Abstract
Coagulation factor V (FV) is a central regulator of the coagulation cascade. Circulating FV is found in plasma and within platelet α granules. The specific functions of these distinct FV pools are uncertain. We now report the generation of transgenic mice with FV gene expression restricted to either the liver or megakaryocyte/platelet lineage using bacterial artificial chromosome (BAC) constructs. Six of 6 independent albumin BAC transgenes rescue the neonatal lethal hemorrhage of FV deficiency. Rescued mice all exhibit liver-specific Fv expression at levels ranging from 6% to 46% of the endogenous Fv gene, with no detectable FV activity within the platelet pool. One of the 3 Pf4 BAC transgenes available for analysis also rescues the lethal FV null phenotype, with FV activity restricted to only the platelet pool (approximately 3% of the wild-type FV level). FV-null mice rescued by either the albumin or Pf4 BAC exhibit nearly normal tail bleeding times. These results demonstrate that Fv expression in either the platelet or plasma FV pool is sufficient for basal hemostasis. In addition, these findings indicate that the murine platelet and plasma FV pools are biosynthetically distinct, in contrast to a previous report demonstrating a plasma origin for platelet FV in humans.
Introduction
Factor V (FV) is a central regulator of hemostasis, serving as an essential component of the prothombinase complex and as a key target of the natural anticoagulant activated protein C (APC).1-5 FV deficiency results in a major bleeding disorder in humans,6 and genetically engineered mice completely deficient in FV exhibit partially penetrant lethality at mid-embryogenesis, with the remaining animals dying of hemorrhage at birth.7 Incomplete embryonic lethality is also observed in tissue factor–deficient,8-10 thrombin-deficient,11,12 and in protease-activated receptor 1 (PAR1)–deficient mice,13 suggesting a common mechanism due to disruption of a critical thrombin-mediated signaling event in the early embryo. Findings in some studies were consistent with embryonic loss due to hemorrhage,8 whereas others implicated an abnormality in the yolk sac vasculature.7,9,13 However, the combination of PAR1 and FV deficiency was recently shown to result in more severe embryonic loss than either deficiency alone, suggesting a more complex role for these molecules in early development, potentially involving more than one pathway.14
Total blood FV is distributed into 2 distinct pools. In humans, approximately 80% of the FV in whole blood circulates in the plasma as a single-chain pro-cofactor and is thought to be derived primarily from synthesis in the liver.15 The remaining 20% of whole blood FV is concentrated within the platelet α granule and stored in a partially cleaved and activated form.16,17 In situ megakaryocytic synthesis was previously thought to be the origin of platelet FV. Isolated guinea pig megakaryocytes were shown to synthesize FV18 and FV expression was also detected in human megakaryocytes.19 However, a more recent study of patients undergoing bone marrow and liver transplantation suggests that human platelet FV is derived primarily or exclusively via uptake from the plasma pool.20 In contrast, data on mouse bone marrow transplantation demonstrate that the murine platelet FV pool is derived exclusively from expression within the megakaryocytes.21
We previously reported the analysis of conventional transgenic mice with Fv transgene expression driven either by the liver-specific albumin (Alb) promoter or the megakaryocyte/platelet-specific platelet factor 4 (Pf4) promoter.22 Rescue of the Fv-null perinatal lethal phenotype by liver-specific Fv expression was observed despite undetectable levels of plasma FV (< 0.1%), suggesting that even trace FV activity in the plasma compartment is sufficient for minimal hemostasis in the mouse. The Pf4 promoter failed to generate detectable Fv transgene expression or rescue the Fv-null phenotype. We now report the application of a bacterial artificial chromosome (BAC) transgene approach to achieve higher and more tissue-specific Fv expression to explore the in vivo functions of the plasma and platelet FV pools in hemostasis.
Materials and methods
Construction ofFvBAC transgenes by homologous recombination inEscherichia coli
BACs containing the murine albumin genes (Alb) and murine platelet factor 4 (Pf4) genes were obtained by screening from a commercially available BAC RPCI-22 mouse BAC genomic library (Research Genetics, Huntsville, AL). A 950-bp PstI/NsiI genomic fragment containing the whole coding sequence of the rat Pf4 gene and a 600-bp reverse transcriptionpolymerase chain reaction (RT-PCR) product from exon 12 to the end of the murine Alb cDNA were used as probes to screen the library, according to the manufacturer's instructions. The positive BACs were obtained from the manufacturer and analyzed by restriction enzyme digestion and pulse field gel electrophoresis. Five murine Alb BACs and one Pf4 BAC were identified in the library screen. The 5 Alb BACs were then analyzed by multiple restriction digests and the BAC with the largest 5′ and 3′ sequences flanking the murine Alb gene was used for all subsequent experiments.
An Fv transgene cassette was inserted by homologous recombination in E coli into the 5′ untranslated region (UTR), immediately downstream of either the Alb or Pf4 promoter. For homologous recombination into the Alb BAC, the Fv transgene cassette was preceded by 619 bp of sequence upstream of the Alb ATG start codon (–630 to –12, A of start codon ATG as +1) and followed by 597 bp of sequences downstream of Alb exon 12 (+45127 to +45725; GenBank NT_039307). For homologous recombination into the Pf4 BAC, the Fv transgene cassette was preceded by 744 bp of Pf4 promoter and 5′UTR sequence (–787 to –43, A of start codon ATG as +1) and followed by 628 bp from the 3′ end of the Pf4 gene (+791 to +1420; GenBank NT_039307).
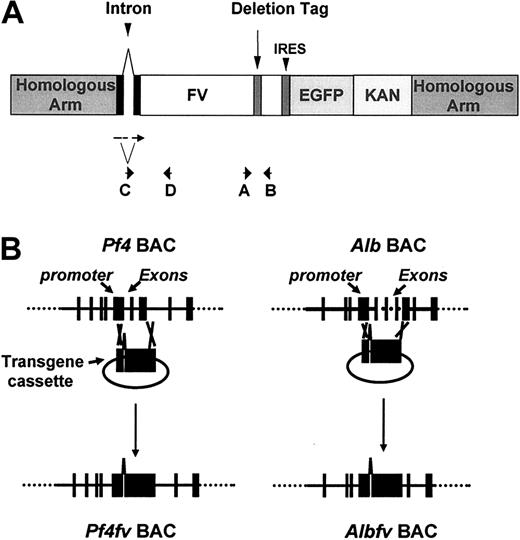
The structure of the transgene cassette is illustrated in Figure 1. The cassette contains the murine Fv cDNA from nucleotide –43 (A of ATG initiation codon as +1) to +6618, including the entire coding sequence, with a 3′ UTR deletion tag (+6564 to +6593) as previously described,22 followed by the enhanced green fluorescent protein (EGFP) coding sequence, preceded by an internal ribosome entry site sequence (IRES; Clontech Laboratories, Palo Alto, CA) and a bacterial kanamycin resistant gene (KAN; generous gift from A. F. Stewart). A chimeric intron with optimized splice signals from the pCI expression vector (Promega, Madison, WI) was introduced upstream of the Fv cDNA in the transgene cassette to enhance gene expression. The EGFP gene was incorporated as a convenient marker for transgene expression. However, GFP fluorescence was not observed in the liver or platelets of the corresponding transgenic mice, probably due to inefficient translation of the EGFP mRNA. Homologous recombination between the flanking sequences of the constructs and corresponding BACs was carried out in E coli by RecA as described by Yang et al.23 The murine Alb gene in the Alb BAC (from nucleotide –12 to exon 12) was replaced with the transgene cassette. A similar strategy in the Pf4 BAC replaced the murine Pf4 gene (from nucleotide –43 to the end of exon 3) with the Fv transgene cassette.
The construction of AlbFv or Pf4Fv BACs. (A) The transgene cassette used for homologous recombination into the Alb or Pf4 BACs. The locations of PCR and RT-PCR primers A to D are illustrated by arrows. The cassette contains the murine Fv cDNA with a chimeric 5′ intron and a 3′ UTR deletion tag, followed by an internal ribosome entry site sequence (IRES) preceding the enhanced green fluorescent protein (EGFP) coding sequence and a bacterial kanamycin resistant gene (KAN). (B) Homologous recombination between the flanking sequences of the constructs and corresponding BACs. Exons 1-12 of the murine Alb gene and exons 1-3 of murine Pf4 gene were replaced with an FV cDNA minigene.
The construction of AlbFv or Pf4Fv BACs. (A) The transgene cassette used for homologous recombination into the Alb or Pf4 BACs. The locations of PCR and RT-PCR primers A to D are illustrated by arrows. The cassette contains the murine Fv cDNA with a chimeric 5′ intron and a 3′ UTR deletion tag, followed by an internal ribosome entry site sequence (IRES) preceding the enhanced green fluorescent protein (EGFP) coding sequence and a bacterial kanamycin resistant gene (KAN). (B) Homologous recombination between the flanking sequences of the constructs and corresponding BACs. Exons 1-12 of the murine Alb gene and exons 1-3 of murine Pf4 gene were replaced with an FV cDNA minigene.
Genotyping for transgene and Fv-null alleles
The Fv BAC DNA was isolated with the Nucleobond Plasmid Maxi Kit (Clontech Laboratories) and microinjected into fertilized oocytes (C57BL/6 × SJL F1). Pronuclear microinjections were performed by the Transgenic Animal Model Core at the University of Michigan. Fertilized eggs obtained by mating (C57BL/6 × SJL/) F1 or C57BL/6 female mice with (C57BL/6 × SJL) F1 male mice were used for injection.
The gDNA was prepared from a tail biopsy sample using the Wizard gDNA purification kit (Promega). Transgene carriers were identified by PCR amplification (annealing temperature 54°C) with primer A and B (Table 1; Figure 1),22 yielding expected products of 160 bp from the transgene and 190 bp from the endogenous Fv gene. The Fv knockout allele was assayed by PCR amplification (annealing temperature 54°C) with primers E, F, and G (Table 1), yielding expected products of 200 bp from the Fv knockout allele and 230 bp from the wild-type Fv allele.
Primers for genotyping and RT-PCR
Name . | Sequence . | GenBank accession no. . | Primer location . |
|---|---|---|---|
| A* | CAAGGGGCACATGAAGAACT | — | — |
| B* | CTTGAAGGAGTTTCTTCAGCTTTTA | — | — |
| C | ATCCCGGGCACTGGGCAGGTGTC | — | — |
| D† | TACTGTTCATACTCTCTGTAGA | NT_039185 | 2568-2588 |
| E† | TGCGATGTTACCAGTGATACAAG | NT_039185 | 30802-30824 |
| F† | CTCCATGTAGCAACCAGGAG | NT_039185 | 31007-31026 |
| G‡ | TATCGCCTTCTTGACGAGTTC | AY147202 | 1798-1778 |
Name . | Sequence . | GenBank accession no. . | Primer location . |
|---|---|---|---|
| A* | CAAGGGGCACATGAAGAACT | — | — |
| B* | CTTGAAGGAGTTTCTTCAGCTTTTA | — | — |
| C | ATCCCGGGCACTGGGCAGGTGTC | — | — |
| D† | TACTGTTCATACTCTCTGTAGA | NT_039185 | 2568-2588 |
| E† | TGCGATGTTACCAGTGATACAAG | NT_039185 | 30802-30824 |
| F† | CTCCATGTAGCAACCAGGAG | NT_039185 | 31007-31026 |
| G‡ | TATCGCCTTCTTGACGAGTTC | AY147202 | 1798-1778 |
— indicates not applicable.
From Yang et al.22
From mouse factor V sequence.
From bacterial kanamycin-resistance gene.
mRNA expression analysis by RT-PCR
Mice were humanely killed and fresh tissues were harvested for RNA preparation using a Trizol kit (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. The tissues used for RT-PCR were liver, spleen, kidney, intestine, lung, heart, muscle, and brain. Bone marrow was also harvested for analysis from rescued mice carrying the Pf4A transgene. Approximately 1 μg total RNA served as template for the SuperScript One-step RT-PCR kit (Invitrogen), using primers C and D (Table 1; Figure 1). The upstream PCR primer (C) is derived from exonic sequences flanking the artificial intron and thus should fail to amplify the unspliced intron (gDNA).
Analysis of FV activity in platelet and plasma pools
Mouse whole blood was drawn by cardiac puncture with one-tenth volume 3.8% sodium citrate. Platelet-rich plasma was prepared by centrifugation at 180g for 10 minutes. Platelet-rich plasma (50 μL) was centrifuged at 2000g for 10 minutes to generate platelet-poor plasma. The platelet pellet was washed once with Tyrode buffer (134 mM NaCl, 2.9 mM KCl, 0.34 mM Na2HPO4, 12 mM NaHCO3, 20 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 1 mM MgCl2, 5 mM glucose) and resuspended in 50 μL Tyrode buffer. Platelet concentration was determined using a Coulter counter (model Z2; Hialeah, FL) for both the original platelet-rich plasma and the final platelet solution suspended in Tyrode buffer. FV activity was determined using a prothrombin time assay with human FV-deficient plasma (George King Biomedical, Overland Park, KS), as previously described.24 The platelets suspended in Tyrode buffer were lysed in 2% Tween 20, followed by a freeze/thaw cycle. The FV activity of the platelet pool was calculated by correcting the FV activity measured in the final Tyrode buffer solution relative to the platelet concentration in the original platelet-rich plasma sample. FV activity measured in human standard plasma was defined as 1000 mU/mL. The sensitivity of the assay is about 1% of the human standard (10 mU/mL), which corresponds to approximately 0.1% to 0.2% of the wild-type mouse plasma FV level.
Measurement of tail vein bleeding times
Tail vein bleeding time was measured using a previously reported method.25 Briefly, mice were anesthetized with pentobarbital sodium and a transverse incision was made with a scalpel in a lateral tail vein at a site where the tail diameter was 2.25 mm. The lacerated tail was immersed in saline (37°C) and the time to cessation of bleeding recorded. Test animals were then observed for an additional 3 hours and the subsequent day for evidence of rebleeding.
Results
Generation of Fv transgenic lines
The correct structure of each engineered BAC transgene was confirmed by PCR, DNA sequencing, and Southern blotting. The modified albumin BAC is about 120 kb in length and contains approximately 40 kb Alb gene and flanking sequences upstream of the transgene cassette and about 70 kb downstream, as determined by pulse field gel electrophoresis (data not shown). The modified Pf4 BAC is about 130 kb in length and contains approximately 60 kb sequences upstream of the transgene cassette and about 60 kb downstream.
Oocyte injections with the Alb and Pf4 Fv BAC constructs yielded 6 and 4 independent transgenic founders, respectively. One of the Pf4 lines was lost early in the genetic cross. All 9 of the remaining Pf4 and Alb transgenic lines were subjected to further analysis.
Tissue-specific pattern of Fv transgene expression
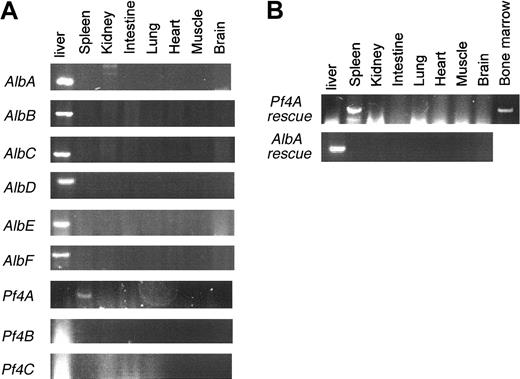
For each independent line, offspring carrying the transgene (Tg+) were screened by RT-PCR to determine the tissue-specific pattern of transgene expression. A signal from contaminating gDNA could be excluded, because PCR primer C is derived from sequences flanking the intron and should only amplify the spliced transgene mRNA product (Figure 1). As shown in Figure 2, only a single Pf4 Fv BAC line (Pf4A) exhibits detectable transgene expression, the latter restricted to megakaryocyte-containing tissues (spleen and bone marrow). In contrast, all 6 Alb Fv BAC lines exhibit transgene expression restricted to the liver.
Expression patterns of Fv transgenes. (A) RT-PCR for Fv transgene expression was performed on tissue samples from 3 Pf4 Fv (Pf4A-C) and 6 Alb Fv (AlbA-F) transgenic mice. (B) RT-PCR for Fv transgene expression was performed on tissue samples from AlbA and Pf4A transgenic rescued FV-null mice (Tg+Fv–/–).
Expression patterns of Fv transgenes. (A) RT-PCR for Fv transgene expression was performed on tissue samples from 3 Pf4 Fv (Pf4A-C) and 6 Alb Fv (AlbA-F) transgenic mice. (B) RT-PCR for Fv transgene expression was performed on tissue samples from AlbA and Pf4A transgenic rescued FV-null mice (Tg+Fv–/–).
Transgene rescue of the Fv-null phenotype
All 9 lines were crossed into the Fv–/– background to test for the ability of each tissue-specific transgene to rescue the uniformly lethal Fv-null phenotype. The results of this analysis are shown in Table 2. As expected, no Fv–/– pups lacking the transgene (Tg–Fv–/–) survived to weaning (age 2-3 weeks). Of the 3 Pf4 BAC transgenic lines, only Pf4A exhibited rescue of the Fv–/– phenotype. In contrast survival of Tg+Fv–/– mice was observed for all 6 Alb transgenic lines. Rescued Tg+Fv–/– mice from lines AlbA and AlbB were observed for up to 1 year, with grossly normal survival and no evidence of spontaneous hemorrhage. The latter observation contrasts with the frequent bleeding observed in the previously reported Tg+Fv–/– mice rescued by a conventional Alb transgene (producing < 0.1% of the wild-type plasma FV level).22 Crosses of Tg+Fv+/– mice with Fv+/– mice yielded the expected approximately 25% Fv–/– pups among the surviving Tg+ offspring for the Alb transgenic lines AlbB, AlbD, AlbE, and AlbF, suggesting that these transgenes rescue both the mid-embryonic developmental defect and the lethal perinatal hemorrhage resulting from complete FV deficiency. In contrast, lines AlbA, AlbC, and Pf4A only rescue about half of the expected number of Tg+Fv+/–, consistent with continued mid-embryonic loss and rescue of only the perinatal hemorrhage,7 similar to the results with the previously reported conventional Alb transgene.22 Surprisingly, more complete rescue is observed for all 7 transgenic lines in a subsequent cross of surviving Tg+Fv–/– mice to Fv+/– (right side of Table 2). These differences could be due to chance or to selection for strain-specific modifier genes among Tg+Fv–/– mice surviving the initial cross. Of note, a similar phenomenon was observed in a recent report of transgene rescue for prothrombin deficiency.26
Genotypic distributions of progeny from crosses of different BAC transgenic founder lines
. | Tg+Fv+/- × Fv+/- . | . | Tg+Fv-/- × Fv+/- . | . | . | ||
|---|---|---|---|---|---|---|---|
. | Tg+, Fv+, %† . | Tg+, Fv-, %† . | Tg+, Fv+, %† . | Tg+, Fv-, %† . | Level* . | ||
| Expected‡ | 75 | 25 | 50 | 50 | - | ||
| AlbA | 86.1 (118) | 13.8 (19) | 53.2 (58) | 46.8 (51) | 31.7 | ||
| AlbB | 74.2 (26) | 25.7 (9) | 48.1 (37) | 51.9 (40) | 13.8 | ||
| AlbC | 83.8 (26) | 16.1 (5) | 50 (7) | 50 (7) | 46.3 | ||
| AlbD | 78.8 (41) | 21.1 (11) | 48.3 (14) | 51.7 (15) | 7.6 | ||
| AlbE | 58.3 (7) | 41.7 (5) | 41.0 (16) | 59 (23) | 14 | ||
| AlbF | 71.1 (37) | 28.8 (15) | 64.3 (18) | 35.7 (10) | 6.6 | ||
| Pf4A | 88.5 (77) | 11.5 (10) | 56.8 (54) | 43.2 (41) | 3.1 | ||
| Pf4B | 100 (49) | 0 (0) | - | - | - | ||
| Pf4C | 100 (46) | 0 (0) | - | - | - | ||
. | Tg+Fv+/- × Fv+/- . | . | Tg+Fv-/- × Fv+/- . | . | . | ||
|---|---|---|---|---|---|---|---|
. | Tg+, Fv+, %† . | Tg+, Fv-, %† . | Tg+, Fv+, %† . | Tg+, Fv-, %† . | Level* . | ||
| Expected‡ | 75 | 25 | 50 | 50 | - | ||
| AlbA | 86.1 (118) | 13.8 (19) | 53.2 (58) | 46.8 (51) | 31.7 | ||
| AlbB | 74.2 (26) | 25.7 (9) | 48.1 (37) | 51.9 (40) | 13.8 | ||
| AlbC | 83.8 (26) | 16.1 (5) | 50 (7) | 50 (7) | 46.3 | ||
| AlbD | 78.8 (41) | 21.1 (11) | 48.3 (14) | 51.7 (15) | 7.6 | ||
| AlbE | 58.3 (7) | 41.7 (5) | 41.0 (16) | 59 (23) | 14 | ||
| AlbF | 71.1 (37) | 28.8 (15) | 64.3 (18) | 35.7 (10) | 6.6 | ||
| Pf4A | 88.5 (77) | 11.5 (10) | 56.8 (54) | 43.2 (41) | 3.1 | ||
| Pf4B | 100 (49) | 0 (0) | - | - | - | ||
| Pf4C | 100 (46) | 0 (0) | - | - | - | ||
Observed percentages of each genotype are shown; numbers within parentheses represent numbers of mice observed. — indicates not applicable.
The level of Fv expression observed in the plasma and platelet pools of the transgenic Fv-null rescued mice (Tg+ Fv-/-) are shown here for all 6 albumin Fv BAC lines and one Pf4 Fv BAC line. The FV level observed in platelets or plasma of a control Fv+/- mouse, corresponding to one endogenous copy of the normal Fv gene, is arbitrarily defined as 50%.
Fv+/+ and Fv+/- genotypes are combined together as Fv+. Only mice carrying transgene are included for analysis.
Expected percentages of each genotype.
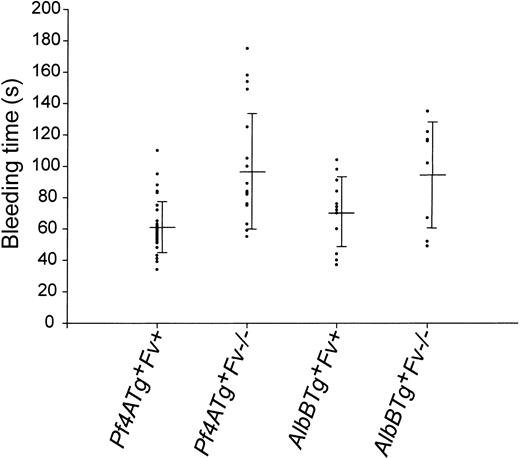
Tail vein bleeding times were measured in AlbB and Pf4A Tg+Fv–/– mice and their Tg+Fv+ littermate controls (Figure 3). Bleeding times were slightly prolonged in both the Pf4A (98 ± 37 seconds) and AlbB Tg+Fv–/– (95 ± 34 seconds) mice compared to Tg+Fv+/– and Tg+Fv+/+ littermate controls (61 ± 16 and 71 ± 22 for the Pf4 and Alb transgenes, respectively). The latter control values are very similar to the tail vein bleeding times previously reported by Broze and coworkers for the C57Bl/6 and 129/Sv strains (51 ± 17 and 62 ± 18 seconds, respectively25 ). No late rebleeding was observed and only one late death occurred (in 1 of 19 Pf4A Tg+Fv–/– mice). In contrast, 5 of 5 mice deficient in factor VIII (FVIII) exhibited uniform exsanguination after the bleeding time procedure.
Tail vein bleeding time. Tail vein bleeding times of Pf4A and AlbB mice Tg+ are shown in seconds. Rescue mice (Tg+Fv–/–) are compared with control littermates carrying the transgene and at least one wild-type Fv allele (Fv+ = Fv+/– or Fv+/+). The mean and SD are also indicated for each genotype.
Tail vein bleeding time. Tail vein bleeding times of Pf4A and AlbB mice Tg+ are shown in seconds. Rescue mice (Tg+Fv–/–) are compared with control littermates carrying the transgene and at least one wild-type Fv allele (Fv+ = Fv+/– or Fv+/+). The mean and SD are also indicated for each genotype.
Fvtransgene expression levels
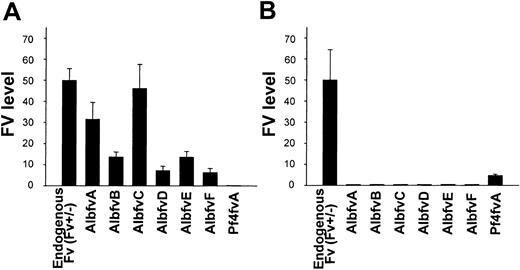
The level of FV protein expression directed by the transgene was determined by measuring plasma and platelet FV levels in Tg+Fv–/– mice for each of the independent founder lines (Table 2; Figure 4). FV activity was detected in the plasma of all 6 Alb lines (AlbA-F) at levels ranging from 6.6% to 46.3% of wild-type mouse plasma. FV activity was undetectable in the platelet pools of all 6 Alb BAC transgenic lines. The Pf4A line exhibited a platelet FV level equivalent to 3.1% of the wild-type FV level, with undetectable levels of FV in the plasma.
Levels of Fv transgene expression. The level of Fv expression observed in the plasma (A) and platelet (B) pools of the transgenic Fv-null rescued mice (Tg+Fv–/–) are shown here for all 6 albumin Fv BAC lines and one Pf4 Fv BAC line. The FV level observed in platelets or plasma of a control Fv+/– mouse, corresponding to one endogenous copy of the normal Fv gene, is arbitrarily defined as 50%. SEs are indicated.
Levels of Fv transgene expression. The level of Fv expression observed in the plasma (A) and platelet (B) pools of the transgenic Fv-null rescued mice (Tg+Fv–/–) are shown here for all 6 albumin Fv BAC lines and one Pf4 Fv BAC line. The FV level observed in platelets or plasma of a control Fv+/– mouse, corresponding to one endogenous copy of the normal Fv gene, is arbitrarily defined as 50%. SEs are indicated.
Discussion
In both humans and mice, approximately 80% of FV circulates free in plasma, with the remaining 20% localized to a discrete pool within the platelet α granule.16,22 The lack of clinical bleeding in some patients with potent circulating anti-FV antibodies,27 along with the severe bleeding observed in factor V Quebec (deficiency of platelet FV),28,29 suggest a critical role for the platelet FV pool. However, significant bleeding in other inhibitor patients, as well the response of congenitally FV-deficient patients to plasma infusion, support a key functional role for the plasma FV pool.30 We have previously explored the relative contributions of the plasma and platelet FV pools to hemostasis using several approaches in genetically engineered mice.21,22 Although transplantation of FV-deficient bone marrow into wild-type mice demonstrated that expression of plasma-derived FV is sufficient for minimal hemostasis, this approach could not directly address the functional role of the platelet FV pool. An earlier study using a conventional transgene approach failed to achieve high levels of FV expression in either the platelet or plasma FV pool, though very low levels of liver-specific Fv gene expression were observed to be sufficient for partial rescue of the lethal FV-null phenotype.22
Because of the disappointingly low levels of Fv expression achieved with conventional Fv transgenes, we have now adapted a new approach based on the use of larger segments of transgenic DNA cloned into BACs. BACs generally contain 100- to 200-kb segments of gDNA and can be readily re-engineered by homologous recombination in E coli, using one of several recently described methods.23,31,32 In addition to the known promoter and enhancer elements incorporated into standard tissue-specific transgene constructs, BAC transgenes also include large segments of flanking sequences that are likely to include numerous additional distant transcriptional regulatory elements, as well as insulator domains inhibiting insertion site position effects. Taken together, these factors should ensure more faithful levels of transgene expression and tissue specificity23,33,34 and this general approach has been successfully applied in a number of other systems.35-37 Consistent with these previous reports, the Fv BAC transgenes described here demonstrated consistently high levels of Fv expression with a faithful tissue-specific pattern, in marked contrast to our earlier conventional transgene experience.22
We observed rescue of the lethal Fv–/– hemorrhagic phenotype by either liver-specific Alb or platelet-specific Pf4 BAC transgenes. All 6 Alb BAC transgene lines were able to reverse the perinatal hemorrhage, confirming our previous transgenic and bone marrow transplantation studies. The spontaneous bleeding previously observed with a conventional Alb transgene22 was not seen with any of the Alb BAC transgenes, probably as a result of the higher levels of plasma FV produced. Similar to the platelet FV-null mice generated by bone marrow transplantation,21 Alb BAC mice exhibited grossly normal hemostasis, even after minor injury such as transection of the tail tip. Bleeding times in Alb Tg+Fv–/– were also only minimally prolonged, though largely overlapping the values observed in controls. These data demonstrate that although the murine platelet FV pool may contribute in the response to serious injury, it is not required for the prevention of spontaneous hemorrhage or for hemostasis following a moderate challenge such as the tail vein bleeding time procedure or tail transection for routine genotyping. These results further suggest that the relatively severe bleeding disorder seen in human patients with FV Quebec is due to the multiple deficiencies of other platelet α-granule proteins also observed in this disease,28,38 rather than deficiency of platelet FV alone.
Our previous failure to rescue the Fv–/– phenotype with conventional Pf4-driven Fv transgenes could have been consistent with an absolute requirement for the plasma FV pool to achieve minimal hemostasis, though simple failure to achieve sufficient levels of platelet Fv expression by this approach could not be excluded.22 The functional significance of the platelet FV pool could also not be addressed by transplantation because the Fv–/– marrow recipients required for this experiment fail to survive past birth. Our current observations with a highly platelet-specific Pf4 BAC Fv transgene indicate that the platelet FV pool is by itself sufficient to prevent spontaneous hemorrhage and to support hemostasis in response to moderate challenge (bleeding time procedure and tail transection for genotyping). Thus, both FV pools are active and can contribute to hemostatic function in vivo, though it is likely that subtle differences in their biologic roles in the response to a range of pathologic challenges remain to be identified.
Two distinct mechanisms have been shown to account for the uniform lethality of complete FV deficiency in mice.7 First, about 50% of null mice, regardless of strain background, die at E9.5. This early embryonic lethality may be due to a defect in yolk sac vasculature formation,7 as also proposed for tissue factor deficiency,9 though this hypothesis is controversial.8 In contrast, the uniform lethality at birth of all Fv–/– that survive the early embryonic developmental block is clearly due defective hemostasis.7,22 The data in Table 2 are consistent with as many as 4 independent Alb Tg+ lines exhibiting full rescue of the E9.5 embryonic lethality, suggesting that Fv gene expression restricted to hepatocytes or embryonic liver precursors may be sufficient for this developmental function. This is consistent with previous evidence of normal survival to term in the absence of platelets.39 The variability in the extent of embryonic lethality among the Alb transgenic lines suggests that this difference may be related to timing of Fv expression rather than to tissue specificity or expression level. In addition, the more complete rescue observed in the second generation of crosses with surviving Tg+Fv–/– mice compared to the initial cross (Table 2) suggests a potentially complex role for strain-specific modifier genes. Possible rescue of the E9.5 embryonic lethality was observed for both Pf4 and Alb BAC transgenes, suggesting that FV in either the platelet or plasma pool can provide this critical activity. However, ectopic transgene expression in the developing embryo cannot be excluded and only limited conclusions about the role of FV in early mammalian development can be drawn from these studies.
The origin of the platelet FV pool in humans has been controversial. Several early studies provided evidence for the presence of FV mRNA19 and direct FV protein synthesis18 with megakaryocytes. The detection of normal platelet FV in a patient with a high titer anti-FV inhibitor also suggested distinct origins for the platelet and plasma FV pools.27 However, the elegant study of Camire et al20 in patients having bone marrow and liver transplants provided convincing evidence that the human platelet FV pool is derived primarily via uptake from the plasma compartment. These latter data are in contrast to the results in a murine bone marrow transplantation model, which demonstrated the exclusive origin of murine platelet FV from biosynthesis within a marrow-derived cell type, presumably the megakaryocyte.21 The transgenic mouse data shown in Figure 4 confirm and extend this latter result, clearly demonstrating that murine platelet FV is derived entirely from synthesis within the megakaryocyte, with no evidence for exchange or uptake from the plasma pool.
These data identify another significant physiologic difference between mouse and human platelets,40,41 and point out the need for caution in extrapolating observations from any heterologous mammalian model system directly to humans. However, this difference also provides a unique opportunity to examine the relative contributions of platelet and plasma FV to hemostasis, studies that could not be performed in humans. Although the biosynthetic origins may be different, the relative sizes of the 2 pools in mouse and human are quite similar, in contrast to some other species.42 Although human FV appears to be derived from liver-specific synthesis with uptake into the megakaryocyte, this process appears to occur early in megakaryocytopoiesis, with no evidence for exchange in mature platelets.20 Thus, the tissue-specific transgenic mice described here may serve as a useful model to dissect the relative contributions of these pools to hemostasis, even though the biosynthetic origins may differ from humans. These BAC transgenes can also be readily modified by homologous recombination in E coli to generate variants carrying specific FV gene mutations for further analysis in vivo.23 Finally, our observations suggest that FV replacement or gene transfer targeted solely to the plasma or platelet compartment may provide adequate hemostasis for FV-deficient patients.
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2003-04-1225.
Supported by National Institutes of Health grants PO1HL57346 (D.G.) and HL39639 (D.G.), and a Judith Graham Pool Research Fellowship from the National Hemophilia Foundation (H.S.). D.G. is a Howard Hughes Medical Institute investigator.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank X.Y. Yang and N. Heintz for providing the BAC recombination reagents, A. F. Stewart for the kanamycin-resistance construct, and H. Rottschafer and S. Pipe for thoughtful review of the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal