the anti-HA antibody confirmed expression of the TEL-AML1(HA) protein in transduced HPCs (Figure 1E).
TEL-AML1 promotes B-cell development in vitro
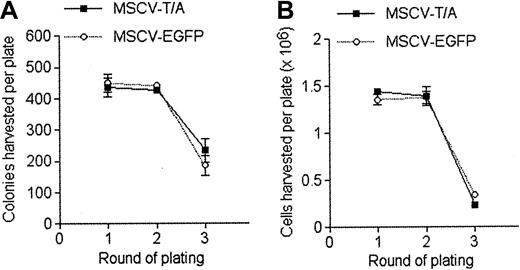
Since the t(12;21) translocation is associated with pre–B-cell ALL, we decided to analyze the effect of TEL-AML1 on B-cell development in vitro. HPCs were transduced with MSCV-T/A or MSCV-EGFP and cultured in methylcellulose media containing growth factors promoting B-cell differentiation.19 After the first round of plating, a small increase was noted in the number of colonies produced by MSCV-T/A–infected cells, compared with the MSCV-EGFP cultures (Figure 2A). However, the absolute number of cells harvested from the first-round plating of MSCV-T/A cultures was much higher than from control MSCV-EGFP cultures (Figure 2B). This was due to an increase in the absolute number of B220+ cells generated by the MSCV-T/A cultures over controls (Figure 2C), with similar numbers of B220– cells being produced by both cultures. Analysis of colony morphology after staining in situ demonstrated that primary MSCV-T/A cultures contained a few very large and dense colonies in addition to the smaller colonies found in both cultures (Figure 2D). These colonies contained a large number of cells, almost exclusively B220+ pre–B cells (data not shown). This probably accounts for the increase in cell number produced by primary MSCV-T/A cultures, compared with control MSCV-EGFP cultures, despite only a relatively small increase in colony number and suggests that TEL-AML1 causes hyperproliferation of B-cell precursors rather than skewing HPC commitment to the B lineage.
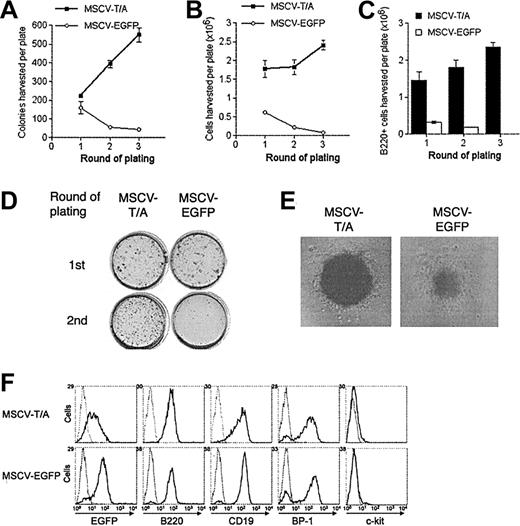
TEL-AML1 enhances development of B-cell precursors in vitro. HPCs transduced with MSCV-T/A or MSCV-EGFP were grown in conditions promoting the development of B-cell progenitors in colony-forming serial replating assays. The number of colonies (A) and the total number of cells (B) produced by MSCV-T/A (▪) and MSCV-EGFP (○) transduced HPCs in each round of plating is shown. Plots show means and SDs of duplicate cultures. The data are representative of 3 independent experiments. (C) Number of B220+ cells harvested from MSCV-T/A (▪) and MSCV-EGFP (□) B-cell methylcellulose cultures from each round of plating. Plots show means and SDs of duplicate cultures. (D) INT-stained methylcellulose MSCV-T/A and MSCV-EGFP cultures from the first and second rounds of plating. (E) Typical pre–B-cell morphology of colonies generated by MSCV-T/A– and MSCV-EGFP–infected cells in the secondary round of plating. Original magnification, × 40. (F) Flow cytometric analysis of EGFP and of lineage-specific marker expression of cells harvested from the secondary round of methylcellulose plating. Gray lines represent the isotype control (or uninfected cells for analysis of EGFP expression), and black lines indicate staining with antibodies specific to the indicated marker, or EGFP expression.
TEL-AML1 enhances development of B-cell precursors in vitro. HPCs transduced with MSCV-T/A or MSCV-EGFP were grown in conditions promoting the development of B-cell progenitors in colony-forming serial replating assays. The number of colonies (A) and the total number of cells (B) produced by MSCV-T/A (▪) and MSCV-EGFP (○) transduced HPCs in each round of plating is shown. Plots show means and SDs of duplicate cultures. The data are representative of 3 independent experiments. (C) Number of B220+ cells harvested from MSCV-T/A (▪) and MSCV-EGFP (□) B-cell methylcellulose cultures from each round of plating. Plots show means and SDs of duplicate cultures. (D) INT-stained methylcellulose MSCV-T/A and MSCV-EGFP cultures from the first and second rounds of plating. (E) Typical pre–B-cell morphology of colonies generated by MSCV-T/A– and MSCV-EGFP–infected cells in the secondary round of plating. Original magnification, × 40. (F) Flow cytometric analysis of EGFP and of lineage-specific marker expression of cells harvested from the secondary round of methylcellulose plating. Gray lines represent the isotype control (or uninfected cells for analysis of EGFP expression), and black lines indicate staining with antibodies specific to the indicated marker, or EGFP expression.
Harvested cells were replated under identical conditions. In secondary assays, MSCV-T/A cultures produced a far greater number of colonies (Figure 2A,D) and cells (Figure 2B) than controls, and the cultures were further enriched for B220+ cells (Figure 2C). Although most of the colonies formed in both the MSCV-T/A and MSCV-EGFP cultures had typical pre–B-cell colony morphology, MSCV-T/A colonies were consistently larger and more dense (Figure 2E). This was presumably a result of increased proliferation of B-cell progenitors within each colony. Harvested cells were mainly pre–B cells (B220+CD19+BP-1+ c-Kit–), and most were positive for EGFP (Figure 2F), the level of expression being lower in MSCV-T/A cells. The phenotype of MSCV-T/A cells was similar to that of control cells, indicating that TEL-AML1 does not cause a block in B-cell development prior to the pre–B-cell stage. While MSCV-EGFP colonies became exhausted of progenitors by the tertiary round of plating, MSCV-T/A cells continued to replate for multiple rounds (Figure 2A-B). Colonies harvested from tertiary cultures were pooled and cells were propagated in liquid culture containing IL-7, Flt-3L, and SCF. Under these conditions, 4 independent MSCV-T/A cell lines were generated and have been maintained for more than 6 months in liquid culture (data not shown). Although MSCV-EGFP cells initially proliferated in liquid culture, they did so at a significantly lower rate than MSCV-T/A cells and failed to establish long-term cultures. Phenotypic analysis established that MSCV-T/A lines were B220+CD19+BP-1+c-Kit– pre–B cells, and Western blotting confirmed that they expressed TEL-AML1(HA) protein (data not shown). These lines remained growth factor dependent (Supplemental Figure S1; see the “Data Set” link at the top of the online article on the Blood website), and withdrawal of growth factors resulted in virtually complete cell death within 48 hours (data not shown).
To test the oncogenic capacity of the MSCV-T/A cell lines, 1 × 106 cells were transplanted into 5 sublethally irradiated C57BL/6-Ly5.1 mice. The transplanted cells failed to grow in vivo and did not induce leukemia in recipient mice over a period of at least 7 months (data not shown). Thus, although TEL-AML1 promotes the generation of long-term pre–B-cell lines in vitro, these cells remain growth factor dependent and are not fully transformed. This is consistent with previous studies demonstrating that TEL-AML1 failed to confer growth factor independence to hematopoietic cell lines.8
TEL-AML1 does not affect myeloid differentiation of HPCs in vitro
To determine whether the in vitro growth promoting activity of TEL-AML1 was specific to B-cell development, we examined myeloid differentiation of MSCV-T/A– and MSCV-EGFP– transduced HPCs in methylcellulose assays. Analysis of primary, secondary, and tertiary cultures demonstrated no significant difference in either colony number (Figure 3A) or cell number (Figure 3B) generated by MSCV-T/A and MSCV-EGFP HPCs. Colony morphology and phenotype of cells produced was not altered by TEL-AML1 expression (data not shown). The number of cells generated in the tertiary MSCV-T/A and MSCV-EGFP cultures was greatly reduced, and cells from both groups failed to replate past the tertiary round. Thus, in these growth conditions TEL-AML1 does not affect the differentiation and self-renewal of myeloid progenitors in vitro.
TEL-AML1 does not affect development of myeloid cells in vitro. HPCs were grown in conditions promoting the development of myeloid cells in colony forming serial replating assays. The number of colonies (A) and the total number of cells (B) produced by MSCV-T/A (▪) and MSCV-EGFP (○) transduced HPCs in each round of plating is shown. Plots show means and SDs of duplicate cultures. The data are representative of 3 independent experiments.
TEL-AML1 does not affect development of myeloid cells in vitro. HPCs were grown in conditions promoting the development of myeloid cells in colony forming serial replating assays. The number of colonies (A) and the total number of cells (B) produced by MSCV-T/A (▪) and MSCV-EGFP (○) transduced HPCs in each round of plating is shown. Plots show means and SDs of duplicate cultures. The data are representative of 3 independent experiments.
TEL-AML1–expressing HPCs do not induce leukemia in mice
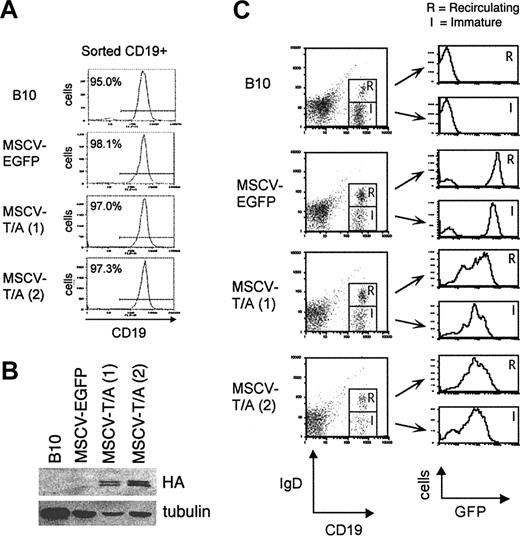
To determine whether expression of TEL-AML1 in HPCs can induce leukemic transformation in vivo, lethally irradiated C57BL/6-Ly5.1 mice received transplants of MSCV-T/A– or MSCV-EGFP–transduced C57BL/10 (Ly5.2+) HPCs. Peripheral blood was analyzed from all mice at 3 and 6 weeks after transplantation to confirm reconstitution with Ly5.2+ donor cells (data not shown). A cohort of 8 MSCV-T/A and 6 MSCV-EGFP mice were examined over a 9-month period, and all have remained viable without developing leukemia (data not shown). Although all MSCV-T/A mice had significant numbers of EGFP+ Ly5.2+ donor cells in their peripheral blood over the time period of our analysis, we wanted to confirm that this correlated with expression of the TEL-AML1 protein. We killed 2 MSCV-T/A and 1 MSCV-EGFP mouse 4 months after receiving transplants and purified CD19+ B cells from their bone marrow by FACS (Figure 4A). Expression of TEL-AML1 protein was detected in CD19+ bone marrow cells from both MSCV-T/A mice but not from MSCV-EGFP mice (Figure 4B).
Analysis of TEL-AML1 expression in bone marrow B cells from mice receiving transplants. (A) CD19+ B cells were sorted from the bone marrow of MSCV-EGFP and MSCV-T/A mice 4 months after transplantation and a C57BL/10 control (B10). The purity of the sorted cells was in excess of 95%. (B) Western blot analysis was performed on protein extracted from the sorted CD19+ bone marrow cells. HA-tagged TEL-AML1 protein was detected using an anti-HA antibody. Anti–alpha tubulin was used to estimate protein loading. (C) Expression of EGFP by (R) CD19+IgD+ recirculating and (I) CD19+IgD– immature bone marrow B cells from MSCV-EGFP, MSCV-T/A, and C57BL/10 mice.
Analysis of TEL-AML1 expression in bone marrow B cells from mice receiving transplants. (A) CD19+ B cells were sorted from the bone marrow of MSCV-EGFP and MSCV-T/A mice 4 months after transplantation and a C57BL/10 control (B10). The purity of the sorted cells was in excess of 95%. (B) Western blot analysis was performed on protein extracted from the sorted CD19+ bone marrow cells. HA-tagged TEL-AML1 protein was detected using an anti-HA antibody. Anti–alpha tubulin was used to estimate protein loading. (C) Expression of EGFP by (R) CD19+IgD+ recirculating and (I) CD19+IgD– immature bone marrow B cells from MSCV-EGFP, MSCV-T/A, and C57BL/10 mice.
TEL-AML1 is associated with pre–B-cell leukemia, and it is possible that expression of TEL-AML1 blocks differentiation of bone marrow progenitor B cells into mature B cells. This is unlikely since we were able to detect mature EGFP+B220+CD19+ B cells in peripheral blood of MSCV-T/A mice. However, it remains possible that TEL-AML1 caused a partial block in B-cell development in the bone marrow. This was investigated by analyzing EGFP expression by immature CD19+IgD– (Hardy Fraction B-E)20 and mature recirculating CD19+IgD+ (Fraction F) B cells in the bone marrow of the 2 MSCV-T/A mice and the control MSCV-EGFP mouse (Figure 4C). The level of EGFP expressed on mature recirculating and immature B cells was equivalent in bone marrow from MSCV-T/A mice, suggesting that TEL-AML1 does not block the development of immature B cells into mature recirculating B cells in vivo. The differences in the level of EGFP expressed by MSCV-T/A– and MSCV-EGFP–transduced cells reflects the different levels of EGFP expressed by the 2 vectors in other cell types, including 3T3 cells and HPCs (Figure 1D). It should be noted that the levels of EGFP expressed on peripheral blood cells from both sets of mice remained constant over the period of our analysis. We also found that there was no evidence for a block in myeloid development of TEL-AML1–expressing cells, since both Mac1+Gr1lo monocytes and Mac1+Gr1hi granulocytes in bone marrow of MSCV-T/A mice expressed equivalently high levels of EGFP (data not shown).
TEL-AML1 confers a selective advantage on hematopoietic repopulation in vivo
Peripheral blood from MSCV-T/A mice contained a significantly higher proportion of donor Ly5.2+ cells than blood from MSCV-EGFP mice, suggesting that TEL-AML1 promoted hematopoietic reconstitution (data not shown). To investigate this possibility, we carried out a further set of experiments transplanting sublethally irradiated mice with MSCV-T/A– or MSCV-EGFP–transduced HPCs. In these experiments, we used lower but equivalent titers of MSCV-T/A and MSCV-EGFP to infect HPCs, such that only a small fraction of donor HPCs were infected (16.6% for MSCV-EGFP and 16.2% for MSCV-T/A for the experiment shown in Figure 5). This allowed us to determine whether the small proportion of EGFP+ LY5.2+ HPCs could outcompete the untransduced EGFP–Ly5.2+ HPCs and recipient Ly5.1+ HPCs in reconstitution of different hematopoietic lineages.
The mean percentage of total donor Ly5.2+ cells in peripheral blood 3 weeks after transplantation was marginally increased in MSCV-T/A mice (60.6% ± 13.1%) compared to mice receiving transplants of untransduced (36.5% ± 8.4%) or MSCV-EGFP (44.9% ± 6.6%) HPCs (P < .05 for both controls, n = 6 for each group). Analysis of EGFP expression by the reconstituting donor cells allowed us to compare the contribution of transduced (EGFP+) cells (filled symbols) and untransduced (EGFP–) cells (open symbols) (Figure 5). EGFP+ cells remained a minor proportion of donor-derived cells in the blood of MSCV-EGFP mice (Figure 5, left panel M-G). However, MSCV-T/A–transduced HPCs demonstrated a striking advantage in repopulating activity, EGFP+ cells becoming a major subpopulation of donor-derived blood cells in MSCV-T/A mice by 3 weeks after transfer (Figure 5, left panel M-T/A). The competitive advantage in repopulating activity by MSCV-T/A HPCs is emphasized by comparing the mean percentage of EGFP+ cells in the blood of MSCV-T/A (35.3% ± 7.0%) and MSCV-EGFP (9.0% ± 3.4%, P < .0001, n = 6) mice. Equivalent numbers of transduced EGFP+ HPCs were originally transferred into mice of each group.
Six weeks after transfer, mice were killed and the contribution of transduced HPCs to donor-derived cells in the spleen (Figure 5, middle panel), and bone marrow (Figure 5, right panel) was evaluated. Consistent with earlier data from peripheral blood, the mean percentage of donor-derived EGFP+ cells in the spleen of MSCV-T/A mice (38.5% ± 4.4%) was much greater than in MSCV-EGFP mice (6.3% ± 3.2%, P < .0001, n = 6). However, although TEL-AML1 expression promoted reconstitution by HPCs, TEL-AML1–expressing cells did not escape the normal homeostatic constraints on the hematopoietic system since there was no evidence of general hyperplasia in MSCV-T/A mice. The mean of absolute splenocyte numbers from MSCV-T/A mice (125 ± 28 × 106) was not significantly different from that of mice that received transplants of untransduced (107 ± 34 × 106, P = .33) or MSCV-EGFP (120 ± 22 × 106, P = .75) HPCs. Enrichment of EGFP+ cells also was evident in the bone marrow of MSCV-T/A mice (Figure 5, right panel).
To establish whether TEL-AML1 expression preferentially promoted reconstitution of specific hematopoietic lineages, the contribution of EGFP+ cells to different lineages was analyzed (Figure 6). EGFP+ cells were clearly enriched in both B220+ B-cell (Figure 6A, left panel) and Mac1+ myeloid (Figure 6B, left panel) lineages in the peripheral blood of MSCV-T/A mice, 3 weeks after transplantation. In contrast, EGFP+ cells in the peripheral blood of MSCV-EGFP mice remained a minor proportion of donor-derived B cells and myeloid cells. Preferential reconstitution of B-cell and myeloid lineages by transduced HPCs also was apparent in splenocytes (Figure 6A-B, middle panel) and bone marrow cells (Figure 6A-B, right panel) from MSCV-T/A mice, 6 weeks after transplantation. However, no enrichment of EGFP+ cells was found in Thy1.2+ T cells from the spleen of MSCV-T/A mice (Figure 6C). There was no significant difference between the mean percentages of donor-derived EGFP+ cells within the T-cell subpopulation of splenocytes from MSCV-T/A (1.4% ± 0.5%) and MSCV-EGFP (1.3% ± 1.3%, P = .8) mice. This contrasts with the differences found between the mean percentages of donor-derived EGFP+ B splenocytes from MSCV-T/A (30.3% ± 4.1%) and MSCV-EGFP (3.9% ± 1.8%, P < .0001) mice, and EGFP+ myeloid splenocytes of MSCV-T/A (2.2% ± 0.6%) and MSCV-EGFP (0.2% ± 0.2%, P < .0001) mice. We were only able to establish the contribution of transduced HPCs to T cells in the spleen because of the very low percentages of Thy1.2+ T cells present in the blood at 3 weeks and bone marrow at 6 weeks after transplantation. Taken together, these data suggest that TEL-AML1 imparts a selective advantage on the hematopoietic reconstitution of the B and myeloid lineages in vivo and does not affect T-cell repopulation.
Discussion
To investigate the effect of TEL-AML1 expression in hematopoeitic progenitor cells, we have generated a murine model of the human t(12;21) chromosomal translocation. We demonstrate that expression of TEL-AML1 enhanced B-cell development and self-renewal of B-cell precursors in vitro and promoted the generation of long-term growth factor–dependent pre–B-cell lines. Furthermore, TEL-AML1 enhanced hematopoietic reconstitution of both B and myeloid lineages in vivo. These data provide the first evidence for specific, lineage-selective alterations in normal hematopoiesis by TEL-AML1. This is entirely consistent with the hypothesis, derived from clinical studies, that TEL-AML1 expression may lead to the generation of covert preleukemic clones, which in some cases produce overt pre–B ALL in patients many years later.13
Mice reconstituted with TEL-AML1–expressing HPCs did not develop leukemia over the time course of our analysis. However, we have only observed the mice that received transplants for 9 months so far, and it is possible that over a longer period of observation some may develop leukemia, as previously reported,9 upon acquisition of secondary mutations by TEL-AML1–expressing cells. Despite the absence of leukemia, our data demonstrate that TEL-AML1 has a dramatic impact on hematopoiesis in the mice that received transplants, which can be clearly seen as early as 3 weeks after transplantation. It is possible that these effects on normal hematopoiesis are responsible for the predisposition to leukemia conferred by TEL-AML1.
Interestingly, cells expressing the AML1-ETO fusion gene from a conditional knock-in allele demonstrated increased replating efficiency in serial methylcellulose assays in the presence of myeloid hematopoietic growth factors, similar to the effect we see with TEL-AML1 in B-cell growth conditions.20 Furthermore, in parallel with our data on TEL-AML1, growth factor–dependent myeloid cell lines could be derived from AML1-ETO knock-in mice, but these cells did not induce leukemia in mice.21
It has been reported that TEL-AML1 blocks the myeloid differentiation of 32Dcl3 cells.15 However, we found that TEL-AML1 had no effect on myeloid differentiation of fetal liver HPCs in vitro. Furthermore, we found that expression of TEL-AML1 was not inconsistent with myeloid development in vivo or the production of bone marrow monocytes and granulocytes. These disparate observations may reflect the different cell types used to analyze the effects of TEL-AML1 expression and the different assays used to assess myeloid differentiation. We have not specifically analyzed differentiation of primary myeloid progenitors in response to granulocyte colony-stimulating factor in vitro and therefore cannot rule out an effect of TEL-AML1 in this assay.
Our data also show that TEL-AML1 promoted myeloid development in vivo (Figure 6B). However, the t(12;21) translocation is not associated with AML, and there are no reports of the TEL-AML1 fusion in preleukemic myeloid cells. Interestingly, in our experiments TEL-AML1 had no effect on myeloid development or the self-renewal of myeloid progenitors in methylcellulose cultures in vitro (Figure 3). This apparent discrepancy may be explained by differences between in vivo hematopoiesis and the culture conditions used to assess myelopoiesis in vitro. Thus, the particular combination of cytokines used in the in vitro myeloid assays may not be appropriate to reveal effects of the TEL-AML1 fusion. It is also possible that B-cell precursors may influence the development of myeloid precursors in vivo, through cell-to-cell contacts or via cytokines. An alternative explanation may be that TEL-AML1 is active only in a specific subset of hematopoietic progenitor cells. Original models of hematopoiesis suggested that B- and T-cell development proceeded from hematopoietic stem cells via a common lymphocyte precursor that had lost the ability to generate myeloid cells.22 This model suggests that the branch point between myeloid and B-cell development would occur relatively early in hematopoiesis. An alternative model has been proposed more recently, suggesting that B-cell progenitors retain the ability to generate myeloid cells after they lose the capacity to generate other lineages.23 Cumano et al found that about half of all B-cell progenitors in day-12 fetal liver (the source of progenitor cells in our study) can give rise to both B and myeloid cells.24 Thus, myeloid potential may accompany early stages of B, T, and erythroid development, and myeloid cells may develop from distinct B/myeloid, T/myeloid, and erythroid/megakaryocyte/myeloid progenitors.23 If this model is accurate, our data could imply that TEL-AML1 is active in bipotential B/myeloid progenitors and therefore promotes both B lymphopoiesis and myelopoiesis through this intermediate in vivo. Conversely, TEL-AML1 may not be active in erythroid/megakaryocyte/myeloid progenitors, the differentiation of which is observed in the in vitro myeloid assays. Furthermore, activity in B/myeloid but not T/myeloid progenitors would be consistent with our data showing that TEL-AML1 did not promote T-cell development in vivo (Figure 6C). The activity of TEL-AML1 in a B/myeloid progenitor cell may also be the explanation for the observation that leukemic blasts from TEL-AML1–positive ALL patients frequently express the myeloid markers CD13 and CD33.25-28 However, it should be noted that the positive effects of TEL-AML1 expression on myeloid development in vivo, in our experiments, cannot be explained by aberrant Mac1 expression on B-cell precursors, since we did not detect cells coexpressing Mac1 and B220 in these mice.
Interestingly, although other TEL fusion proteins such as TEL-JAK2 and TEL-ABL2 have been found in cases of T-lineage ALL, TEL-AML1 appears only to occur in B-precursor ALL.29,30 This is consistent with our data showing that TEL-AML1 did not affect T-cell repopulation and suggests that both TEL and AML1 portions of the fusion protein contribute to the lineage specificity of leukemias with the t(12;21) translocation.
It is still unclear how the effects of TEL-AML1 on normal hematopoiesis predispose to leukemia. Our data do show that TEL-AML1 does not contribute to leukemogenesis by causing a block in B-cell development, since cells expressing the fusion gene were able to mature into recirculating B cells. Thus, in contrast to TEL-AML1–positive leukemic cells, B-cell progenitors expressing TEL-AML1 are capable of terminal differentiation. Studies of relapse in pre–B-cell ALL have demonstrated that distinct leukemias, derived from the same preleukemic clone, are often present at diagnosis and relapse.31,32 These observations suggest that both the chromosomal translocation and preleukemic expansion occur in early B-cell progenitors, before rearrangement of antigen receptor genes, and that the progeny of these cells may become fully transformed at later stages of B-cell development. Our data provide the first experimental evidence for such a preleukemic expansion driven by expression of TEL-AML1. This supports the hypothesis that predisposition to leukemia by the TEL-AML1 fusion is associated with production of covert preleukemic cells, which require further mutations to develop into clinically overt leukemia.13 Characterization of these preleukemic cells is fundamental to gaining insight into the cellular and molecular mechanisms that underlie leukemogenesis. We are currently investigating how TEL-AML1 causes selective expansion of hematopoietic lineages and which secondary mutations cause leukemic transformation of TEL-AML1–expressing cells.
Prepublished online as Blood First Edition Paper, January 15, 2004; DOI 10.1182/blood-2003-10-3695.
Supported by a grant from the Leukaemia Research Fund, United Kingdom.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Trisha Norton for excellent animal husbandry and to Chris Atkins for performing cell sorting. We thank Giuseppina Nucifora (Loyola University) for kindly providing the pCMV-HA-Tel/Aml1 plasmid and Mel Greaves (Institute of Cancer Research) and Alexandre Ptocnik (National Institute for Medical Research) for interesting and useful discussions about our work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal