Abstract
Erythroid progenitors undergo renewal (proliferation without apparent differentiation) in response to erythropoietin (Epo), stem cell factor (SCF), and glucocorticoids (dexamethasone) (Dex). SCF and Dex cooperate with Epo to promote proliferation and inhibit differentiation of erythroid progenitors, while Epo alone is required to protect erythroid cells from apoptosis during terminal red cell maturation. To examine the mechanism of the synergistic interactions of Epo, SCF, and Dex, we analyzed gene expression patterns using DNA chip–based large-scale comparative gene profiling using microarrays enriched in hematopoietic transcripts or containing randomly selected genes. Differentially regulated genes were validated by real-time reverse transcription–polymerase chain reaction (RT-PCR). The results reveal cooperative regulation of gene expression by glucocorticoids and Epo/SCF on a number of genes, such as CIS, BTG1, VDUP1, CXCR4, GILZ, and RIKEN29300106B05. While Epo and SCF never showed opposite effects on gene expression, Dex either enhanced or attenuated the effect of Epo and/or SCF. Several glucocorticoid receptor (GR)–target genes were regulated by Dex only in the presence of Epo and/or SCF, suggesting that the GR functions in the context of a larger transactivation complex to regulate these genes. The data also suggest that modulation of cytokine-induced signals by the GR is an important mechanism in erythroid progenitor renewal.
Introduction
Hematopoiesis requires tight control of survival, proliferation, and differentiation of progenitors to generate and maintain correct numbers of mature cells in the periphery. This process is dependent on cooperation between lineage-specific and more general growth factors and hormones (for review, see Lapidot and Petit1 ). Steady-state erythropoiesis involves a limited number of “differentiation divisions”2,3 and is controlled by erythropoietin (Epo), while stress erythropoiesis, induced by hypoxia, is coregulated by Epo, stem cell factor (SCF), and glucocorticoid receptor (GR) ligands, for example, hydrocortisone or dexamethasone (Dex).4,5 Epo signaling is required for survival and terminal differentiation of erythroid progenitors, since mice lacking Epo or the Epo receptor (EpoR) die at midgestation with a lack of definitive erythrocytes.6,7 In the absence of SCF or its receptor c-Kit, erythrocytes are generated, but the mice die at birth with severe anemia, and their bone marrow contains reduced numbers of erythroid progenitors (erythroid burst-forming units [BFU-Es] and erythroid colony-forming units [CFU-Es]).8-11 Mice carrying a transactivation-deficient mutation of the GR (GRdim/dim)12 are viable, show normal erythropoiesis under normal conditions, but fail to respond to hypoxia and show no stress-induced erythropoiesis in spleen.4
In cell culture, the cooperative action of Epo, SCF, and Dex induces prolonged expansion of primary, proerythroblast-like, c-Kit–expressing erythroid progenitors, which appears to mimic stress erythropoiesis.4,13 Additionally, immortal cultures can reproducibly be established from p53–/– erythroid progenitors that remain fully factor dependent and differentiation competent and retain a stable diploid genome, which allowed unlimited expansion (eg, line I/11).3,13,14 In these models, Epo is strictly required both for terminal differentiation and for expansion. In the absence of Dex, SCF promotes additional cell divisions and delays red cell differentiation, but is unable to cause a complete differentiation arrest.3,13,15,16 On the other hand, activation of the GR upon induction of terminal differentiation by Epo hardly affects differentiation except for reduced hemoglobin accumulation.14 Thus, the cooperation of SCF, Epo, and Dex is required to allow expansion of erythroid progenitors and arrest differentiation. The receptors for Epo and SCF are located in a joint signaling complex at the plasma membrane and activate common signaling pathways, such as the phosphatidylinositol-3-kinase (PI3K) and the mitogen-activated protein kinase/extracellular signal–regulated kinase (MAPK/ERK) cascade,3,17,18 albeit to a different extent. In addition, the EpoR is solely responsible for signal transducer and activator of transcription 5 (Stat5) activation, a pathway known to cooperate with the GR through synergistic and/or antagonistic effects on transcription.19-21
To analyze how cooperative signaling by Epo, SCF, and Dex may control gene expression relevant for progenitor renewal, we used expression profiling using cDNA microarrays. Since the existing libraries of expressed sequence tag (EST) sequences derived from mouse tissues may lack cDNA sequences derived from erythroid-specific genes, we also generated a microarray specific for transcripts from hematopoietic cells to complement the genome-wide arrays. We show that combining this novel microarray with microarrays containing known genes/ESTs was suitable to characterize erythroid-specific gene expression patterns. Thus, a set of genes could be identified (ie, BTG1, VDUP1, CXCR4, and RIKEN29300106B05) whose regulation by Epo or Epo/SCF was counteracted by Dex while a similar antagonistic effect of Epo versus SCF was never observed.
Materials and methods
Cells and culture conditions
Primary erythroid progenitors were expanded from fetal livers of wild-type or GRdim/dim (C57Bl/6 × 129 mixed background) E12.5 embryos as described.12,13 Briefly, dissected fetal livers were suspended in serum-free medium (StemPro-34; Life Technologies, Gibco BRL, Carlsbad, CA) containing Nutrient supplement according to the instructions of the manufacturer, and connective tissue was removed by filtering through a cell strainer (70 μM) (Becton Dickinson, Franklin Lakes, NJ). These primary cell suspensions, as well as the immortal I/11 erythroblast line3 were cultivated in StemPro-34 medium supplemented with 2 U/mL human recombinant Epo (Erypo; Cilag, Baar, Switherland), 100 ng/mL murine recombinant SCF (R&D Systems, Minneapolis, MN), 10–6 M Dex (Sigma, St Louis, MO), and 40 ng/mL insulin-like growth factor 1 (IGF-1) (Promega, Madison, WI). Cultures were maintained at densities between 2 and 4 × 106 cells per milliliter and analyzed daily for cell number and cell size distribution by means of an electronic cell counter (CASY-1; Schärfe-System, Reutlingen, Germany). Primary cell populations were used for experiments until day 10 after seeding.
Factor induction
For acute stimulation with cytokines and hormones, proliferating I/11 erythroid progenitors or cells from primary cultures were washed twice in phosphate-buffered saline (PBS) and seeded at 4 × 106 cells per milliliter in plain Iscove modified Dulbecco medium (IMDM) or StemPro-34, containing Nutrient supplement, without factors. After 4 hours of starvation in this medium, factors were added as indicated (2 U/mL Epo; 100 ng/mL SCF; 10–6 M Dex; or 3 × 10–6 M GR antagonist ZK112.993 [ZK], a kind gift from Schering, Berlin, Germany). Cells were harvested after the indicated times.
Northern blotting
Total RNA was prepared from 2 to 5 × 107 cells with the use of the Trizol LS reagent according to the manufacturer's instructions (Life Technologies, Gibco BRL). For each sample, 15 μg RNA was separated in 1.4% agarose gels containing formaldehyde, transferred to a nylon membrane (Gene Screen; NEN DuPont, Wilmington, DE), fixed by ultraviolet (UV) irradiation, and hybridized sequentially with various 32P-labeled cDNA probes as described.14
DNA array generation, hybridization, and analysis
The custom-made hematopoietic microarray contains approximately 9000 cDNA spots enriched for erythroid- and T-cell–specific cDNAs. RNA isolated from 3T3 fibroblasts and EpH4 epithelial cells was subtracted from expanding I/11 cells and quiescent CD4+ T-cell RNA by suppression-subtractive hybridization (SSH) (by means of PCR-Select cDNA Subtraction Kit; Clontech, Palo Alto, CA). Polymerase chain reaction (PCR) products of the enriched 9000 cDNAs were spotted onto poly-lysine–coated glass slides by a chip-spotting robot developed by A. Massimi for the microarray facility at the Albert Einstein College of Medicine of Yeshiva University (New York, NY) and built by Promedia Association (Antwerp, Belgium). The protocols used for the complete process can be found at http://sequence.aecom.yu.edu/bioinf/microarray/protocol.html (accessed July 23, 2003) and in Cheung et al.22 Prior to hybridization with total RNA, the quality of this RNA was determined by means of a bioanalyzer (2100 Bioanalyzer; Agilent), according to the manufacturer's instructions. For a single hybridization, 30 μg total RNA was reverse transcribed into cDNA by means of oligo(deoxythymidine)12-18 (oligo(dT)12-18) primer (Life Technologies) and 4 μL cyanin 5–uridine 5′-triphosphate (Cy5-UTP) (CyDye; Amersham Biosciences, Freiburg, Germany), while control total RNA was labeled with Cy3-UTP. The microarrays were hybridized overnight with the labeled probes. The scanning was performed with a GenePix 4000A scanner (Axon Instruments, Union City, CA), and the analysis used the GenePix program (Axon Instruments). For the cluster analysis, the data were prefiltered by excluding all genes that did not show any signal after all hybridizations. All spots have been included that showed signals in at least 2 experiments out of 5 or 6 and analyzed by means of GeneSpring (Silicon Genetics, Redwood City, CA).
RNA isolation and cDNA synthesis
RNA was treated with DNase I (Stratagene, Amsterdam, Netherlands), 1 U/μg RNA, incubated 30 minutes at 37°C, and heat-inactivated 10 minutes at 70°C. RNA concentration was determined on a Pharmacia LKB Ultrospec III UV-visible spectrophotometer (Pharmacia, Uppsala, Sweden). Poly(A)+ mRNA was purified from isolated total RNA (10 to 20 μg) with the oligotex mRNA minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The cDNA was synthesized from 1 μg total or poly(A)+ RNA in a 40 μL reverse-transcription reaction mixture containing 2 μg random hexamers (Amersham Pharmacia Biotech, Piscataway, NJ) or 0.25 μg oligo(dT)12-18 (Life Technologies) and 40 mM deoxynucleoside triphosphates (dNTPs), heated 5 minutes at 65°C, and incubated 10 minutes at room temperature. Then, 20 μL 5 × first-stranded buffer (250 mM Tris-HCl [tris(hydroxymethyl)aminomethane-HCl], pH 8.3; 375 mM KCl; 15 mM MgCl2), 200 mM dithiothreitol, 40 U RNasin and 200 U Superscript II RNase H– reverse transcriptase (Life Technologies) were added. The reaction was incubated 2 hours at 42°C, heated 3 minutes at 95°C, and quick-chilled on ice. The cDNA was diluted 1:10 to 1:400 prior to PCR amplification.
Primer pairs
Gene-specific primers corresponding to BTG1 (L16846), CXCR4 (D87747), glucocorticoid-induced leucine zipper (GILZ,NM_010286), MMP-2 (XM_125113), ribonuclease inhibitor (IMAGE:1366946), RIKEN9230106B05 (AK020317), and VDUP1 (BC011212) were obtained from Invitrogen Life Technologies or Sigma-Genosys (The Woodlands, TX). The sequence of the primers used for the amplification were as follows: BTG1, forward 5′TGCAGGAGCTGCTGGCAG3′, reverse 5′TGCTACCTCCTGCTGGTGA3′; CXCR4, forward 5′TGACTTCGAGAGCATTGTGC3′, reverse 5′TGACTCCGTGGAGACGGAAG3′; GILZ, forward 5′TGTATCAGACCCCCATGGAG3′, reverse 5′TCCATGGCCTGCTCAATCTTG3′; MMP-2, forward 5′TGACCTTGACCAGAACACCA3′, reverse 5′TGCCAGGAGTCCATCCTTG3′; ribonuclease inhibitor, forward 5′TCCAGTGTGAGCAGCTGAG3′, reverse 5′TGCAGGCACTGAAGCACCA3′; RIKEN9230106B05, forward 5′TAGGCTAAAGTGCCACGGAC3′, reverse 5′TACATGCCATTCCTGTGGCA3′; and VDUP1, forward 5′TCCTTGCTGATCTACGTCAG3′, reverse 5′TGTCTTGAGAGTCGTCCACA3′.
Real-time PCR
The real-time reverse-transcription PCR (RT-PCR) assay involves TaqMan technology (PE Applied Biosystems model 7900 sequence detector; Applied Biosystems, Weiterstadt, Germany), which combines rapid thermo-cycling with online fluorescence detection of the PCR products. The reactions were performed in a volume of 25 μL of a mixture containing 4 μL cDNA dilution, primers at 5 μM, and 12.5 μL 2 × SYBR Green PCR Master mix (PE Biosystems, Warmington, United Kingdom) containing Amplitaq Gold DNA polymerase, reaction buffer, dNTP mix with dUTP, passive reference, and the double-stranded DNA (dsDNA)–specific fluorescence dye SYBR Green I. Samples were placed in a 96-well plate, capped, and placed in the TaqMan sequence detector. The amplification program consisted of 50°C with 2-minute hold (AmpErase urasil-DNA glycosylase [UNG] incubation), followed by 95°C with 10-minute hold (AmpliTaq Gold Activation), and then by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 62°C for 30 seconds, and extension at 62°C for 30 seconds. All the different primer pairs have a similar optimal PCR annealing temperature. Acquisition of the fluorescence signal from the samples was carried out at the end of the elongation step. To confirm amplification specificity, the PCR products from each primer pair were subjected to melting curve analysis.
Results
While Epo is required for both expansion and differentiation of erythroid progenitors, SCF and Dex cooperate with Epo to promote expansion and arrest differentiation (Figure 1A). To examine the synergistic interactions and the molecular events downstream of the respective cooperating signaling pathways, we first optimized the conditions to detect joint gene regulation by these factors in I/11 erythroid progenitors. Cells were deprived of growth factors for 4 hours and restimulated by Epo (2 U/mL) and SCF (100 ng/mL) plus an agonist of the GR (10–6 M Dex) or an antagonist of the GR (3 × 10–6 M ZK). The expression levels of potential target genes for cooperative Epo/SCF/Dex signaling were then analyzed by Northern blot (Figure 1B). While expression of the Epo-target genes PIM-1 and CIS was enhanced and/or prolonged by exposure to ZK as compared with Dex treatment, activation of suppressor of cytokine signaling 1 (SOCS1) was almost completely repressed in the absence of Dex. However, Dex alone did not induce SOCS1 (data not shown). As observed in other cells,23 Dex reduced expression of its own receptor in cooperation with Epo and SCF. The expression kinetics of GATA1 and IκBα were initially similar in the presence or absence of Dex, but persisted in the presence of ZK at late time points. Given that c-MYB expression is rapidly down-regulated at the onset of differentiation, while GATA-1 expression is even transiently increased (Figure 1A; also Dolznig et al13 ), the decreasing expression of c-MYB compared with GATA1 in the presence of Epo, SCF, and ZK (18 and 24 hours) indicates that the differentiation program has been activated (Figure 1B). In contrast, the ratio between c-MYB and GATA1 remains constant in the presence of Epo, SCF, and Dex. Steady-state activation of EpoR and c-Kit in the presence of either Dex or ZK showed much weaker expression changes in these genes (data not shown). This suggested that acute stimulation of EpoR/c-Kit signaling in the presence or absence of Dex is the method of choice to analyze cooperative effects on gene expression by the 3 receptors required for renewal.
Cooperation of Dex with Epo and SCF to induce renewal divisions of erythroid progenitors. (A) Erythroid progenitor cells can be expanded in vitro in the presence of Epo, SCF, and Dex. In the presence of Epo alone, they undergo terminal differentiation into mature enucleated erythrocytes, while addition of Dex and SCF leads to an expansion of immature erythroid progenitors (renewal). The expression of c-MYB and GATA1 was detected by Northern blot analysis on RNA isolated from erythroid progenitors differentiated from 0 to 72 hours in the presence of Epo (10 U/mL). Hybridization with NF-E2 was taken as a control for differentiation. (B) The I/11 erythroid progenitors were factor deprived for 4 hours (4h starvation) and subsequently restimulated with Epo (2 U/mL), SCF (100 ng/mL), and either Dex (10–6 M) or the GR antagonist ZK (3 × 10–6 M). Cells were harvested at the indicated times, and the expression of the indicated genes was analyzed by Northern blot analysis. Hybridization with GAPDH cDNA was performed as a loading control.
Cooperation of Dex with Epo and SCF to induce renewal divisions of erythroid progenitors. (A) Erythroid progenitor cells can be expanded in vitro in the presence of Epo, SCF, and Dex. In the presence of Epo alone, they undergo terminal differentiation into mature enucleated erythrocytes, while addition of Dex and SCF leads to an expansion of immature erythroid progenitors (renewal). The expression of c-MYB and GATA1 was detected by Northern blot analysis on RNA isolated from erythroid progenitors differentiated from 0 to 72 hours in the presence of Epo (10 U/mL). Hybridization with NF-E2 was taken as a control for differentiation. (B) The I/11 erythroid progenitors were factor deprived for 4 hours (4h starvation) and subsequently restimulated with Epo (2 U/mL), SCF (100 ng/mL), and either Dex (10–6 M) or the GR antagonist ZK (3 × 10–6 M). Cells were harvested at the indicated times, and the expression of the indicated genes was analyzed by Northern blot analysis. Hybridization with GAPDH cDNA was performed as a loading control.
Generating a hematopoietic microarray
Although a large choice of EST libraries is available for gene expression studies, these libraries are still likely to be devoid of genes specifically expressed in red blood cells or their progenitors. Therefore, we generated a novel microarray, enriched for specific hematopoietic transcripts. Messenger RNA prepared from proliferating erythroid progenitors (I/11 cells)3 and resting CD4+ T-cells (freshly isolated by fluorescence-activated cell sorter [FACS]) was used to generate an erythroblast- and a T-cell–specific library by SSH, by means of mRNA from a mixture of fibroblasts (NIH 3T3) and epithelial cells (EpH4) for subtraction. Individual cDNA clones from the 2 subtracted libraries were spotted onto glass slides, which were hybridized with Cy5-labeled cDNA derived from I/11 erythroid cells (red) and Cy3-labeled cDNA derived from CD4+ T-cells (green) (Figure 2A). As expected, most of the spotted clones were specific either for erythroid progenitors (red) or mature T-cells (green) (Figure 2A), while only a minority of the genes represented generally expressed ones.
Generation of a hematopoietic cDNA microarray. (A) Two cDNA libraries enriched for T-cell– and erythroid-specific transcripts, respectively, by SSH were spotted onto glass slides (hematopoietic microarray) and hybridized with Cy5-labeled cDNA derived from I/11 erythroid cells (red) and Cy3-labeled cDNA derived from CD4+ T cells (green). Only a small, typical section of the microarray containing approximately 9000 clones is shown. (B) The I/11 erythroid progenitors were factor deprived for 4 hours and subsequently restimulated with Epo, SCF, and either Dex or the GR antagonist ZK for 2 hours. Cy3-labeled cDNA was generated from Epo/SCF/ZK–treated cell samples; Cy5-labeled cDNA from Epo/SCF/Dex–treated cells and both labeled cDNAs were hybridized to the hematopoietic microarray. In a control experiment, the cyanin dyes for labeling of the cDNAs were exchanged (Dex-Cy3 and ZK-Cy5). Some typical clones up- or down-regulated by Epo/SCF/Dex treatment as compared with Epo/SCF/ZK are boxed.
Generation of a hematopoietic cDNA microarray. (A) Two cDNA libraries enriched for T-cell– and erythroid-specific transcripts, respectively, by SSH were spotted onto glass slides (hematopoietic microarray) and hybridized with Cy5-labeled cDNA derived from I/11 erythroid cells (red) and Cy3-labeled cDNA derived from CD4+ T cells (green). Only a small, typical section of the microarray containing approximately 9000 clones is shown. (B) The I/11 erythroid progenitors were factor deprived for 4 hours and subsequently restimulated with Epo, SCF, and either Dex or the GR antagonist ZK for 2 hours. Cy3-labeled cDNA was generated from Epo/SCF/ZK–treated cell samples; Cy5-labeled cDNA from Epo/SCF/Dex–treated cells and both labeled cDNAs were hybridized to the hematopoietic microarray. In a control experiment, the cyanin dyes for labeling of the cDNAs were exchanged (Dex-Cy3 and ZK-Cy5). Some typical clones up- or down-regulated by Epo/SCF/Dex treatment as compared with Epo/SCF/ZK are boxed.
To evaluate the quality of the libraries produced and to identify the genes represented by the erythroid- or T-cell–specific clones, 192 randomly selected clones from the erythroid-specific library and 96 from the T-cell specific library were sequenced (Table 1). Despite the SSH protocol, the erythroid library was redundant; for instance, beta-globin was found 59 times in the 192 erythroid-specific sequences analyzed. Other erythroid-specific genes like the carbonic anhydrase I or II (CAI, CAII), or Rhag (Rhesus-associated antigen) were also found more than once (Table 1). However, less abundantly expressed genes like p53, eEF1α1, p-selectin, PKCδ, or RNA helicases were also present on the microarray. In addition, we found several sequences without significant homology to sequences in the public databases, most likely representing unknown genes.
Identification of randomly selected erythroid and T-cell—specific cDNA clones
Accession no. . | Short description . | Frequency . |
|---|---|---|
| Erythroid clones | ||
| AK012755 | beta-globin | 59 |
| NA | ESTs, no homolog | 18 |
| NM_009799 | CAI | 9 |
| NM_009801 | CAII | 7 |
| NM_011269 | Rhag | 7 |
| NM_009414 | Tph | 5 |
| NM_011157 | Prg | 3 |
| NM_022994 | CED3 | 2 |
| NM_007638 | chaperonin 7 | 2 |
| AB031387 | CLAST3 | 2 |
| NM_010581 | Itgp | 2 |
| D13695 | ST2L | 2 |
| NM_007961 | TEL | 2 |
| NM_011738 | Ywhah | 2 |
| L31397 | Dynamin | 1 |
| BC005660 | eEF1a1 | 1 |
| BC003433 | eEF2-kinase | 1 |
| BC004801 | IPP delta isomerase | 1 |
| NM_000546 | p53 | 1 |
| NM_008774 | PABP | 1 |
| NM_008859 | PKCteta | 1 |
| NM_007452 | Prdx3 | 1 |
| AK007805 | RNA-helicase | 1 |
| AK019178 | RpL41 | 1 |
| AV269247 | rsca3 | 1 |
| NM_011372 | Sialyltransferase | 1 |
| NM_026506 | SMG | 1 |
| NM_012032 | Tde1 | 1 |
| AF220124 | TRIM11 | 1 |
| NM_011671 | Ucp2 | 1 |
| AK019157 | V-tRNA ligase | 1 |
| NM_011738 | Ywhae | 1 |
| AK018370 | BCL-6 | 1 |
| S71213 | Gi2 alpha (Gnai2) | 1 |
| NM_008160 | glutathione peroxidase | 1 |
| AK013048 | msp1 | 1 |
| NA | low quality, no hit | 36 |
| AF259074 | virrep | 12 |
| T-cell specific | ||
| BC006834 | BTG1 | 1 |
| M38381 | Clk | 1 |
| NM_018813 | Cpsf3 | 2 |
| NM_016811 | Dka | 1 |
| U01138 | EST | 1 |
| NM_010634 | Fabp5 | 1 |
| NM_008080 | Galgt1 | 1 |
| NM_016849 | IRF3 | 1 |
| MMP40GPRT | LANCL 1 | 1 |
| NM_011210 | LY-5 | 7 |
| NM_010444 | Nr4a1 | 1 |
| NM_016389 | NS1-BP | 2 |
| NM_011103 | PKCdelta | 1 |
| NM_009151 | p-selectin | 2 |
| NM_000975 | Rpl11 | 2 |
| NM_009438 | RpL13 | 2 |
| NM_021338 | Rpl35a | 1 |
| NM_009122 | Satb1 | 2 |
| NM_011756 | Zfp36 | 1 |
| NA | Low quality, no hit | 25 |
| AC021063 | Virrep | 40 |
Accession no. . | Short description . | Frequency . |
|---|---|---|
| Erythroid clones | ||
| AK012755 | beta-globin | 59 |
| NA | ESTs, no homolog | 18 |
| NM_009799 | CAI | 9 |
| NM_009801 | CAII | 7 |
| NM_011269 | Rhag | 7 |
| NM_009414 | Tph | 5 |
| NM_011157 | Prg | 3 |
| NM_022994 | CED3 | 2 |
| NM_007638 | chaperonin 7 | 2 |
| AB031387 | CLAST3 | 2 |
| NM_010581 | Itgp | 2 |
| D13695 | ST2L | 2 |
| NM_007961 | TEL | 2 |
| NM_011738 | Ywhah | 2 |
| L31397 | Dynamin | 1 |
| BC005660 | eEF1a1 | 1 |
| BC003433 | eEF2-kinase | 1 |
| BC004801 | IPP delta isomerase | 1 |
| NM_000546 | p53 | 1 |
| NM_008774 | PABP | 1 |
| NM_008859 | PKCteta | 1 |
| NM_007452 | Prdx3 | 1 |
| AK007805 | RNA-helicase | 1 |
| AK019178 | RpL41 | 1 |
| AV269247 | rsca3 | 1 |
| NM_011372 | Sialyltransferase | 1 |
| NM_026506 | SMG | 1 |
| NM_012032 | Tde1 | 1 |
| AF220124 | TRIM11 | 1 |
| NM_011671 | Ucp2 | 1 |
| AK019157 | V-tRNA ligase | 1 |
| NM_011738 | Ywhae | 1 |
| AK018370 | BCL-6 | 1 |
| S71213 | Gi2 alpha (Gnai2) | 1 |
| NM_008160 | glutathione peroxidase | 1 |
| AK013048 | msp1 | 1 |
| NA | low quality, no hit | 36 |
| AF259074 | virrep | 12 |
| T-cell specific | ||
| BC006834 | BTG1 | 1 |
| M38381 | Clk | 1 |
| NM_018813 | Cpsf3 | 2 |
| NM_016811 | Dka | 1 |
| U01138 | EST | 1 |
| NM_010634 | Fabp5 | 1 |
| NM_008080 | Galgt1 | 1 |
| NM_016849 | IRF3 | 1 |
| MMP40GPRT | LANCL 1 | 1 |
| NM_011210 | LY-5 | 7 |
| NM_010444 | Nr4a1 | 1 |
| NM_016389 | NS1-BP | 2 |
| NM_011103 | PKCdelta | 1 |
| NM_009151 | p-selectin | 2 |
| NM_000975 | Rpl11 | 2 |
| NM_009438 | RpL13 | 2 |
| NM_021338 | Rpl35a | 1 |
| NM_009122 | Satb1 | 2 |
| NM_011756 | Zfp36 | 1 |
| NA | Low quality, no hit | 25 |
| AC021063 | Virrep | 40 |
Total clone frequencies: 192 (erythroid) and 96 (T-cell specific). NA indicates not applicable; Virrep, viral repeat sequences.
Expression of cytokine-responsive genes modulated by Dex
First, we used the hematopoietic microarray to study gene regulation by Dex in I/11 erythroid progenitors during acute Epo/SCF stimulation. After factor withdrawal for 4 hours, cells were incubated in the presence of Epo, SCF, and either Dex or the GR antagonist ZK for 2, 6, 18, and 24 hours and processed for mRNA isolation. Cy3-labeled cDNA was generated from Epo/SCF/ZK–treated cell samples, while Cy5-labeled cDNA was prepared from the Epo/SCF/Dex–treated cells. From each time point, the mixed Cy5/Cy3 probes were hybridized to the hematopoietic microarrays. To test for reproducibility, the microarrays were also hybridized with cDNAs labeled with the opposite dye combination, yielding similar results (Figure 2B).
After quantification, the results from the 2 hybridizations were combined and analyzed by means of the clustering program GeneSpring, applying a hierarchic clustering algorithm and a pseudo–color visualization matrix. Data for Dex–up-regulated and Dex–down-regulated genes are shown in Tables 2 and 3. As mentioned above, the SSH-derived cDNA arrays contained multiple copies of hematopoietic-specific, abundantly expressed genes. Rhag and BTG1 (B-cell translocation gene 1) were represented at least 48 and 18 times, respectively, always up-regulated by Dex (Tables 2 and 3). A large cluster of down-regulated sequences represented mainly beta-globin cDNAs (617 times on the microarray). The redundancy combined with the microarray hybridization data performed in duplicate allowed a direct estimation of the reproducibility of the data.
Genes differentially regulated upon exposure to Epo/SCF/Dex or Epo/SCF/ZK for 2, 6, 18, or 24 hours, including their regulation in GRdim/dim versus wt erythroid progenitors
. | . | . | 2 hours . | . | 6 hours . | . | 18 hours . | . | 24 hours . | . | dim/wt . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession no. . | Gene description . | Frequency . | fc . | SD . | fc . | SD . | Fc . | SD . | fc . | SD . | fc . | SD . | |||||
| Dex up-regulated | |||||||||||||||||
| NM_011269 | Rhesus blood group-associated A glycoprotein | 48 | 1.08 | 0.27 | 2.02 | 0.19 | 1.26 | 0.29 | 1.77 | 0.48 | 0.91 | 0.36 | |||||
| NM_011157 | Proteoglycan, secretory granule | 19 | 1.03 | 0.26 | 0.93 | 0.08 | 1.45 | 0.22 | 1.95 | 0.37 | 1.21 | 0.25 | |||||
| BC006834 | B-cell translocation gene 1, antiproliferative | 18 | 0.98 | 0.19 | 2.41 | 0.28 | 3.35 | 2.22 | 1.29 | 0.27 | 0.67 | 0.15 | |||||
| NM_009735 | Beta-2 microglobulin (B2m) | 21 | 0.99 | 0.18 | 1.34 | 0.12 | 1.92 | 0.43 | 2.48 | 0.95 | 0.65 | 0.15 | |||||
| NM_018813 | Cleavage and polyadenylation specificity factor 3 | 11 | 0.97 | 0.14 | 1.06 | 0.10 | 2.78 | 1.35 | 2.45 | 0.94 | 0.70 | 0.15 | |||||
| NM_010634 | Fatty acid binding protein 5, epidermal | 10 | 1.04 | 0.26 | 0.89 | 0.07 | 1.73 | 0.41 | 3.17 | 1.57 | 0.84 | 0.19 | |||||
| NM_008529 | Lymphocyte antigen 6 complex, locus E | 10 | 1.00 | 0.17 | 1.15 | 0.11 | 2.00 | 0.61 | 2.48 | 0.98 | 0.76 | 0.15 | |||||
| AK003942 | RIKEN:1110028E10 | 7 | 1.05 | 0.15 | 1.61 | 0.12 | 3.78 | 1.86 | 3.19 | 1.44 | 0.50 | 0.09 | |||||
| AK020317 | RIKEN:9230106B05 | 6 | 1.17 | 0.18 | 2.27 | 0.26 | 1.58 | 0.66 | 1.73 | 0.69 | 0.72 | 0.20 | |||||
| NM_023719 | Thioredoxin-interacting protein | 4 | 1.16 | 0.27 | 1.82 | 0.17 | 1.08 | 0.24 | 1.21 | 0.44 | 1.10 | 0.33 | |||||
| NM_013627 | Paired box gene 6 (Pax6), mRNA | 4 | 1.10 | 0.19 | 1.84 | 0.18 | 1.07 | 0.24 | 0.96 | 0.16 | 0.84 | 0.19 | |||||
| NM_008722 | Nucleophosmin 1 (Npm1) | 3 | 0.99 | 0.26 | 0.75 | 0.14 | 1.68 | 0.34 | 3.28 | 1.46 | 0.75 | 0.14 | |||||
| AF260927 | D4Cole 1e | 2 | 1.32 | 0.25 | 2.07 | 0.25 | 2.93 | 2.61 | 3.65 | 1.13 | 0.96 | 0.23 | |||||
| NM_011755 | Zinc finger protein 35 | 2 | 1.09 | 0.28 | 1.15 | 0.11 | 1.30 | 0.37 | 1.97 | 0.34 | 0.69 | 0.09 | |||||
| NM_009429 | Tumor protein, translationally controlled 1 (Tpl1) | 2 | 1.06 | 0.34 | 0.98 | 0.05 | 1.32 | 0.18 | 2.26 | 0.61 | 0.66 | 0.21 | |||||
| NM_011050 | Programmed cell death 4 (Pdcd4) | 2 | 0.98 | 0.31 | 0.85 | 0.09 | 1.57 | 0.28 | 2.37 | 0.70 | 0.65 | 0.15 | |||||
| AW912233 | Soares mammary gland, IMAGE:1514107 5′ | 2 | 1.05 | 0.15 | 1.25 | 0.12 | 2.20 | 0.24 | 3.75 | 2.01 | 0.55 | 0.25 | |||||
| NM_008379 | Karyopherin (importin) beta 1 | 2 | 0.90 | 0.13 | 0.86 | 0.17 | 1.41 | 0.13 | 2.55 | 1.00 | 1.07 | 0.29 | |||||
| Dex repressed | |||||||||||||||||
| AB020015 | Beta-1-globin | 98 | 1.05 | 0.25 | 0.84 | 0.18 | 0.67 | 0.24 | 0.62 | 0.20 | 1.57 | 0.39 | |||||
| NM_009414 | Tryptophan hydroxylase | 30 | 1.44 | 0.33 | 0.64 | 0.30 | 1.30 | 0.42 | 1.71 | 0.57 | 6.37 | 2.66 | |||||
| NM_016633 | Erythroid differentiatiation factor | 15 | 0.99 | 0.21 | 0.59 | 0.06 | 0.65 | 0.11 | 0.87 | 0.32 | 1.42 | 0.37 | |||||
| NM_009801 | Carbonic anhydrase 2 (Car2) | 4 | 1.01 | 0.24 | 0.65 | 0.01 | 0.85 | 0.08 | 1.22 | 0.56 | 1.76 | 0.41 | |||||
. | . | . | 2 hours . | . | 6 hours . | . | 18 hours . | . | 24 hours . | . | dim/wt . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession no. . | Gene description . | Frequency . | fc . | SD . | fc . | SD . | Fc . | SD . | fc . | SD . | fc . | SD . | |||||
| Dex up-regulated | |||||||||||||||||
| NM_011269 | Rhesus blood group-associated A glycoprotein | 48 | 1.08 | 0.27 | 2.02 | 0.19 | 1.26 | 0.29 | 1.77 | 0.48 | 0.91 | 0.36 | |||||
| NM_011157 | Proteoglycan, secretory granule | 19 | 1.03 | 0.26 | 0.93 | 0.08 | 1.45 | 0.22 | 1.95 | 0.37 | 1.21 | 0.25 | |||||
| BC006834 | B-cell translocation gene 1, antiproliferative | 18 | 0.98 | 0.19 | 2.41 | 0.28 | 3.35 | 2.22 | 1.29 | 0.27 | 0.67 | 0.15 | |||||
| NM_009735 | Beta-2 microglobulin (B2m) | 21 | 0.99 | 0.18 | 1.34 | 0.12 | 1.92 | 0.43 | 2.48 | 0.95 | 0.65 | 0.15 | |||||
| NM_018813 | Cleavage and polyadenylation specificity factor 3 | 11 | 0.97 | 0.14 | 1.06 | 0.10 | 2.78 | 1.35 | 2.45 | 0.94 | 0.70 | 0.15 | |||||
| NM_010634 | Fatty acid binding protein 5, epidermal | 10 | 1.04 | 0.26 | 0.89 | 0.07 | 1.73 | 0.41 | 3.17 | 1.57 | 0.84 | 0.19 | |||||
| NM_008529 | Lymphocyte antigen 6 complex, locus E | 10 | 1.00 | 0.17 | 1.15 | 0.11 | 2.00 | 0.61 | 2.48 | 0.98 | 0.76 | 0.15 | |||||
| AK003942 | RIKEN:1110028E10 | 7 | 1.05 | 0.15 | 1.61 | 0.12 | 3.78 | 1.86 | 3.19 | 1.44 | 0.50 | 0.09 | |||||
| AK020317 | RIKEN:9230106B05 | 6 | 1.17 | 0.18 | 2.27 | 0.26 | 1.58 | 0.66 | 1.73 | 0.69 | 0.72 | 0.20 | |||||
| NM_023719 | Thioredoxin-interacting protein | 4 | 1.16 | 0.27 | 1.82 | 0.17 | 1.08 | 0.24 | 1.21 | 0.44 | 1.10 | 0.33 | |||||
| NM_013627 | Paired box gene 6 (Pax6), mRNA | 4 | 1.10 | 0.19 | 1.84 | 0.18 | 1.07 | 0.24 | 0.96 | 0.16 | 0.84 | 0.19 | |||||
| NM_008722 | Nucleophosmin 1 (Npm1) | 3 | 0.99 | 0.26 | 0.75 | 0.14 | 1.68 | 0.34 | 3.28 | 1.46 | 0.75 | 0.14 | |||||
| AF260927 | D4Cole 1e | 2 | 1.32 | 0.25 | 2.07 | 0.25 | 2.93 | 2.61 | 3.65 | 1.13 | 0.96 | 0.23 | |||||
| NM_011755 | Zinc finger protein 35 | 2 | 1.09 | 0.28 | 1.15 | 0.11 | 1.30 | 0.37 | 1.97 | 0.34 | 0.69 | 0.09 | |||||
| NM_009429 | Tumor protein, translationally controlled 1 (Tpl1) | 2 | 1.06 | 0.34 | 0.98 | 0.05 | 1.32 | 0.18 | 2.26 | 0.61 | 0.66 | 0.21 | |||||
| NM_011050 | Programmed cell death 4 (Pdcd4) | 2 | 0.98 | 0.31 | 0.85 | 0.09 | 1.57 | 0.28 | 2.37 | 0.70 | 0.65 | 0.15 | |||||
| AW912233 | Soares mammary gland, IMAGE:1514107 5′ | 2 | 1.05 | 0.15 | 1.25 | 0.12 | 2.20 | 0.24 | 3.75 | 2.01 | 0.55 | 0.25 | |||||
| NM_008379 | Karyopherin (importin) beta 1 | 2 | 0.90 | 0.13 | 0.86 | 0.17 | 1.41 | 0.13 | 2.55 | 1.00 | 1.07 | 0.29 | |||||
| Dex repressed | |||||||||||||||||
| AB020015 | Beta-1-globin | 98 | 1.05 | 0.25 | 0.84 | 0.18 | 0.67 | 0.24 | 0.62 | 0.20 | 1.57 | 0.39 | |||||
| NM_009414 | Tryptophan hydroxylase | 30 | 1.44 | 0.33 | 0.64 | 0.30 | 1.30 | 0.42 | 1.71 | 0.57 | 6.37 | 2.66 | |||||
| NM_016633 | Erythroid differentiatiation factor | 15 | 0.99 | 0.21 | 0.59 | 0.06 | 0.65 | 0.11 | 0.87 | 0.32 | 1.42 | 0.37 | |||||
| NM_009801 | Carbonic anhydrase 2 (Car2) | 4 | 1.01 | 0.24 | 0.65 | 0.01 | 0.85 | 0.08 | 1.22 | 0.56 | 1.76 | 0.41 | |||||
The ratio between Epo/SCF/Dex over Epo/SCF/ZK at any time point is given as fold change (fc). When the frequency of a sequence was 2 or more (n > 1) the data points were averaged and the standard deviation (SD) is shown. wt indicates wild type.
Genes with a frequency of 1 and differentially regulated upon exposure to Epo/SCF/Dex or Epo/SCF/ZK for 2, 6, 18, or 24 hours, including their regulation in GRdim/dim versus wt erythroid progenitors
. | . | 2 hours . | . | 6 hours . | . | 18 hours . | . | 24 hours . | . | dim/wt . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession no. . | Gene description . | First . | Second . | First . | Second . | First . | Second . | First . | Second . | First . | Second . | |||||
| Dex up-regulated | ||||||||||||||||
| AA020552 | IMAGE:455550 5 | 0.86 | 1.20 | 1.15 | 1.27 | 1.70 | 2.12 | 1.56 | 3.44 | 1.11 | 2.10 | |||||
| AW553415 | Mouse newborn ovary cDNA library | 0.70 | 1.60 | 0.76 | 0.80 | 1.10 | 1.70 | 2.02 | 2.06 | 0.61 | 1.03 | |||||
| NM_007475 | Acidic ribosomal phosphoprotein PO | 0.88 | 1.36 | 2.20 | 2.55 | 1.50 | 1.41 | 1.78 | 1.43 | 0.69 | 0.77 | |||||
| NM_025840 | Basic leucine zipper and W2 domains 2 | 0.77 | 1.46 | 1.02 | 1.07 | 0.69 | 1.96 | 2.05 | 2.31 | 0.48 | 0.93 | |||||
| AK003679 | RIKEN:1110014C11 | 0.91 | 1.33 | 1.13 | 1.25 | 0.94 | 1.57 | 1.95 | 2.88 | 0.76 | 0.91 | |||||
| D87747 | CXCR-4 | 1.07 | 1.66 | 1.51 | 1.29 | 1.16 | 1.18 | 1.58 | 1.90 | 0.91 | 1.15 | |||||
| NM_011722 | Dynactin 6 | 1.29 | 1.28 | 1.87 | 1.96 | 1.25 | 3.00 | 2.88 | 4.64 | 0.68 | 0.89 | |||||
| BC005660 | Eukaryotic translation elongation factor 1 alpha 1 | 0.72 | 1.36 | 1.06 | 0.92 | 0.88 | 1.46 | 2.13 | 2.27 | 0.52 | 0.89 | |||||
| BG080849 | CDNA clone H3058D01-5 | 0.78 | 1.55 | 1.92 | 1.62 | 1.01 | 0.70 | 0.74 | 0.94 | 0.62 | 0.80 | |||||
| AA655703 | IMAGE:1149396 | 0.78 | 1.49 | 2.03 | 2.03 | 0.90 | 1.17 | 1.70 | 2.42 | 0.94 | 1.17 | |||||
| BE630992 | IMAGE:3375525 | 1.25 | 0.81 | 2.10 | 2.06 | 1.90 | 1.79 | 1.26 | 1.15 | 0.88 | 0.83 | |||||
| AK020317 | RIKEN 9230106805 | 1.21 | 0.74 | 1.52 | 1.93 | 2.93 | 1.79 | 1.02 | 1.84 | 0.74 | 0.64 | |||||
| NM_021099 | kit oncogene | 1.32 | 1.02 | 1.09 | 1.23 | 2.12 | 4.71 | 2.06 | 3.61 | 0.60 | 0.53 | |||||
| BC006720 | Karyopherin (importin) alpha 2 | 0.62 | 1.42 | 0.99 | 0.86 | 0.57 | 0.93 | 1.61 | 1.55 | 0.67 | 1.56 | |||||
| K02061 | L32-4A pseudogene | 0.96 | 1.20 | 1.13 | 1.01 | 1.42 | 1.52 | 1.88 | 3.30 | 0.57 | 0.75 | |||||
| NM_010686 | Laptm5 | 1.17 | 0.77 | 1.14 | 1.36 | 2.04 | 3.88 | 1.40 | 0.98 | 2.34 | 1.37 | |||||
| NM_010708 | Lectin, galactose binding, soluble 9 | 1.08 | 0.73 | 0.80 | 0.81 | 1.49 | 2.67 | 1.56 | 3.06 | 1.20 | 0.85 | |||||
| NM_020573 | Oxysterol binding proteinlike 1A | 0.88 | 1.10 | 1.10 | 1.14 | 1.21 | 1.71 | 1.98 | 2.14 | 0.99 | 1.09 | |||||
| BC005779 | RIKEN cDNA 0610010E03 gene | 0.75 | 1.21 | 1.20 | 0.93 | 0.87 | 1.16 | 1.67 | 1.93 | 0.57 | 0.91 | |||||
| AK002923 | RIKEN cDNA 0610043A03 gene | 0.97 | 1.10 | 1.45 | 1.21 | 1.74 | 2.04 | 1.61 | 2.92 | 0.55 | 0.78 | |||||
| AK004571 | RIKEN cDNA 1200003I10 gene | 0.82 | 1.47 | 1.65 | 1.65 | 0.57 | 1.02 | 2.15 | 2.11 | 0.71 | 1.38 | |||||
| NM_023270 | RIKEN cDNA 1300002C13 gene | 1.36 | 1.04 | 1.15 | 1.42 | 1.22 | 1.27 | 1.57 | 2.30 | 0.96 | 0.67 | |||||
| AK010014 | RIKEN cDNA 2310061N23 gene | 0.87 | 1.44 | 2.15 | 2.27 | 2.16 | 1.00 | 1.15 | 1.49 | 0.70 | 0.78 | |||||
| NM_011291 | Similar to ribosomal protein L7 | 0.93 | 1.10 | 1.18 | 1.12 | 1.19 | 1.67 | 1.61 | 3.17 | 1.09 | 0.97 | |||||
| NM_016959 | Ribosomal protein S3a (Rps3a) | 1.41 | 0.68 | 1.03 | 1.12 | 1.54 | 1.74 | 1.35 | 3.15 | 1.05 | 0.45 | |||||
| NM_009096 | Ribosomal protein S6 (Rps6) | 1.50 | 0.74 | 1.08 | 1.01 | 1.56 | 1.68 | 1.37 | 4.36 | 1.05 | 0.46 | |||||
| NM_011300 | Similar to ribosomal protein S7 | 0.94 | 1.22 | 1.03 | 1.03 | 1.00 | 1.13 | 1.55 | 2.50 | 0.78 | 0.95 | |||||
| NM_023380 | Samsn1 | 1.18 | 0.84 | 1.15 | 1.46 | 1.58 | 1.61 | 1.57 | 3.02 | 0.64 | 0.59 | |||||
| NM_026506 | Small nuclear ribonucleoprotein polypeptide G | 0.65 | 1.31 | 1.21 | 0.81 | 0.75 | 1.37 | 1.87 | 2.49 | 0.37 | 0.57 | |||||
| D13695 | ST2L protein | 0.72 | 1.51 | 1.69 | 2.05 | 0.78 | 1.08 | 1.92 | 1.04 | 0.58 | 1.19 | |||||
| AA199458 | UDP-glucose ceramide glucosyltransferase | 0.91 | 1.61 | 1.63 | 2.23 | 0.87 | 1.01 | 3.00 | 3.03 | 0.65 | 1.01 | |||||
| Dex repressed | ||||||||||||||||
| AK010947 | RIKEN clone 2510009J19 | 1.37 | 0.57 | 0.89 | 0.69 | 0.99 | 0.42 | 0.50 | 0.64 | 2.32 | 1.30 | |||||
| BC005803 | IMAGE:3154023 | 1.14 | 0.78 | 0.84 | 0.84 | 1.76 | 1.46 | 0.57 | 0.52 | 1.25 | 1.08 | |||||
| BC006617 | IMAGE:3967600 | 1.48 | 0.92 | 1.20 | 1.01 | 1.30 | 0.72 | 0.57 | 0.51 | 0.88 | 0.63 | |||||
| AF290209 | Podocalyxin | 1.12 | 1.81 | 0.57 | 0.54 | 1.13 | 1.24 | 1.60 | 1.90 | 9.41 | 7.04 | |||||
| NM_030680 | Regulator of nonsense transcripts 1 | 1.22 | 1.04 | 0.89 | 0.75 | 0.86 | 0.41 | 0.49 | 0.60 | 1.87 | 1.44 | |||||
| AK003374 | RIKEN clone 1110003L06 | 1.13 | 0.89 | 0.86 | 0.77 | 0.84 | 0.48 | 0.54 | 0.64 | 1.63 | 1.59 | |||||
| AK010608 | RIKEN clone:2410029N08 | 1.46 | 0.87 | 1.30 | 0.91 | 1.43 | 0.41 | 0.58 | 0.46 | 1.48 | 1.06 | |||||
| NM_011403 | Solute carrier family 4 (anion exchanger), Slc4a1 | 1.32 | 0.93 | 0.65 | 1.05 | 0.83 | 0.49 | 0.61 | 0.48 | 2.02 | 1.28 | |||||
| NM_031878 | Smarcd2 | 1.67 | 0.72 | 0.92 | 0.77 | 1.04 | 0.52 | 0.53 | 0.64 | 2.35 | 1.25 | |||||
| U69135 | UCP2 | 0.71 | 1.49 | 1.03 | 0.80 | 1.33 | 1.37 | 0.58 | 0.26 | 1.67 | 2.46 | |||||
. | . | 2 hours . | . | 6 hours . | . | 18 hours . | . | 24 hours . | . | dim/wt . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession no. . | Gene description . | First . | Second . | First . | Second . | First . | Second . | First . | Second . | First . | Second . | |||||
| Dex up-regulated | ||||||||||||||||
| AA020552 | IMAGE:455550 5 | 0.86 | 1.20 | 1.15 | 1.27 | 1.70 | 2.12 | 1.56 | 3.44 | 1.11 | 2.10 | |||||
| AW553415 | Mouse newborn ovary cDNA library | 0.70 | 1.60 | 0.76 | 0.80 | 1.10 | 1.70 | 2.02 | 2.06 | 0.61 | 1.03 | |||||
| NM_007475 | Acidic ribosomal phosphoprotein PO | 0.88 | 1.36 | 2.20 | 2.55 | 1.50 | 1.41 | 1.78 | 1.43 | 0.69 | 0.77 | |||||
| NM_025840 | Basic leucine zipper and W2 domains 2 | 0.77 | 1.46 | 1.02 | 1.07 | 0.69 | 1.96 | 2.05 | 2.31 | 0.48 | 0.93 | |||||
| AK003679 | RIKEN:1110014C11 | 0.91 | 1.33 | 1.13 | 1.25 | 0.94 | 1.57 | 1.95 | 2.88 | 0.76 | 0.91 | |||||
| D87747 | CXCR-4 | 1.07 | 1.66 | 1.51 | 1.29 | 1.16 | 1.18 | 1.58 | 1.90 | 0.91 | 1.15 | |||||
| NM_011722 | Dynactin 6 | 1.29 | 1.28 | 1.87 | 1.96 | 1.25 | 3.00 | 2.88 | 4.64 | 0.68 | 0.89 | |||||
| BC005660 | Eukaryotic translation elongation factor 1 alpha 1 | 0.72 | 1.36 | 1.06 | 0.92 | 0.88 | 1.46 | 2.13 | 2.27 | 0.52 | 0.89 | |||||
| BG080849 | CDNA clone H3058D01-5 | 0.78 | 1.55 | 1.92 | 1.62 | 1.01 | 0.70 | 0.74 | 0.94 | 0.62 | 0.80 | |||||
| AA655703 | IMAGE:1149396 | 0.78 | 1.49 | 2.03 | 2.03 | 0.90 | 1.17 | 1.70 | 2.42 | 0.94 | 1.17 | |||||
| BE630992 | IMAGE:3375525 | 1.25 | 0.81 | 2.10 | 2.06 | 1.90 | 1.79 | 1.26 | 1.15 | 0.88 | 0.83 | |||||
| AK020317 | RIKEN 9230106805 | 1.21 | 0.74 | 1.52 | 1.93 | 2.93 | 1.79 | 1.02 | 1.84 | 0.74 | 0.64 | |||||
| NM_021099 | kit oncogene | 1.32 | 1.02 | 1.09 | 1.23 | 2.12 | 4.71 | 2.06 | 3.61 | 0.60 | 0.53 | |||||
| BC006720 | Karyopherin (importin) alpha 2 | 0.62 | 1.42 | 0.99 | 0.86 | 0.57 | 0.93 | 1.61 | 1.55 | 0.67 | 1.56 | |||||
| K02061 | L32-4A pseudogene | 0.96 | 1.20 | 1.13 | 1.01 | 1.42 | 1.52 | 1.88 | 3.30 | 0.57 | 0.75 | |||||
| NM_010686 | Laptm5 | 1.17 | 0.77 | 1.14 | 1.36 | 2.04 | 3.88 | 1.40 | 0.98 | 2.34 | 1.37 | |||||
| NM_010708 | Lectin, galactose binding, soluble 9 | 1.08 | 0.73 | 0.80 | 0.81 | 1.49 | 2.67 | 1.56 | 3.06 | 1.20 | 0.85 | |||||
| NM_020573 | Oxysterol binding proteinlike 1A | 0.88 | 1.10 | 1.10 | 1.14 | 1.21 | 1.71 | 1.98 | 2.14 | 0.99 | 1.09 | |||||
| BC005779 | RIKEN cDNA 0610010E03 gene | 0.75 | 1.21 | 1.20 | 0.93 | 0.87 | 1.16 | 1.67 | 1.93 | 0.57 | 0.91 | |||||
| AK002923 | RIKEN cDNA 0610043A03 gene | 0.97 | 1.10 | 1.45 | 1.21 | 1.74 | 2.04 | 1.61 | 2.92 | 0.55 | 0.78 | |||||
| AK004571 | RIKEN cDNA 1200003I10 gene | 0.82 | 1.47 | 1.65 | 1.65 | 0.57 | 1.02 | 2.15 | 2.11 | 0.71 | 1.38 | |||||
| NM_023270 | RIKEN cDNA 1300002C13 gene | 1.36 | 1.04 | 1.15 | 1.42 | 1.22 | 1.27 | 1.57 | 2.30 | 0.96 | 0.67 | |||||
| AK010014 | RIKEN cDNA 2310061N23 gene | 0.87 | 1.44 | 2.15 | 2.27 | 2.16 | 1.00 | 1.15 | 1.49 | 0.70 | 0.78 | |||||
| NM_011291 | Similar to ribosomal protein L7 | 0.93 | 1.10 | 1.18 | 1.12 | 1.19 | 1.67 | 1.61 | 3.17 | 1.09 | 0.97 | |||||
| NM_016959 | Ribosomal protein S3a (Rps3a) | 1.41 | 0.68 | 1.03 | 1.12 | 1.54 | 1.74 | 1.35 | 3.15 | 1.05 | 0.45 | |||||
| NM_009096 | Ribosomal protein S6 (Rps6) | 1.50 | 0.74 | 1.08 | 1.01 | 1.56 | 1.68 | 1.37 | 4.36 | 1.05 | 0.46 | |||||
| NM_011300 | Similar to ribosomal protein S7 | 0.94 | 1.22 | 1.03 | 1.03 | 1.00 | 1.13 | 1.55 | 2.50 | 0.78 | 0.95 | |||||
| NM_023380 | Samsn1 | 1.18 | 0.84 | 1.15 | 1.46 | 1.58 | 1.61 | 1.57 | 3.02 | 0.64 | 0.59 | |||||
| NM_026506 | Small nuclear ribonucleoprotein polypeptide G | 0.65 | 1.31 | 1.21 | 0.81 | 0.75 | 1.37 | 1.87 | 2.49 | 0.37 | 0.57 | |||||
| D13695 | ST2L protein | 0.72 | 1.51 | 1.69 | 2.05 | 0.78 | 1.08 | 1.92 | 1.04 | 0.58 | 1.19 | |||||
| AA199458 | UDP-glucose ceramide glucosyltransferase | 0.91 | 1.61 | 1.63 | 2.23 | 0.87 | 1.01 | 3.00 | 3.03 | 0.65 | 1.01 | |||||
| Dex repressed | ||||||||||||||||
| AK010947 | RIKEN clone 2510009J19 | 1.37 | 0.57 | 0.89 | 0.69 | 0.99 | 0.42 | 0.50 | 0.64 | 2.32 | 1.30 | |||||
| BC005803 | IMAGE:3154023 | 1.14 | 0.78 | 0.84 | 0.84 | 1.76 | 1.46 | 0.57 | 0.52 | 1.25 | 1.08 | |||||
| BC006617 | IMAGE:3967600 | 1.48 | 0.92 | 1.20 | 1.01 | 1.30 | 0.72 | 0.57 | 0.51 | 0.88 | 0.63 | |||||
| AF290209 | Podocalyxin | 1.12 | 1.81 | 0.57 | 0.54 | 1.13 | 1.24 | 1.60 | 1.90 | 9.41 | 7.04 | |||||
| NM_030680 | Regulator of nonsense transcripts 1 | 1.22 | 1.04 | 0.89 | 0.75 | 0.86 | 0.41 | 0.49 | 0.60 | 1.87 | 1.44 | |||||
| AK003374 | RIKEN clone 1110003L06 | 1.13 | 0.89 | 0.86 | 0.77 | 0.84 | 0.48 | 0.54 | 0.64 | 1.63 | 1.59 | |||||
| AK010608 | RIKEN clone:2410029N08 | 1.46 | 0.87 | 1.30 | 0.91 | 1.43 | 0.41 | 0.58 | 0.46 | 1.48 | 1.06 | |||||
| NM_011403 | Solute carrier family 4 (anion exchanger), Slc4a1 | 1.32 | 0.93 | 0.65 | 1.05 | 0.83 | 0.49 | 0.61 | 0.48 | 2.02 | 1.28 | |||||
| NM_031878 | Smarcd2 | 1.67 | 0.72 | 0.92 | 0.77 | 1.04 | 0.52 | 0.53 | 0.64 | 2.35 | 1.25 | |||||
| U69135 | UCP2 | 0.71 | 1.49 | 1.03 | 0.80 | 1.33 | 1.37 | 0.58 | 0.26 | 1.67 | 2.46 | |||||
The ratio between Epo/SCF/Dex over Epo/SCF/ZK at any time point is given as fold change of the duplicate experiments. First indicates first hybridization; second, second hybridization.
Four genes up-regulated by Dex in the presence of Epo/SCF (RIKEN9230106B05, VDUP1, CXCR4, and BTG1; represented on the microarray 6, 4, 1, and 18 times, respectively) were analyzed in more detail. Dex-dependent up-regulation of these genes is shown in Figure 3A, plotted as 2log values of the fold changes between (1) Epo/SCF plus Dex and (2) Epo/SCF plus ZK. The reproducibility of the data is evidenced by the small standard deviation obtained in 2 independent experiments plus concomitant color switch of the hybridization probes (Figure 3A). To validate Dex-controlled expression, the amount of transcript before and after induction by Epo/SCF/Dex or Epo/SCF/ZK was determined by real-time RT-PCR in 2 independent experiments, again in duplicate determinations (Figure 3B).
Expression regulation of the putative GR-target genes RIKEN9230106B05, VDUP1, BTG1, and CXCR4 by Epo/SCF and Dex. (A) The fold changes (2log values) of gene expression detected on the hematopoietic microarrays comparing cDNA of factor-depleted I/11 erythroid progenitors treated with Epo/SCF/Dex (ESD) or Epo/SCF/ZK (ESZK) for the indicated times are shown (ESD compared with ESZK). The number of clones (n) representing each of the above genes on the hematopoietic microarray is indicated. (B) Expression of the indicated genes was subsequently determined by real-time RT-PCR. RNA levels of factor-depleted I/11 erythroid progenitors restimulated with ESD or ESZK for indicated times were compared with RNA levels of control cells factor deprived for 4 hours (NF). The fold changes of cytokine/hormone restimulation versus factor deprivation alone are shown as 2log values. (C) RNA levels of proliferating erythroid progenitors derived from either GRdim/dim fetal livers or wt littermates were compared by real-time RT-PCR. Fold changes of GRdim/dim over wt are idicated as 2log-values. The –fold regulation is shown as 2log values of fold induction or repression; for instance, a 2-fold induction and 4-fold repression yield values of 1 and –2, respectively. Mean values and standard deviations from 2 separate experiments repeated in duplicate are shown.
Expression regulation of the putative GR-target genes RIKEN9230106B05, VDUP1, BTG1, and CXCR4 by Epo/SCF and Dex. (A) The fold changes (2log values) of gene expression detected on the hematopoietic microarrays comparing cDNA of factor-depleted I/11 erythroid progenitors treated with Epo/SCF/Dex (ESD) or Epo/SCF/ZK (ESZK) for the indicated times are shown (ESD compared with ESZK). The number of clones (n) representing each of the above genes on the hematopoietic microarray is indicated. (B) Expression of the indicated genes was subsequently determined by real-time RT-PCR. RNA levels of factor-depleted I/11 erythroid progenitors restimulated with ESD or ESZK for indicated times were compared with RNA levels of control cells factor deprived for 4 hours (NF). The fold changes of cytokine/hormone restimulation versus factor deprivation alone are shown as 2log values. (C) RNA levels of proliferating erythroid progenitors derived from either GRdim/dim fetal livers or wt littermates were compared by real-time RT-PCR. Fold changes of GRdim/dim over wt are idicated as 2log-values. The –fold regulation is shown as 2log values of fold induction or repression; for instance, a 2-fold induction and 4-fold repression yield values of 1 and –2, respectively. Mean values and standard deviations from 2 separate experiments repeated in duplicate are shown.
Except for minor differences, real-time RT-PCR confirmed the data obtained by the cDNA microarray analysis. In fact, the fold-change values were mostly larger when measured by real-time RT-PCR. Following factor deprivation, all 4 genes were down-regulated by Epo/SCF in absence of Dex (ie, in ZK) while coactivation of the GR by Dex diminished this down-modulation or, in case of RIKEN9230106B05, even resulted in up-regulation relative to the factor-deprived control.
Expression of Dex-modulated Epo/SCF–target genes is altered in erythroid progenitors derived from GRdim/dim mice
To validate our data by an independent approach, we analyzed the expression of putative GR-target genes in erythroid progenitors derived from fetal livers of GRdim/dim mice. These mice carry a glucocorticoid receptor mutated in the dimerization domain, rendering it unable to contribute to gene transactivation.12 Erythroid progenitors from fetal livers of GRdim/dim embryos or wt littermates were expanded for 7 days. RNA was directly isolated from proliferating cells without factor depletion or stimulation. Microarray analysis showed that all genes up-regulated in the presence of Dex were expressed at lower levels in the GRdim/dim erythroid progenitors compared with their wt counterpart (Tables 2 and 3). On the other hand, beta-globin, which is strongly down-regulated by the addition of Dex, was up-regulated in the GRdim/dim erythroid progenitors as compared with wt cells (Tables 2 and 3). Furthermore, real-time RT-PCR analysis confirmed that the selected genes RIKEN9230106B05, VDUP1, BTG1, and CXCR4, found to be up-regulated in the presence of Dex plus Epo/SCF, were down-regulated in GRdim/dim when compared with wt erythroid progenitors (Figure 3C).
Cooperation of Epo, SCF, and Dex in regulation of the erythroid gene expression program
To unravel how the individual factors Epo, SCF, and Dex regulate gene expression and how they cooperate in various combinations, we obviously had to analyze a larger number of potential target genes than were present on our hematopoietic microarray. For that purpose, we used cDNA microarrays containing approximately 17 000 different EST sequences (represented once or maximally twice [17K]; details available at: http://www.imp.univie.ac.at/2003/Beug/erythroblast_renewal/. Accessed on July 23, 2003). The I/11 erythroid progenitors were factor deprived for 4 hours and treated for 2 hours with Epo, Dex, or SCF alone, as well as with combinations of Epo/SCF (ES), Epo/SCF and Dex (ESD) and Epo/SCF and ZK (ESZK). The cDNA from these cells was labeled with Cy5 and hybridized to both the hematopoietic and the 17K cDNA microarray. In parallel, Cy3-labeled cDNA from erythroid progenitors factor deprived for 6 hours was included as a control. First, we defined prefiltering conditions by determining to what extent the different factors affected overall gene expression. Upon stimulation by Epo or SCF, 1.29% and 1.24% of the genes on the 17K cDNA microarray were more than 1.75-fold up- or down-regulated (Table 4). Yet a similar number of Dex-regulated genes was obtained only when genes up- or down-regulated by a ratio exceeding 1.5 were included (Table 4). Therefore, we set the prefiltering threshold to 1.5 (fold induction or repression) for Dex targets and to 1.75 for Epo and SCF targets, being aware of the risk of incorporating a larger number of nonrelevant and false-positive genes in the resulting gene lists.
Epo, SCF, and Dex in the regulation of the erythroid gene expression program
. | Down-regulation . | . | . | Up-regulation . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | Fold change less than -2 . | Fold change -2.0 to -1.75 . | Fold change -1.75 to -1.50 . | Fold change 1.50 to 1.75 . | Fold change 1.75 to 2.0 . | Fold change 2.0 to 2.5 . | Fold change more than 2.5 . | |||||
| Epo, ‰ | 0.9 | 4.8 | 10.2 | 12.7 | 4.2 | 1.8 | 1.2 | |||||
| Dex, ‰ | 0.2 | 1.1 | 3.6 | 1.8 | 0.9 | 0.4 | 0.1 | |||||
| SCF, ‰ | 1.2 | 3.9 | 8.9 | 14.1 | 4.1 | 2.1 | 1.1 | |||||
| Ratio ESD/ESZK, ‰ | 0.2 | 1.2 | 4.1 | 5.9 | 1.8 | 0.8 | 0.3 | |||||
. | Down-regulation . | . | . | Up-regulation . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | Fold change less than -2 . | Fold change -2.0 to -1.75 . | Fold change -1.75 to -1.50 . | Fold change 1.50 to 1.75 . | Fold change 1.75 to 2.0 . | Fold change 2.0 to 2.5 . | Fold change more than 2.5 . | |||||
| Epo, ‰ | 0.9 | 4.8 | 10.2 | 12.7 | 4.2 | 1.8 | 1.2 | |||||
| Dex, ‰ | 0.2 | 1.1 | 3.6 | 1.8 | 0.9 | 0.4 | 0.1 | |||||
| SCF, ‰ | 1.2 | 3.9 | 8.9 | 14.1 | 4.1 | 2.1 | 1.1 | |||||
| Ratio ESD/ESZK, ‰ | 0.2 | 1.2 | 4.1 | 5.9 | 1.8 | 0.8 | 0.3 | |||||
Erythroid progenitors were factor deprived and restimulated with Epo, Dex, SCF, Epo/SCF, Epo/SCF/Dex, or Epo/SCF/ZK. Cy5-labeled cDNA synthesized from mRNA of restimulated cells was hybridized in parallel with Cy3-labeled cDNA synthesized from factor-deprived cells to the hematopoietic and the 17K EST arrays. Values in table represent the fraction (given in permillage [‰]) of cDNAs present on the microarrays that shows a certain fold change of expression in stimulated versus unstimulated cells.
Applying these thresholds to the combined results of the 9K hematopoietic microarray and the 17K microarray, we performed cluster analysis by means of the GeneSpring program. Epo and SCF induced similar patterns of gene expression changes: genes up-regulated by Epo were never down-regulated by SCF or vice versa (Figure 4A-B). In addition, expression changes induced by Epo but not by SCF were always present in the Epo/SCF (ES) combination, suggesting that effects of the differentiation-inducer Epo are not neutralized by the renewal factor SCF. While a small number of genes were differentially regulated by Dex plus Epo/SCF (Figure 4A), Dex alone caused a completely different pattern of gene regulation (Figure 4B).
Cooperation of Epo, SCF, and Dex in the regulation of the erythroid gene expression program. Erythroid progenitors were factor deprived and restimulated with Epo, Dex, SCF, Epo/SCF, Epo/SCF/Dex, or Epo/SCF/ZK. Cy5-labeled cDNA synthesized from mRNA of restimulated cells was hybridized in parallel with Cy3-labeled cDNA synthesized from factor-deprived cells to the hematopoietic and the 17K EST arrays. (A) Hierarchic clustering (obtained with the use of Pearson correlation function) of 1200 clones that showed an alteration in gene expression greater than 1.75-fold up- or down-regulation for Epo or SCF, or greater than 1.5-fold up- or down-regulation for Dex. The color scale representing –fold up-regulation (red) or down-regulation (blue) of gene expression is shown on the left. (B) Bar graphs of clones selected for up- or down-regulation by Epo, Dex, or SCF showing the gene regulation by all other factor combinations versus factor deprivation. Fold changes are indicated on the vertical axes as 2log values. Left column shows down-regulated genes; right column, up-regulated genes. The numbers of preselected, regulated genes/ESTs are indicated in the different panels.
Cooperation of Epo, SCF, and Dex in the regulation of the erythroid gene expression program. Erythroid progenitors were factor deprived and restimulated with Epo, Dex, SCF, Epo/SCF, Epo/SCF/Dex, or Epo/SCF/ZK. Cy5-labeled cDNA synthesized from mRNA of restimulated cells was hybridized in parallel with Cy3-labeled cDNA synthesized from factor-deprived cells to the hematopoietic and the 17K EST arrays. (A) Hierarchic clustering (obtained with the use of Pearson correlation function) of 1200 clones that showed an alteration in gene expression greater than 1.75-fold up- or down-regulation for Epo or SCF, or greater than 1.5-fold up- or down-regulation for Dex. The color scale representing –fold up-regulation (red) or down-regulation (blue) of gene expression is shown on the left. (B) Bar graphs of clones selected for up- or down-regulation by Epo, Dex, or SCF showing the gene regulation by all other factor combinations versus factor deprivation. Fold changes are indicated on the vertical axes as 2log values. Left column shows down-regulated genes; right column, up-regulated genes. The numbers of preselected, regulated genes/ESTs are indicated in the different panels.
According to their regulation in response to either single agents (ie, Epo, SCF, Dex), Epo/SCF, or Epo/SCF/Dex in comparison with Epo/SCF/ZK, genes were clustered into various profile groups (Table 5). Profile group 1 shows that 54 genes were up-regulated by Epo (Table 5) (group 1A), whereas 36 genes were down-regulated by Epo (group 1B). Similarly, with SCF 63 genes were up-regulated (group 2A) and 58 genes down-regulated (2B). In profile group 3, we distinguished between genes regulated by Dex alone (3A has 21 genes up; 3B, 33 genes down), or by Dex alone and by Dex in combination with Epo and SCF (3A′, 18 up; 3B′, 2 down), or only by Dex in combination with Epo and SCF (3A″, 106 up; 3B″, 72 down). Although genes coregulated by Dex and Epo/SCF were a minority, we considered them to be the most interesting, since genes subject to this coregulation seemed to be the best candidates to give insight into the mechanism of renewal. Table 6, therefore, lists the genes showing up- or down-regulation by Dex, Epo/Dex (profile group 5), SCF/Dex (profile group 6), or Epo/SCF/Dex (profile group 7).
Cooperative signaling among Dex, Epo, and SCF: existence of multiple profile groups
Profile Group . | Dex . | ESD/ESZK . | Dex (± Epo, SCF) . | Epo . | SCF . | No. genes . |
|---|---|---|---|---|---|---|
| 1A | — | — | — | Up | — | 54 |
| 1B | — | — | — | Down | — | 36 |
| 2A | — | — | — | — | Up | 63 |
| 2B | — | — | — | — | Down | 58 |
| 3A | Up | — | Up | — | — | 21 |
| 3A′ | Up | Up | Up | — | — | 18 |
| 3A″ | — | Up | Up | — | — | 106 |
| 3B | Down | — | Down | — | — | 33 |
| 3B′ | Down | Down | Down | — | — | 2 |
| 3B″ | — | Down | Down | — | — | 72 |
| 4A1 | — | — | — | Up | Up | 51 |
| 4B1 | — | — | — | Down | Down | 16 |
| 5A1 | NS | NS | Up | Up | — | 5 |
| 5A2 | NS | NS | Up | Down | — | 2 |
| 5B1 | NS | NS | Down | Up | — | 3 |
| 6A1 | NS | NS | Up | — | Up | 3 |
| 6A2 | NS | NS | Up | — | Down | 4 |
| 6B1 | NS | NS | Down | — | Up | 1 |
| 7A1 | NS | NS | Up | Up | Up | 3 |
| 7A3 | NS | NS | Up | Down | Down | 5 |
| 7B1 | NS | NS | Down | Up | Up | 3 |
Profile Group . | Dex . | ESD/ESZK . | Dex (± Epo, SCF) . | Epo . | SCF . | No. genes . |
|---|---|---|---|---|---|---|
| 1A | — | — | — | Up | — | 54 |
| 1B | — | — | — | Down | — | 36 |
| 2A | — | — | — | — | Up | 63 |
| 2B | — | — | — | — | Down | 58 |
| 3A | Up | — | Up | — | — | 21 |
| 3A′ | Up | Up | Up | — | — | 18 |
| 3A″ | — | Up | Up | — | — | 106 |
| 3B | Down | — | Down | — | — | 33 |
| 3B′ | Down | Down | Down | — | — | 2 |
| 3B″ | — | Down | Down | — | — | 72 |
| 4A1 | — | — | — | Up | Up | 51 |
| 4B1 | — | — | — | Down | Down | 16 |
| 5A1 | NS | NS | Up | Up | — | 5 |
| 5A2 | NS | NS | Up | Down | — | 2 |
| 5B1 | NS | NS | Down | Up | — | 3 |
| 6A1 | NS | NS | Up | — | Up | 3 |
| 6A2 | NS | NS | Up | — | Down | 4 |
| 6B1 | NS | NS | Down | — | Up | 1 |
| 7A1 | NS | NS | Up | Up | Up | 3 |
| 7A3 | NS | NS | Up | Down | Down | 5 |
| 7B1 | NS | NS | Down | Up | Up | 3 |
— indicates no regulation; NS, not specified.
Examples of genes coregulated by Epo, Dex, and SCF in expansion of erythroid progenitors (from the indicated profile groups shown inTable 5)
Profile group, accession no. . | Gene description . | Epo . | SCF . | Dex . | ES . | ESD . | ESZK . | ESD/ESZK . |
|---|---|---|---|---|---|---|---|---|
| 3A ESD/ESZK up, Dex up | ||||||||
| AI853706 | Rho guanine nucleotide exchange factor (GEF)3 | 1.10 | 0.91 | 2.25 | 1.30 | 3.49 | 1.24 | 2.80 |
| AI847541 | zinc finger protein 61 | 0.69 | 0.77 | 2.15 | 0.68 | 1.07 | 0.58 | 1.85 |
| AI845829 | Asialoglycoprotein receptor 1 | 1.26 | 1.44 | 2.10 | 1.40 | 2.02 | 1.23 | 1.65 |
| AI415662 | Mm.23097 | 1.02 | 0.95 | 2.09 | 1.14 | 2.07 | 0.91 | 2.28 |
| AI840878 | RIKEN cDNA 2300002F06 gene | 1.04 | 1.26 | 2.07 | 1.16 | 2.50 | 1.08 | 2.32 |
| AI840515 | EST | 0.60 | 0.76 | 2.06 | 0.63 | 1.14 | 0.72 | 1.59 |
| AI844317 | Mm.41137 | 1.23 | 0.64 | 1.97 | 1.01 | 1.45 | 0.82 | 1.76 |
| AI854349 | Serum/glucocorticoid regulated kinase | 0.84 | 0.87 | 1.95 | 0.63 | 2.56 | 0.79 | 3.24 |
| AI465375 | RIKEN cDNA 2510025F08 gene | 0.99 | 0.89 | 1.88 | 0.89 | 1.27 | 0.78 | 1.62 |
| AI841954 | Mm.38924 | 0.90 | 0.68 | 1.86 | 0.76 | 1.18 | 0.58 | 2.04 |
| AI837598 | RIKEN cDNA 1300007C21 gene | 0.91 | 0.62 | 1.82 | 0.66 | 0.96 | 0.50 | 1.91 |
| AI846411 | Mm.218636 | 1.40 | 0.79 | 1.73 | 1.50 | 2.45 | 1.11 | 2.20 |
| AI447893 | Mm.30874 | 0.98 | 1.12 | 1.61 | 0.95 | 2.03 | 1.10 | 1.85 |
| AI846077 | Sec7 and coiled/coil domains 3 | 0.78 | 0.92 | 1.58 | 0.60 | 0.97 | 0.52 | 1.85 |
| AI850091 | CD59a antigen | 1.36 | 0.82 | 1.57 | 1.04 | 1.75 | 1.10 | 1.58 |
| AI326787 | lipoprotein lipase | 0.98 | 1.12 | 1.52 | 1.08 | 1.68 | 0.94 | 1.78 |
| AA199458 | IMAGE:638536 | 0.96 | 0.75 | 1.65 | 0.71 | 0.82 | 0.51 | 1.63 |
| D87747 | CXCR-4 | 0.69 | 0.86 | 1.93 | 0.46 | 0.82 | 0.44 | 1.89 |
| 3B ESD/ESZK down, Dex down | ||||||||
| AI853422 | EST cluster Mm.259384 | 1.39 | 0.95 | 0.64 | 1.65 | 1.27 | 2.05 | 0.62 |
| AI413887 | EST cluster Mm.22863 | 0.71 | 1.10 | 0.67 | 0.62 | 0.60 | 0.99 | 0.61 |
| 5A1 Epo up, Dex up | ||||||||
| AI528522 | Protein kinase C, theta | 2.52 | 0.62 | 1.11 | 1.91 | 2.92 | 1.53 | 1.91 |
| AI385650 | Beta-galactosidase alpha-2,3-sialytransferase | 2.38-2.63 | 1.22-1.07 | 1.32-0.99 | 3.05-3.07 | 3.82-3.86 | 1.88-1.90 | 2.03-2.03 |
| AI323712 | ATPase, beta 56/58 kDa, isoform 2 | 1.75 | 0.92 | 1.00 | 2.06 | 2.55 | 1.21 | 2.11 |
| AI838311 | Matrix metalloproteinase 2 | 2.22-2.14 | 1.18-1.08 | 0.96-0.96 | 2.07-2.43 | 3.70-7.52 | 1.46-1.80 | 2.54-4.19 |
| AI834916 | EST, cluster Mm.24036 | 2.35 | 1.04 | 1.38 | 1.98 | 3.24 | 1.89 | 1.72 |
| 5B1 Epo up, Dex down | ||||||||
| AI850575 | Phospholipase C, epsilon 2 | 1.93-2.16 | 1.11-0.99 | 1.27-1.47 | 2.31-2.27 | 1.66-1.61 | 2.18-2.53 | 0.76-0.64 |
| AI385595 | Cytokine inducible SH2-containing protein | 4.49 | 1.15 | 1.03 | 2.87 | 2.15 | 4.44 | 0.48 |
| AI854137 | EST AA408729 | 2.19 | 1.39 | 0.96 | 2.87 | 1.85 | 3.04 | 0.61 |
| 5A2 Epo down, Dex up | ||||||||
| AI843911 | Glucocorticoid-induced leucine zipper | 0.46-0.53 | 0.72-0.76 | 2.93-2.46 | 0.42-0.52 | 1.06-1.08 | 0.82-0.53 | 1.72-0.77 |
| AI450157 | Chloride intracellular channel 1 | 0.52 | 0.74 | 0.88 | 0.49 | 0.99 | 0.63 | 1.57 |
| 6A1 SCF up, Dex up | ||||||||
| AI847904 | SEC22 vesicle trafficking proteinlike 1 | 1.38 | 1.85 | 1.09 | 0.97 | 2.02 | 0.78 | 2.60 |
| AI836926 | Melanoma X-actin | 1.73 | 2.00 | 1.19 | 1.80 | 2.15 | 1.12 | 1.92 |
| AI893440 | RAB4A, member RAS oncogene family | 1.04 | 1.81 | 0.98 | 0.67 | 0.93 | 0.47 | 1.98 |
| 6B1 SCF up, Dex down | ||||||||
| AI836530 | RIKEN cDNA 3110038L01 gene | 1.48 | 1.99 | 0.94 | 2.17 | 1.38 | 2.21 | 0.62 |
| 6A2 SCF down, Dex up | ||||||||
| AI851500 | RIKEN cDNA 1110046J11 gene | 0.70 | 0.52 | 1.25 | 0.44 | 0.53 | 0.33 | 1.58 |
| AI849362 | EST, clusterMm.41089 | 0.91 | 0.59 | 1.02 | 0.40 | 0.75 | 0.41 | 1.82 |
| AI848411 | B-cell translocation gene 1 | 0.71-0.88 | 0.48-0.50 | 1.68-1.45 | 0.45-0.60 | 1.07-1.07 | 0.41-0.57 | 2.63-1.86 |
| AI846731 | RIKEN cDNA 2700017106 | 1.08 | 0.41 | 1.83 | 0.90 | 1.76 | 0.75 | 2.33 |
| 7A1 ES up, Dex up | ||||||||
| A1844751 | Translation releasing factor eRF-1 | 1.91 | 2.06 | 0.93 | 2.92 | 1.83 | 0.94 | 1.94 |
| AI850110 | WD repeat domain 1 | 1.83 | 2.51 | 1.04 | 2.35 | 2.08 | 0.94 | 2.20 |
| AI323629 | Low-density lipoprotein receptor | 1.82 | 2.56 | 1.00 | 2.42 | 2.35 | 1.44 | 1.63 |
| 7B1 ES up, Dex down | ||||||||
| AI323806 | Cyclin-dependent kinase inhibitor 1A(P21) | 1.91 | 1.12 | 0.76 | 3.13 | 2.00 | 4.22 | 0.47 |
| AI840156 | RIKEN cDNA 2310076D10 gene | 1.78 | 2.54 | 1.10 | 2.87 | 2.00 | 3.19 | 0.63 |
| 7A3 ES down, Dex up | ||||||||
| AI428195 | RIKEN cDNA 2410024N18 gene | 0.55 | 0.44 | 1.54 | 0.27 | 0.50 | 0.32 | 1.56 |
| AB022209 | Ribonucleoprotein F | 0.61-0.65 | 0.51-0.48 | 1.75-1.60 | 0.49-0.52 | 0.56-0.63 | 0.45-0.52 | 1.24-1.21 |
| AI846040 | RIKEN cDNA 1500004A08 gene | 0.56 | 0.39 | 1.43 | 0.25 | 0.70 | 0.37 | 1.90 |
| D87747 | CXCR-4 | 0.69 | 0.86 | 1.93 | 0.46 | 0.82 | 0.44 | 1.89 |
Profile group, accession no. . | Gene description . | Epo . | SCF . | Dex . | ES . | ESD . | ESZK . | ESD/ESZK . |
|---|---|---|---|---|---|---|---|---|
| 3A ESD/ESZK up, Dex up | ||||||||
| AI853706 | Rho guanine nucleotide exchange factor (GEF)3 | 1.10 | 0.91 | 2.25 | 1.30 | 3.49 | 1.24 | 2.80 |
| AI847541 | zinc finger protein 61 | 0.69 | 0.77 | 2.15 | 0.68 | 1.07 | 0.58 | 1.85 |
| AI845829 | Asialoglycoprotein receptor 1 | 1.26 | 1.44 | 2.10 | 1.40 | 2.02 | 1.23 | 1.65 |
| AI415662 | Mm.23097 | 1.02 | 0.95 | 2.09 | 1.14 | 2.07 | 0.91 | 2.28 |
| AI840878 | RIKEN cDNA 2300002F06 gene | 1.04 | 1.26 | 2.07 | 1.16 | 2.50 | 1.08 | 2.32 |
| AI840515 | EST | 0.60 | 0.76 | 2.06 | 0.63 | 1.14 | 0.72 | 1.59 |
| AI844317 | Mm.41137 | 1.23 | 0.64 | 1.97 | 1.01 | 1.45 | 0.82 | 1.76 |
| AI854349 | Serum/glucocorticoid regulated kinase | 0.84 | 0.87 | 1.95 | 0.63 | 2.56 | 0.79 | 3.24 |
| AI465375 | RIKEN cDNA 2510025F08 gene | 0.99 | 0.89 | 1.88 | 0.89 | 1.27 | 0.78 | 1.62 |
| AI841954 | Mm.38924 | 0.90 | 0.68 | 1.86 | 0.76 | 1.18 | 0.58 | 2.04 |
| AI837598 | RIKEN cDNA 1300007C21 gene | 0.91 | 0.62 | 1.82 | 0.66 | 0.96 | 0.50 | 1.91 |
| AI846411 | Mm.218636 | 1.40 | 0.79 | 1.73 | 1.50 | 2.45 | 1.11 | 2.20 |
| AI447893 | Mm.30874 | 0.98 | 1.12 | 1.61 | 0.95 | 2.03 | 1.10 | 1.85 |
| AI846077 | Sec7 and coiled/coil domains 3 | 0.78 | 0.92 | 1.58 | 0.60 | 0.97 | 0.52 | 1.85 |
| AI850091 | CD59a antigen | 1.36 | 0.82 | 1.57 | 1.04 | 1.75 | 1.10 | 1.58 |
| AI326787 | lipoprotein lipase | 0.98 | 1.12 | 1.52 | 1.08 | 1.68 | 0.94 | 1.78 |
| AA199458 | IMAGE:638536 | 0.96 | 0.75 | 1.65 | 0.71 | 0.82 | 0.51 | 1.63 |
| D87747 | CXCR-4 | 0.69 | 0.86 | 1.93 | 0.46 | 0.82 | 0.44 | 1.89 |
| 3B ESD/ESZK down, Dex down | ||||||||
| AI853422 | EST cluster Mm.259384 | 1.39 | 0.95 | 0.64 | 1.65 | 1.27 | 2.05 | 0.62 |
| AI413887 | EST cluster Mm.22863 | 0.71 | 1.10 | 0.67 | 0.62 | 0.60 | 0.99 | 0.61 |
| 5A1 Epo up, Dex up | ||||||||
| AI528522 | Protein kinase C, theta | 2.52 | 0.62 | 1.11 | 1.91 | 2.92 | 1.53 | 1.91 |
| AI385650 | Beta-galactosidase alpha-2,3-sialytransferase | 2.38-2.63 | 1.22-1.07 | 1.32-0.99 | 3.05-3.07 | 3.82-3.86 | 1.88-1.90 | 2.03-2.03 |
| AI323712 | ATPase, beta 56/58 kDa, isoform 2 | 1.75 | 0.92 | 1.00 | 2.06 | 2.55 | 1.21 | 2.11 |
| AI838311 | Matrix metalloproteinase 2 | 2.22-2.14 | 1.18-1.08 | 0.96-0.96 | 2.07-2.43 | 3.70-7.52 | 1.46-1.80 | 2.54-4.19 |
| AI834916 | EST, cluster Mm.24036 | 2.35 | 1.04 | 1.38 | 1.98 | 3.24 | 1.89 | 1.72 |
| 5B1 Epo up, Dex down | ||||||||
| AI850575 | Phospholipase C, epsilon 2 | 1.93-2.16 | 1.11-0.99 | 1.27-1.47 | 2.31-2.27 | 1.66-1.61 | 2.18-2.53 | 0.76-0.64 |
| AI385595 | Cytokine inducible SH2-containing protein | 4.49 | 1.15 | 1.03 | 2.87 | 2.15 | 4.44 | 0.48 |
| AI854137 | EST AA408729 | 2.19 | 1.39 | 0.96 | 2.87 | 1.85 | 3.04 | 0.61 |
| 5A2 Epo down, Dex up | ||||||||
| AI843911 | Glucocorticoid-induced leucine zipper | 0.46-0.53 | 0.72-0.76 | 2.93-2.46 | 0.42-0.52 | 1.06-1.08 | 0.82-0.53 | 1.72-0.77 |
| AI450157 | Chloride intracellular channel 1 | 0.52 | 0.74 | 0.88 | 0.49 | 0.99 | 0.63 | 1.57 |
| 6A1 SCF up, Dex up | ||||||||
| AI847904 | SEC22 vesicle trafficking proteinlike 1 | 1.38 | 1.85 | 1.09 | 0.97 | 2.02 | 0.78 | 2.60 |
| AI836926 | Melanoma X-actin | 1.73 | 2.00 | 1.19 | 1.80 | 2.15 | 1.12 | 1.92 |
| AI893440 | RAB4A, member RAS oncogene family | 1.04 | 1.81 | 0.98 | 0.67 | 0.93 | 0.47 | 1.98 |
| 6B1 SCF up, Dex down | ||||||||
| AI836530 | RIKEN cDNA 3110038L01 gene | 1.48 | 1.99 | 0.94 | 2.17 | 1.38 | 2.21 | 0.62 |
| 6A2 SCF down, Dex up | ||||||||
| AI851500 | RIKEN cDNA 1110046J11 gene | 0.70 | 0.52 | 1.25 | 0.44 | 0.53 | 0.33 | 1.58 |
| AI849362 | EST, clusterMm.41089 | 0.91 | 0.59 | 1.02 | 0.40 | 0.75 | 0.41 | 1.82 |
| AI848411 | B-cell translocation gene 1 | 0.71-0.88 | 0.48-0.50 | 1.68-1.45 | 0.45-0.60 | 1.07-1.07 | 0.41-0.57 | 2.63-1.86 |
| AI846731 | RIKEN cDNA 2700017106 | 1.08 | 0.41 | 1.83 | 0.90 | 1.76 | 0.75 | 2.33 |
| 7A1 ES up, Dex up | ||||||||
| A1844751 | Translation releasing factor eRF-1 | 1.91 | 2.06 | 0.93 | 2.92 | 1.83 | 0.94 | 1.94 |
| AI850110 | WD repeat domain 1 | 1.83 | 2.51 | 1.04 | 2.35 | 2.08 | 0.94 | 2.20 |
| AI323629 | Low-density lipoprotein receptor | 1.82 | 2.56 | 1.00 | 2.42 | 2.35 | 1.44 | 1.63 |
| 7B1 ES up, Dex down | ||||||||
| AI323806 | Cyclin-dependent kinase inhibitor 1A(P21) | 1.91 | 1.12 | 0.76 | 3.13 | 2.00 | 4.22 | 0.47 |
| AI840156 | RIKEN cDNA 2310076D10 gene | 1.78 | 2.54 | 1.10 | 2.87 | 2.00 | 3.19 | 0.63 |
| 7A3 ES down, Dex up | ||||||||
| AI428195 | RIKEN cDNA 2410024N18 gene | 0.55 | 0.44 | 1.54 | 0.27 | 0.50 | 0.32 | 1.56 |
| AB022209 | Ribonucleoprotein F | 0.61-0.65 | 0.51-0.48 | 1.75-1.60 | 0.49-0.52 | 0.56-0.63 | 0.45-0.52 | 1.24-1.21 |
| AI846040 | RIKEN cDNA 1500004A08 gene | 0.56 | 0.39 | 1.43 | 0.25 | 0.70 | 0.37 | 1.90 |
| D87747 | CXCR-4 | 0.69 | 0.86 | 1.93 | 0.46 | 0.82 | 0.44 | 1.89 |
Since the values of up- or down-regulation of the genes represented in Table 6 were often marginal, validation of these values was required. For 2 genes taken as examples, the glucocorticoid-induced leucine zipper (GILZ) and VDUP1, the results from microarray analysis could be clearly verified by real-time RT-PCR (Figure 5A). Transcripts for GILZ (represented 2 times on microarray) were up-regulated by Dex alone and down-regulated by Epo and Epo/SCF, while coregulation by Epo, SCF, and Dex abolished Epo-dependent down-regulation, as also confirmed by real-time RT-PCR. Similarly VDUP1 (4 times on microarray) was suppressed in response to Epo/SCF, while Dex reduced this down-regulation although Dex by itself had no effect (Figure 5A). Another pattern of gene regulation, again clearly confirmed by real-time RT-PCR, is represented by MMP-2, present 2 times on the microarrays. MMP-2 was up-regulated by Epo and Epo/SCF, but only slightly by SCF alone. Dex enhanced the up-regulation by Epo and Epo/SCF, but again, had little effect on its own (Figure 5A). For MMP-2 and GILZ, the up-regulation by Dex in the presence of Epo/SCF (ie, difference between ESD and ESZK) could also be confirmed by real-time RT-PCR in samples of cells that had been exposed to Epo/SCF/Dex or Epo/SCF/ZK for longer time periods (4, 6, or 18 hours) (Figure 5B). The modulation of Epo/SCF gene regulation by Dex was detectable at all times, but showed different kinetics for the 2 genes. In line with the up-regulation of both genes by Dex in the presence of Epo/SCF, the expression levels in the GR transactivation-deficient GRdim/dim cells were strongly reduced in comparison with the wt cells (Figure 5C). These comparisons between microarray data and verification data obtained by real-time RT-PCR, yielding close similarity of regulation patterns in all examples analyzed, clearly strengthen the significance of the results shown in Tables 5 and 6: that Dex modulates the effects of Epo, SCF, or Epo/SCF differently in a large number of genes.
Expression profiles of the putative GR-target genes VDUP1, MMP-2, and GILZ in response to Epo, SCF, Dex, and combinations thereof. (A, microarray) Mean fold changes (2log values) of gene expression detected on the microarrays for the genes indicated, comparing cDNA of factor-depleted I/11 erythroid progenitors with cells subsequently treated with Epo (E), SCF (S), Dex (D), Epo/SCF (ES), Epo/SCF/Dex (ESD), or Epo/SCF/ZK (ESZK) for 2 hours. The number of clones representing each of the genes on the microarrays is indicated (n). (A, RT-PCR) Expression of the indicated genes was subsequently determined by real-time RT-PCR. RNA levels of factor-depleted and restimulated cells (as indicated) were compared with RNA levels of factor-deprived cells. Fold changes of restimulation versus factor deprivation are indicated as 2log values. (B) RNA levels of factor-depleted I/11 erythroid progenitors restimulated with Epo/SCF/Dex (ESD) or Epo/SCF/ZK (ESZK) for the indicated time periods were compared with RNA levels of cells factor deprived for 4 hours (NF). Fold changes of restimulation versus factor deprivation are indicated as 2log values. (C) RNA levels of proliferating erythroid progenitors derived from either GRdim/dim fetal livers or wt littermates (used either as primary cells [prim] or after immortalization in culture [imm]) were determined by real-time RT-PCR. Fold changes of GRdim/dim over wt are indicated as 2log values. Mean values and standard deviations from 2 separate experiments repeated in duplicate are shown.
Expression profiles of the putative GR-target genes VDUP1, MMP-2, and GILZ in response to Epo, SCF, Dex, and combinations thereof. (A, microarray) Mean fold changes (2log values) of gene expression detected on the microarrays for the genes indicated, comparing cDNA of factor-depleted I/11 erythroid progenitors with cells subsequently treated with Epo (E), SCF (S), Dex (D), Epo/SCF (ES), Epo/SCF/Dex (ESD), or Epo/SCF/ZK (ESZK) for 2 hours. The number of clones representing each of the genes on the microarrays is indicated (n). (A, RT-PCR) Expression of the indicated genes was subsequently determined by real-time RT-PCR. RNA levels of factor-depleted and restimulated cells (as indicated) were compared with RNA levels of factor-deprived cells. Fold changes of restimulation versus factor deprivation are indicated as 2log values. (B) RNA levels of factor-depleted I/11 erythroid progenitors restimulated with Epo/SCF/Dex (ESD) or Epo/SCF/ZK (ESZK) for the indicated time periods were compared with RNA levels of cells factor deprived for 4 hours (NF). Fold changes of restimulation versus factor deprivation are indicated as 2log values. (C) RNA levels of proliferating erythroid progenitors derived from either GRdim/dim fetal livers or wt littermates (used either as primary cells [prim] or after immortalization in culture [imm]) were determined by real-time RT-PCR. Fold changes of GRdim/dim over wt are indicated as 2log values. Mean values and standard deviations from 2 separate experiments repeated in duplicate are shown.
Discussion
While Epo is required for both expansion and differentiation of erythroid progenitors, the synergistic action of SCF and Dex is required to induce progenitor expansion and arrest differentiation. To investigate the molecular processes behind the cooperation of these factors, we analyzed how Epo, SCF, and Dex signaling affects target gene expression, applying mRNA expression profiling. Since an effect of Dex on expansion and differentiation is obvious only in the presence of SCF, we particularly focused on the cross-talk between the GR and the signaling pathways emanating from EpoR and c-Kit.
Significance of microarray profiling
The interpretation of expression-profiling results is subject to various considerations regarding the significance of the data. By choosing thresholds of 1.75-fold regulation for Epo and SCF and 1.5-fold regulation for Dex, we were able to consider the approximately 1% most regulated genes. The redundancy of transcripts on the 9K hematopoietic microarray provided a neat control for the high reproducibility of the data (compare Figures 3A and 5A). Moreover, validation of data by real-time RT-PCR invariably showed an equal or even more prominent regulation compared with the microarray data, and the expression changes mostly increased upon extended stimulation. Nevertheless, we cannot exclude that the compiled lists may contain false-positive data. Another point of consideration is the significance, which is difficult to predict. For some gene products, an up-regulation of 1.4-fold may be highly significant as the resulting increase in available protein could be many-fold if, for example, a negatively regulating binding protein is outbalanced. For other genes, a 3-fold increase in mRNA may be irrelevant when protein expression and processing are regulated. Although these considerations are relevant to every expression-profiling experiment, our main aim was to determine whether cooperation of SCF and Dex also results in the coregulation of certain genes and whether SCF-induced delay of differentiation would involve inhibition of Epo-induced, differentiation-associated genes. Therefore, we considered the 1% most strongly regulated genes for every factor, accepting that not all those genes may be of direct physiologic relevance.
Importantly, all microarray screens were performed with the use of p53-deficient erythroid progenitors. Some of the target genes, like the SCF–down-regulated gene BTG2 or the Epo–up-regulated gene p21WAF are also known as p53-target genes. However, comparisons between p53-deficient and p53 wt cultures showed that cytokine regulation of these genes is independent of p53 (data not shown), suggesting that p53-mediated up-regulation following, for example, DNA damage occurs independently of cytokine stimulation. It may be different for genes regulated by glucocorticoids, as p53 and glucocorticoids may act antagonistically in erythroid progenitors.24 However, all GR-target genes from this screen analyzed in more detail were validated by PCR on RNA isolated from primary, p53 wt cells. This does not rule out that the lists of nonvalidated genes shown may contain genes that are not regulated or that are less regulated by glucocorticoids in p53 wt cells, but so far we did not detect antagonism between p53 and glucocorticoids or cytokines on expression levels of single genes.
Gene regulation by Epo, SCF, and Dex
The data in Table 5 show that each cytokine, Epo and SCF, targets a distinct group of genes while the 2 also have a set of common target genes, which is more extended, as shown in Figure 4. We observed that many SCF-specific targets showed a similar regulation in response to Epo, yet just below the threshold level. We previously described that SCF/c-Kit–dependent signaling leads to a much stronger activation of PI3kinase than Epo/EpoR signaling, as evidenced by phosphorylation of the downstream target PKB3 (W.B. et al, unpublished data, March 2002). Accordingly, known PI3kinase targets such as BTG1 occur in Table 5 only as SCF-specific targets, albeit they are regulated by both SCF and Epo. Importantly, SCF never suppresses genes that were up-regulated by Epo or vice versa. This shows that SCF does not reverse Epo-induced gene regulation. Recently, we showed that the main Epo-signaling pathway required for differentiation is STAT5-dependent up-regulation of BclXL,14 while Epo-dependent progenitor expansion involves recruitment of the tyrosine kinase receptor RON plus the associated docking molecule Grb-associated binder 1 (Gab1) (van den Akker et al, unpublished data, November 2001). Direct activation of the Epo-target RON induced PI3kinase and MAPK kinase 1 (MEK1)/ERK activation, which was sufficient to substitute for Epo in progenitor expansion and delayed differentiation in the presence of Epo. These findings, together with the observation that Epo and SCF show a large overlap in target genes, suggest that many of the Epo-target genes found may contribute to induction of progenitor expansion rather than the induction of differentiation.
Remarkably, the effects of Dex on gene transcription do not parallel the effects of Epo and SCF. Among the genes up-regulated by Dex, many are down-modulated by Epo and/or SCF. In some cases, this counteraction still implies mutual enhancement. For instance, Epo-induced expression of the Janus-activated kinase 2 (JAK2) inhibitor CIS is attenuated by Dex, resulting in enhanced Epo signaling. In general, however, the different patterns of Epo/SCF– and Dex-induced gene expression are in line with the fact that Epo/SCF enhances proliferation of erythroid progenitors without blocking differentiation,16,25 while Dex arrests differentiation (eg, hemoglobin accumulation) without affecting the differentiation-induced proliferation arrest.14
A number of GR-target genes were regulated by Dex only in the presence of Epo and/or SCF. As the GR is known to associate with several transcription factor complexes that are themselves regulated by signal transduction, for example, STAT5, activator protein 1 (AP-1), nuclear factor κB (NFκB),26-30 it is likely that this type of transcription factor interactions may be involved in the regulation of genes coregulated by Dex and Epo/SCF signaling.
The role of GR-target genes in erythropoiesis.
Analysis of the GR-target genes shows that many of them regulate signal transduction or cell cycle progression. This implies that activation of the GR can modify the cell's response to environmental factors, as target genes include the stem-cell–derived factor 1 (SDF-1) receptor CXC chemokine receptor 4 (CXCR4), which is an important homing factor of hematopoietic cells; tryptophane hydroxylase, a key enzyme in serotonine synthesis; VDUP1, which controls reactive oxygen species (ROS) and thereby modulates phosphatase activity; the receptor tyrosine phosphatase CD45 (LY-6), GILZ, which interferes with signaling-controlled activity of NFκB and AP-1 and the tyrosine kinase JAK2 inhibitor CIS.31,32
Glucocorticoids are known to induce rapid apoptosis in certain cell types (eg, lymphocytes or thymocytes).33-35 This effect of glucocorticoids, however, may depend on the cellular context and on the presence of different factors. In erythroid progenitors, activation of the GR in absence of Epo and SCF causes cell cycle arrest.14,36 The screening results show that Dex treatment indeed leads to up-regulation of antiproliferative genes, for example, BTG1 or GILZ. Markedly, these genes are down-regulated by SCF or Epo. Thus, the cooperation between SCF and Dex at least partly consists of SCF/Epo–mediated attenuation of growth-inhibitory effects. Possibly, Dex induces expression of a set of genes crucial for induction of renewal, while attenuation of growth-inhibitory genes prevents effects of Dex that interfere with renewal. However, it is equally possible that this balance between cell cycle inhibition and cell cycle stimulation constitutes the mechanism of renewal induction. Notably, an extended G1 phase of 11 hours is characteristic for a “renewal cell cycle,” while the G1 phase of differentiating cells is reduced to 5 hours. We previously showed that the prime trigger allowing erythroid progenitors to differentiate into mature erythrocytes is the removal of signals that induce renewal.14,15 One of the genes found to be regulated in our screen is BTG1, a very labile protein. In proliferating 3T3 fibroblasts, BTG1 is rapidly up-regulated at the beginning of the G1 cell cycle phase. Expression is gradually reduced and only when BTG1 expression is lost do the cells progress to S phase.37,38 Thus, it is possible that a function of Dex-induced and SCF-modulated BTG1 expression during renewal is to maintain a long G1 phase required for erythroblast to enter S phase at a size compatible with renewal.
Prepublished online as Blood First Edition Paper, July 17, 2003; DOI 10.1182/blood-2003-03-0923.
Supported by grants from the European Union (ERBFMRXT980197 and HPRN-CT-2000-00083) (H.B.) and from the Dutch Cancer Society (EUR 2000-2230) (M v. L.), and by a fellowship of the Dutch Academy of Arts and Sciences (M. v. L.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Gabriele Litos and Herbert Auer for technical support; Drs Holger Reichardt and Guenther Schuetz for the GRdim/dim mice; and Drs Helmut Dolznig, Stefan Gruenert, and Marina Schorpp-Kistner for critically reading this manuscript.

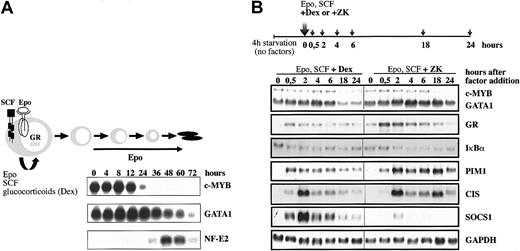
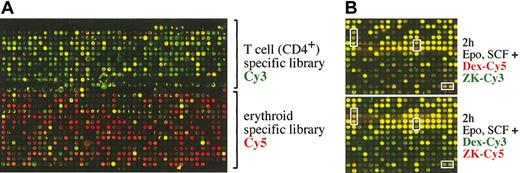
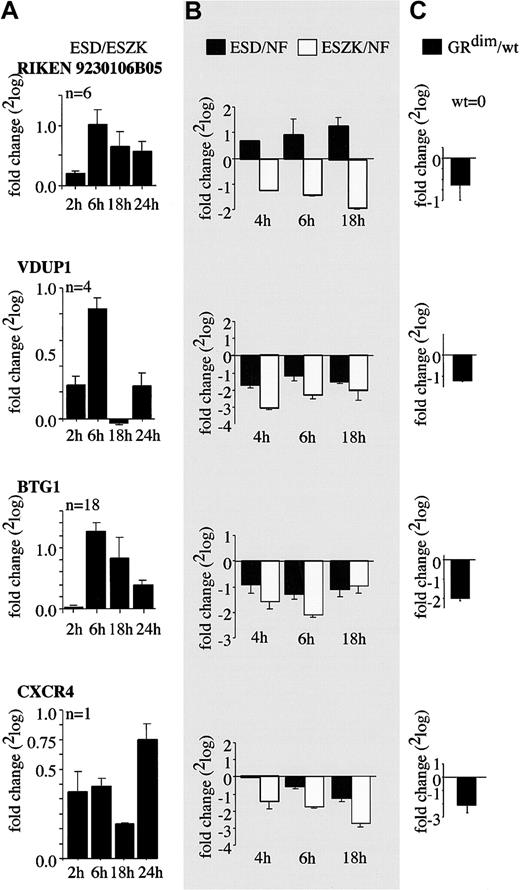
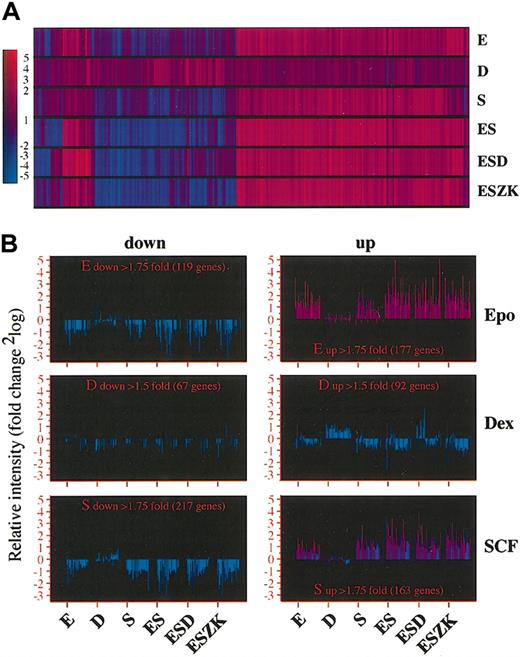
![Figure 5. Expression profiles of the putative GR-target genes VDUP1, MMP-2, and GILZ in response to Epo, SCF, Dex, and combinations thereof. (A, microarray) Mean fold changes (2log values) of gene expression detected on the microarrays for the genes indicated, comparing cDNA of factor-depleted I/11 erythroid progenitors with cells subsequently treated with Epo (E), SCF (S), Dex (D), Epo/SCF (ES), Epo/SCF/Dex (ESD), or Epo/SCF/ZK (ESZK) for 2 hours. The number of clones representing each of the genes on the microarrays is indicated (n). (A, RT-PCR) Expression of the indicated genes was subsequently determined by real-time RT-PCR. RNA levels of factor-depleted and restimulated cells (as indicated) were compared with RNA levels of factor-deprived cells. Fold changes of restimulation versus factor deprivation are indicated as 2log values. (B) RNA levels of factor-depleted I/11 erythroid progenitors restimulated with Epo/SCF/Dex (ESD) or Epo/SCF/ZK (ESZK) for the indicated time periods were compared with RNA levels of cells factor deprived for 4 hours (NF). Fold changes of restimulation versus factor deprivation are indicated as 2log values. (C) RNA levels of proliferating erythroid progenitors derived from either GRdim/dim fetal livers or wt littermates (used either as primary cells [prim] or after immortalization in culture [imm]) were determined by real-time RT-PCR. Fold changes of GRdim/dim over wt are indicated as 2log values. Mean values and standard deviations from 2 separate experiments repeated in duplicate are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/9/10.1182_blood-2003-03-0923/6/m_h82135136005.jpeg?Expires=1769103810&Signature=kWTDjUey4DNb6EFUgyABzW4Bx-JXJcW1JEhwpfRon27g6-gDy1yMXAdo9FMLsYCPGDw8UCNfvWjm-mtcEIuY2dvCj8l2zxWxIrOcla3N8cj4F5PvNDH-XAJHl3Fs4mrRwkOpnwG22P21hLG-DPaprHx1FRjRd7bQJJeA0EwccBssMagFcv8IC0EGFtugNH-~ZaPglKESHnQKFt4CnkviVvBLDelH01Kw2gxDiPQuCOiHTrEHjtx~s41iELMi-x1gmvsOEYJLNrz8qj6i-gShSrKq3E9t0CX8m9CwbNzd~-9aKU-zz3thf91wcbvl2MNOQyIZu369Tj4tx4GRMA~qGg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal