Abstract
MLL rearrangements in acute myeloid leukemia (AML) include translocations and intragenic abnormalities such as internal duplication and breakage induced by topoisomerase II inhibitors. In adult AML, FLT3 internal tandem duplications (ITDs) are more common in cases with MLL intragenic abnormalities (33%) than those with MLL translocation (8%). Mutation/deletion involving FLT3 D835 are found in more than 20% of cases with MLL intragenic abnormalities compared with 10% of AML with MLL translocation and 5% of adult AML with normal MLL status. Real-time quantification of FLT3 in 141 cases of AML showed that all cases with FLT3 D835 express high level transcripts, whereas FLT3-ITD AML can be divided into cases with high-level FLT3 expression, which belong essentially to the monocytic lineage, and those with relatively low-level expression, which predominantly demonstrate PML-RARA and DEK-CAN. FLT3 abnormalities in CBF leukemias with AML1-ETO or CBFβ-MYH11 were virtually restricted to cases with variant CBFβ-MYH11 fusion transcripts and/or atypical morphology. These data suggest that the FLT3 and MLL loci demonstrate similar susceptibility to agents that modify chromatin configuration, including topoisomerase II inhibitors and abnormalities involving PML and DEK, with consequent errors in DNA repair. Variant CBFβ-MYH11 fusions and bcr3 PML-RARA may also be initiated by similar mechanisms.
Introduction
FLT3 is a class III receptor tyrosine kinase that is normally expressed by hematopoietic stem cells, early myeloid and lymphoid precursors, and immature and mature monocytic cells.1 FLT3 abnormalities on chromosome 13 represent one of the most common genetic aberrations in de novo adult AML (reviewed in Gilliland and Griffin2 ). FLT3 internal tandem duplications (ITDs)3 occur in approximately 20% of cases and lead to variable, in-frame duplications within the juxtamembrane domain, resulting in constitutive tyrosine kinase activity. Their highest frequency occurs in AML with normal cytogenetics (30%-40%), PML-RARA (40%),4 or DEK-CAN (more than 50%).5 An additional 7% of AML patients demonstrate “activation loop” mutations involving the aspartic acid, D835 residue, which leads to an activated configuration and transformation of 32D cells.6 These are predominantly point mutations but microdeletions also occur5 ; they are collectively referred to as D835 here. Both D835 and FLT3-ITD are more common in monocytic AML.5 Patients with FLT3-ITD have a poor prognosis in most reported series,2 with a high mutant-to-wild-type ratio being particularly pejorative.5 D835 does not have clear prognostic impact.7 Both are thought to represent secondary genetic events.8,9 Development of AML is increasingly considered to involve oncogenic cooperation, whereby FLT3 abnormalities provide a proliferative advantage to a population in which normal maturation is blocked.2 In keeping with this, several studies, describe FLT3-ITD in subclones of variable intensity, with 10%-25% also demonstrating loss of the wild-type allele.5
Chromosome 11q23 abnormalities are found in approximately 5% of adult and in most infants with AML.10,11 Most involve the MLL gene, with a variety of partner genes/chromosomes (reviewed in Ayton and Cleary12 ). MLL internal tandem duplication is common in adult, but not pediatric, AML. It is more frequent in older patients13 and in those with trisomy 1114 and also has been identified at low levels in healthy adults.15 MLL abnormalities are frequently screened for by Southern blotting, fluorescent in situ hybridization (FISH), and/or reverse transcriptase–polymerase chain reaction (RT-PCR), in addition to classical karyotyping.11 Only Southern blotting allows detection of a further category of MLL abnormalities, which involves DNA double-strand breakage at a topoisomerase II site within the BamHI 8.2-kilobase MLL translocation breakpoint cluster region (bcr),16 referred to here as MLL topoII breakage. Such breakage can be detected after treatment by the topoII inhibitors commonly used in induction chemotherapy, including etoposide/VP16 and anthracyclins, but also are found in fewer than 5% of AMLs analyzed prior to chemotherapy,17 when they may reflect exposure to environmental topoisomerase inhibitory activity.18 Their recognition is important in order not to misclassify them as MLL translocations.
We have compared the distribution of FLT3-ITD and D835 in a series of AML patients with known MLL status. FLT3 abnormalities are common in patients with MLL duplication or topoII breakage (> 50%), but, particularly FLT3-ITD, are relatively rare in cases with MLL translocation (15%). Accurate quantification of FLT3 transcript levels showed that D835 mutations occur exclusively in patients with high-level FLT3 expression, whereas FLT3-ITD are either associated with high-level FLT3 expression and found predominantly in monocytic AML or associated with lower level FLT3 expression and found predominantly in PML-RARA and DEK-CAN AML.
Patients, materials, and methods
Patient samples
Patients were treated according to the European Organisation for the Research and Treatment of Cancer (EORTC) AML10 protocol in 5 French centers. Approval was obtained from the participating institutional review boards for these studies. Informed consent was provided according to the Declaration of Helsinki. Cells were ficolled prior to DNA and total RNA extraction. Diagnostic samples of peripheral blood and bone marrow origin from 168 de novo adult AML patients (mean age, 40.3 years) were prospectively screened for AML1-ETO and CBFβ-MYH11 fusion transcripts by RT-PCR as described19,20 and were subsequently analyzed for FLT3-ITD and D835 mutations. Cytogenetic data were available for 155 AML patients; data were considered normal if at least 20 mitoses could be analyzed, and MLL abnormal in the presence of molecular evidence of rearrangement, regardless of chromosome 11q23 status. All patients with available material were prospectively screened for MLL rearrangement by Southern blotting11 using an exon 8-12 cDNA probe, thus allowing detection of MLL translocations, MLL duplication, and MLL breakage at the topoII site located 1.5 kilobase upstream to the 3′ BamHI site delimiting the MLL bcr.16 MLL rearranged cases were analyzed by RT-PCR for MLL duplication and the AF6, AF9, AF10, ENL, EEN, and ELL fusion transcripts, as described,11 complemented by FISH when possible. An additional 48 AML patients (7 infants, 9 children, and 32 adults) with MLL abnormalities treated on a variety of clinical protocols were included. The incidence of MLL duplication and topoII breakage in pediatric AML was not evaluated and most were submitted for analysis because of an MLL/11q23 abnormality. We also analyzed 16 PML-RARA and 9 DEK-CAN AML and 31 B-lineage ALL patients, including 14 with MLL translocations, from a variety of clinical protocols, for FLT3 mutation and expression. Control bone marrow samples were taken from patients with BCR-ABL–negative essential thrombocythemia.
Detection of FLT3-ITD
One microgram of total RNA was reverse transcribed as described,21 and RNA quality and quantity were assessed relative to the ABL housekeeping gene, as designed within the Europe against Cancer network on an ABI PRISM 7700 (Applied Biosystems, Foster City, CA). Samples with CtABL values above 32 (fluorescent threshold, 0.1) were considered uninterpretable. PCR was performed from DNA (500 ng) and cDNA (0.2 μg RNA equivalent) using 0.3 μM exon 13 (FLT3E (S) TGGTGTTTGTCTCCTCTTCATTGT) and exon 15 (FLT3Q (AS) GTTGCGTTCATCACTTTTCCAA) primers22 in 50 μL with 5 mM MgCl2, 0.75 mM deoxynucleoside triphosphate (dNTP), 1.25 U AmpliTaq GOLD (Applera) at 94°C for 8 minutes, followed by 35 cycles at 94°C for 30 seconds, 60°C for 1 minute, 72°C for 1 minute, and a final elongation step at 72°C for 10 minutes. PCR products were analyzed on 3% agarose gel or by Genescan analysis using a 5′HEX-Flt3Q–labeled primer, analyzed on an ABI PRISM 310.
PCR detection of FLT3 exon 20 D835 mutations
Mutations within FLT3 exon 20 were detected as described.23 Twenty microliters of PCR product was digested with 5U EcoRV (New England Biolabs, Frankfurt, Germany) for 1 hour at 37°C and D835 mutations were detected by 12% polyacrylamide gel electrophoresis (PAGE) of digested PCR products. Cases with heteroduplex formation were further characterized by excision of nondigested and heteroduplex bands, reamplification, and direct sequencing using BigDye terminator cycle kits (Applied Biosystems) on an ABI 3700.
Quantification of FLT3 transcript
Real-time quantitative (RQ)–PCR was performed using an ABI PRISM 7700 in a total volume of 25 μL in the presence of 300 nM primers, 100 ng cDNA, and 200 nM probe, using Europe Against Cancer standardized conditions.24 Each run included 4 nontemplate controls to exclude false-positive results. Results were normalized for RNA quality relative to ABL using delta Ct (ΔCt = CtABL–CtFLT3). Exon 20-21 FLT3 primers and probes were designed using Primer Express (Applied Biosystems). FLT3 ex20 sense: CAATCACATCCAAATTCCAGCA; FLT3 ex21 antisense: GCCCTGAGATTTGATCCGAG; FLT3 ex21 probe: CCTGGTTCAAGAGAAGTTCAGATACACCCG.
Statistical analysis
Comparison between median FLT3 expression in different groups was performed using ANOVA analysis of variation and Pearson χ2 frequency analysis. A P value less than .05 was considered statistically significant. All statistical calculations were performed using version 5.97 STATISTICA software (StatSoft, Krakow, Poland).
Results
Detection of FLT3-ITD from DNA versus RNA
Screening of 168 adult patients with de novo AML treated on the EORTC AML10 protocol showed FLT3-ITD in 18% and D835 in 6% (Table 1), in keeping with previous, albeit larger, series.5,7 FLT3-ITD varied in length from 24 to 90 bp. Six patients with AML demonstrated loss of the wild-type allele and were considered as homozygous FLT3-ITD; these correspond to cases classified as having a high mutant-to-wild-type ratio in certain series.5 The remaining 25 demonstrated ITD of variable intensity relative to wild-type FLT3, suggestive of subclones.9 From DNA were screened 106 patients; from cDNA, 42; and from both, 20. In contrast to Schnittger et al,25 comparison of the sensitivity of detection of FLT3-ITD from diagnostic DNA and cDNA in 4 cases showed a 1-2 log higher sensitivity (0.1%-0.5%) from cDNA compared with DNA (10%; Figure 1A). Parallel analysis of diagnostic samples showed similar profiles, although detection of additional, minor FLT3-ITD alleles was more frequent from cDNA (Figure 1B), when loss of the wild-type allele was also more common (Figure 1C). Only patients with easily detected peaks were considered ITD positive, and all patients who were classified as positive from cDNA were also positive from DNA. We therefore considered that FLT3-ITD detection from cDNA is likely to be more sensitive, at least for the detection of minor subclones, and is more likely to detect apparent homozygosity, since transcriptionally silent, nonleukemic DNA will be detected from DNA, but not RNA. No case demonstrated both FLT3-ITD and D835.
Distribution of FLT3-ITD, D835, and transcript expression by FAB morphological and cytogenetics subgroup in AML
. | Distribution, n (%) . | . | FLT3* expression (n) ITD/D835- . | |
|---|---|---|---|---|
. | FLT3 ITD+ . | FLT3 D835+ . | . | |
| Total AML† | 31 of 168 (18) | 10 of 164 (6) | 2.6 ± 1.4 (100) | |
| FAB | ||||
| M0 | 0 of 7 (0) | 0 of 6 (0) | 4 ± 1.5 (4) | |
| M1‡ | 7 of 35 (20) | 2 of 34 (6) | 2.2 ± 1.7 (16) | |
| M2, AMLI-ETO+ | 0 of 16 | 0 of 16 | 2.2 ± 1.3 (8) | |
| M2, AMLI-ETO- | 3 of 22 (14) | 1 of 22 (5) | 2.5 ± 1 (9) | |
| M4eo, CBFβ-MYH11+ | 1 of 15 (7) | 1 of 15 (7) | 2.1 ± 1.2 (8) | |
| M4, CBFβ-MYH11- | 7 of 25 (28) | 3 of 24 (13) | 2.5 ± 1.2 (11) | |
| M5‡ | 11 of 38 (30) | 3 of 38 (8) | 3.4 ± 0.9 (27) | |
| M6 | 1 of 3 (33) | 0 of 3 (0) | 1.7 (1) | |
| CG‡ | ||||
| Normal | 19 of 67 (28) | 2 of 65 (3) | 3.0 ± 1.2 (24) | |
| Complex, 5, 7 | 1 of 14 (7) | 1 of 12 (8) | 1.9 ± 1.9 (8) | |
| Abnormal§ | 2 of 22 (9) | 1 of 22 (5) | 2.9 ± 1.2 (12) | |
| AML1-ETO | 2 of 20 (10) | 0 of 20 (0) | 2.2 ± 1.2 (10) | |
| CBFβ-MYH1∥ | 1 of 17 (6) | 2 of 17 (12) | 2.1 ± 1.2 (8) | |
| DEK-CAN† | 6 of 9 (67) | 0 of 9 (0) | 2.9 ± 0.8 (2) | |
| PML-RARA† | 6 of 16 (38) | 1 of 16 (6) | 2.4 ± 1.8 (7) | |
| MLL† | ||||
| Dupl.¶ | 5 of 16 (31)¶ | 3 of 16 (19) | 2.1 ± 2.1 (6) | |
| Topo∥ | 3 of 8 (38) | 2 of 8 (25) | 2.9 ± 1 (2) | |
| Translocation | ||||
| Total | 2 of 40 (5) | 4 of 39 (10) | 2.8 ± 1.3 (22) | |
| Age | ||||
| Adults | 2 of 25 (8) | 2 of 25 (8) | ND | |
| Younger than 1 y# | 0 of 7 (0) | 1 of 7 (14) | ND | |
| 1-15 y** | 0 of 8 (0) | 1 of 7 (14) | ND | |
| MLL-partner | ||||
| AF6 | 0 of 8 (0) | 1 of 8 (13) | ND | |
| AF9 | 0 of 9 (0) | 1 of 9 (11) | ND | |
| AF10 | 0 of 7 (0) | 1 of 7 (14) | ND | |
| ELL | 0 of 6 (0) | 1 of 6 (17) | ND | |
| ENL | 1 of 3 (33) | 0 of 3 (0) | ND | |
| Other†† | 0 of 6 (0) | 0 of 5 (0) | ND | |
| t(11,17) | 1 of 1 (100) | 0 of 1 (0) | ND | |
. | Distribution, n (%) . | . | FLT3* expression (n) ITD/D835- . | |
|---|---|---|---|---|
. | FLT3 ITD+ . | FLT3 D835+ . | . | |
| Total AML† | 31 of 168 (18) | 10 of 164 (6) | 2.6 ± 1.4 (100) | |
| FAB | ||||
| M0 | 0 of 7 (0) | 0 of 6 (0) | 4 ± 1.5 (4) | |
| M1‡ | 7 of 35 (20) | 2 of 34 (6) | 2.2 ± 1.7 (16) | |
| M2, AMLI-ETO+ | 0 of 16 | 0 of 16 | 2.2 ± 1.3 (8) | |
| M2, AMLI-ETO- | 3 of 22 (14) | 1 of 22 (5) | 2.5 ± 1 (9) | |
| M4eo, CBFβ-MYH11+ | 1 of 15 (7) | 1 of 15 (7) | 2.1 ± 1.2 (8) | |
| M4, CBFβ-MYH11- | 7 of 25 (28) | 3 of 24 (13) | 2.5 ± 1.2 (11) | |
| M5‡ | 11 of 38 (30) | 3 of 38 (8) | 3.4 ± 0.9 (27) | |
| M6 | 1 of 3 (33) | 0 of 3 (0) | 1.7 (1) | |
| CG‡ | ||||
| Normal | 19 of 67 (28) | 2 of 65 (3) | 3.0 ± 1.2 (24) | |
| Complex, 5, 7 | 1 of 14 (7) | 1 of 12 (8) | 1.9 ± 1.9 (8) | |
| Abnormal§ | 2 of 22 (9) | 1 of 22 (5) | 2.9 ± 1.2 (12) | |
| AML1-ETO | 2 of 20 (10) | 0 of 20 (0) | 2.2 ± 1.2 (10) | |
| CBFβ-MYH1∥ | 1 of 17 (6) | 2 of 17 (12) | 2.1 ± 1.2 (8) | |
| DEK-CAN† | 6 of 9 (67) | 0 of 9 (0) | 2.9 ± 0.8 (2) | |
| PML-RARA† | 6 of 16 (38) | 1 of 16 (6) | 2.4 ± 1.8 (7) | |
| MLL† | ||||
| Dupl.¶ | 5 of 16 (31)¶ | 3 of 16 (19) | 2.1 ± 2.1 (6) | |
| Topo∥ | 3 of 8 (38) | 2 of 8 (25) | 2.9 ± 1 (2) | |
| Translocation | ||||
| Total | 2 of 40 (5) | 4 of 39 (10) | 2.8 ± 1.3 (22) | |
| Age | ||||
| Adults | 2 of 25 (8) | 2 of 25 (8) | ND | |
| Younger than 1 y# | 0 of 7 (0) | 1 of 7 (14) | ND | |
| 1-15 y** | 0 of 8 (0) | 1 of 7 (14) | ND | |
| MLL-partner | ||||
| AF6 | 0 of 8 (0) | 1 of 8 (13) | ND | |
| AF9 | 0 of 9 (0) | 1 of 9 (11) | ND | |
| AF10 | 0 of 7 (0) | 1 of 7 (14) | ND | |
| ELL | 0 of 6 (0) | 1 of 6 (17) | ND | |
| ENL | 1 of 3 (33) | 0 of 3 (0) | ND | |
| Other†† | 0 of 6 (0) | 0 of 5 (0) | ND | |
| t(11,17) | 1 of 1 (100) | 0 of 1 (0) | ND | |
ND indicates not done.
Mean ± standard deviation ΔCt levels of expression are given for each subcategory of FLT3-ITD/D835-negative AML. Levels of expression for positive cases are indicated in Figure 3.
FAB classification is restricted to the 168 AML 10 patients (FAB and karyotypic details were not available for 7 and 13 patients, respectively), whereas the cytogenetic classification (CG) also includes 9 DEK-CAN, 16 PML-RARA, and 48 non-AML 10 AML cases with MLL abnormalities, which were treated on a variety of different protocols.
Includes 2 AML1-ETO, 1 CBFβ-MYH11 in each FAB group.
Includes trisomies (+2, 8, 11, 21) and all other structural chromosomal translocations.
Includes one patient with CBFβ-MYH11 and MLL duplication and one inv(16) for which the type of CBFβ-MYH11 fusion transcript could not be identified.
Includes one 11-year-old child with hyperdiploid AML M1 (51, XY, +5, +6, +9, +10, +13).
Includes 3 MLL-AF10, 4 MLL-ELL.
Includes 4 MLL-AF9, 3 MLL-AF10, one unidentified t(9; 11).
Includes 3 t(9, 11), 2 t(11, 19), and one t(11,22).
Fluorescent genescan detection of FLT3-ITD. (A) Sensitivity of detection from cDNA (left column) versus DNA (right column). PCR products from a representative case (unique patient number [UPN] 529), after dilution into PBL DNA/RNA. The size of the ITD is given relative to the wild-type allele and the fluorescent scale indicated. (B) Subclone detection: minor subclones were more frequently detected from cDNA, as demonstrated for 2 representative patients. (C) Differences in relative intensity of the wild-type compared with FLT3-ITD alleles for cDNA versus DNA in a representative AML.
Fluorescent genescan detection of FLT3-ITD. (A) Sensitivity of detection from cDNA (left column) versus DNA (right column). PCR products from a representative case (unique patient number [UPN] 529), after dilution into PBL DNA/RNA. The size of the ITD is given relative to the wild-type allele and the fluorescent scale indicated. (B) Subclone detection: minor subclones were more frequently detected from cDNA, as demonstrated for 2 representative patients. (C) Differences in relative intensity of the wild-type compared with FLT3-ITD alleles for cDNA versus DNA in a representative AML.
FLT3 abnormalities in CBF leukemias
FLT3-ITD and, to a lesser extent, D835 were most frequent in M5 and CBF-negative M4 AML (Table 1). As expected, there was a high incidence of FLT3-ITD in AML10 patients with a normal karyotype, and a low incidence in those with complex cytogenetic abnormalities or with CBF leukemias, demonstrating a t(8;21) and/or AML1-ETO (n = 20) or inv16 and/or CBFβ-MYH11 (n = 17, including 12 type A, 4 variants, and one for which molecular subtype was not available). None of the type A CBFβ-MYH11 AML cases showed FLT3 abnormalities, whereas 3 of 4 cases with variant CBFβ-MYH1126-28 did so, including an M4Eo with FLT3-ITD and a type E CBFβ-MYH11, an M4 with D835 and type I, and an M1 with D835 and a type D fusion transcript. The fourth case, an M4Eo with type E CBFβ-MYH11 demonstrated no FLT3 abnormalities but showed MLL duplication. Neither of the AML1-ETO+ AML cases with abnormal FLT3 were FAB M2 (one M1, one M5). FLT3 abnormalities therefore correlate with atypical CBF leukemias.
AML cases with MLL duplication or topoII breakage frequently demonstrate FLT3 abnormalities
Among adult EORTC AML10 patients, 158 of 168 were screened for MLL abnormalities by Southern blotting, followed by RT-PCR and/or FISH.11 This allowed identification of 6 MLL translocations (3.8%), 5 MLL duplications (3.2%), and 5 AML with MLL topoII breakage (3.2%). One patient with a trisomy 11 (48, XY,+8,+11) but no evidence of MLL translocation by FISH demonstrated a major rearranged band on DNA digestion with BamHI but not with HindIII, XbaI, BglII, SacI, and as such was considered to represent a BamHI polymorphism. No patient demonstrated multiple MLL abnormalities.
Of the 10 patients with MLL duplication or topoII breakage, 7 showed FLT3 abnormalities, whereas only 1 of 6 patients with MLL translocations did so. We therefore extended our FLT3 analysis to a total of 64 AML cases with MLL abnormalities by Southern blotting, karyotype, or FISH (Table 1). FLT3-ITD or D835 occurred in 13 of 24 (54%) of patients with MLL duplication or topoII breakage, compared with only 6 of 40 (15%; P = .000 58) AML cases with MLL translocations. FLT3-ITD was less frequent in adults with MLL translocation than in AML patients overall and was absent in pediatric AML patients with MLL translocation, although not from the single pediatric case of MLL duplication (Table 1). The frequency of D835 in AML patients with MLL translocation was not decreased but frequently corresponded to minor deletions, as evidenced by heteroduplex formation (Figure 2). A 4-month-old infant with MLL-AF10 (UPN1344) demonstrated a deletion of R834 and D835 (CGAGAT). A 6-year-old child with MLL-AF9 AML (UPN507) showed a classical D835H point mutation and deletion of I836 (ATC), as described in infant ALL.29 It was not possible to determine whether these occurred in different subclones. A 74-year-old patient (UPN1236) with AML-M4 and MLL duplication also demonstrated a 3-bp deletion of D835. Deletion therefore accounted for 3 of 9 FLT3 mutations in MLL AML, compared with 0 of 8 non-MLL adult AML (Table 1). These data extend recent evidence29 suggesting that the D835/I836 abnormalities, which occur in leukemias with MLL translocation, preferentially correspond to deletions rather than point mutations, at least in pediatric cases.
Detection of FLT3 D835 mutations. Undigested and EcoRV-digested PCR products from UPN1344 with a 6-bp deletion of R834 and D835 and UPN507 with D835H mutation and deletion of I836. The upper doublets correspond to heteroduplexes. UPN306 and UPN3162 are D835 mutated and normal AML cases, respectively. MW indicates molecular weight markers.
Detection of FLT3 D835 mutations. Undigested and EcoRV-digested PCR products from UPN1344 with a 6-bp deletion of R834 and D835 and UPN507 with D835H mutation and deletion of I836. The upper doublets correspond to heteroduplexes. UPN306 and UPN3162 are D835 mutated and normal AML cases, respectively. MW indicates molecular weight markers.
Quantitation of FLT3 expression in AML
Since both types of FLT3 abnormalities were more common in monocytic AML, when FLT3 expression is maintained1 we hypothesized that FLT3 and MLL intragenic abnormalities could result from increased or prolonged accessibility to a common genotoxic stress. We therefore quantified both wild-type and mutated FLT3 expression by RQ-PCR, using exon 20 and 21 primers. We also analyzed PML-RARA and DEK-CAN AML since FLT3 abnormalities are particularly common in these categories (Table 1 and Table 2). As expected,4 FLT3-ITDs were more common in PML-RARA M3 with 5′ bcr3 breakpoints (4 of 7, 57%) compared to cases with bcr1/2 3′ breakpoints (2 of 9, 22%). FLT3 expression was quantified in 11 PML-RARA and 7 DEK-CAN cases.
Distribution of FLT3-ITD+ cases according to FLT3 expression level
. | High . | Low . |
|---|---|---|
| FAB | ||
| M1 | 2 | 5 |
| M2 | 2 | 0 |
| M4 | 3 | 1 |
| M5 | 8 | 0 |
| Cytogenetics | ||
| Norm | 9 | 5 |
| CBF | 1 | 0 |
| Comp/abn | 2 | 0 |
| PML/RARA | 0 | 3 |
| DEK/CAN | 2 | 3 |
| MLL trans. | 0 | 1 |
| MLL topoll/dupl. | 5 | 1 |
. | High . | Low . |
|---|---|---|
| FAB | ||
| M1 | 2 | 5 |
| M2 | 2 | 0 |
| M4 | 3 | 1 |
| M5 | 8 | 0 |
| Cytogenetics | ||
| Norm | 9 | 5 |
| CBF | 1 | 0 |
| Comp/abn | 2 | 0 |
| PML/RARA | 0 | 3 |
| DEK/CAN | 2 | 3 |
| MLL trans. | 0 | 1 |
| MLL topoll/dupl. | 5 | 1 |
“High” indicates cases with FLT3 expression higher than ΔCt2.7; “low,” cases with FLT3 expression lower than ΔCt2.7); CG, cytogenetic category; and comp/abn, cases with complex or other abnormalities.
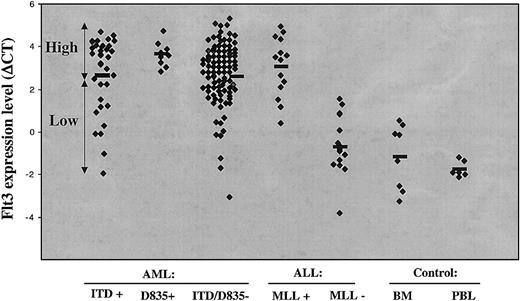
Mean FLT3 expression in control bone marrows (Figure 3) was similar to that seen in B-lineage ALL with no evidence of MLL abnormality, but was, as expected,30 higher in cases with MLL translocation, which were predominantly MLL-AF4 (12 of 14). FLT3 was transcribed at similar levels in ITD/D835-negative AML, at 1-1.5 log higher levels than controls. The highest levels were seen in M0 and M5 (Table 1). FLT3 expression was significantly higher in cases of AML with D835 compared with ITD/D835-negative cases (P = .036), suggesting that high-level FLT3 expression and/or accessibility predisposes to D835.
Real-time FLT3 quantitation.FLT3 transcript levels are expressed as ΔCt relative to ABL transcripts in different acute leukemia and normal control samples. Bar indicates mean expression level.
Real-time FLT3 quantitation.FLT3 transcript levels are expressed as ΔCt relative to ABL transcripts in different acute leukemia and normal control samples. Bar indicates mean expression level.
The impact of FLT3-ITD on FLT3 expression was more complex, since mean levels (ΔCt = 2.7) were not different from ITD/D835-negative cases and were heterogeneous (Figure 3). The morphological and genotypic profiles of FLT3-ITD+ AML with high level FLT3 expression (ΔCt > 2.7) was therefore compared to cases with FLT3 levels less than 2.7 (Table 2). The former included most AML M2, M4, and M5 and/or those with a normal karyotype or intragenic MLL abnormalities. As with D835, FLT3-ITD in these cases is likely to be due, at least in part, to accessibility of the FLT3 locus. In contrast, FLT3 levels with ΔCt less than 2.7 were found in most AML with AML M1 or M3 and/or cases with PML-RARA or DEK-CAN and in a proportion of AML with normal cytogenetics. FLT3 levels were exceptionally low in ITD-positive M3, reflecting lower level expression in bcr3 (0.4 ± 1.8, n = 4) versus bcr1/2 cases (2.9 ± 0.9, n = 7).
Discussion
In this study we show that AML with MLL duplication or topoII induced breakage demonstrate FLT3-ITD or D835 in more than 50% of cases, suggesting that the FLT3 and MLL loci present similar susceptibility to agents inducing DNA damage and/or DNA repair defects. FLT3 abnormalities in AML are either associated with high-level FLT3 expression and probably result from prolonged exposure of the locus to a variety of genotoxic insults, or occur in the presence of lower-level FLT3 expression and may result from modified DNA/chromatin configuration. FLT3 D835 are restricted to cases with high-level FLT3 expression, whereas FLT3-ITDs are found in both categories. We also show that FLT3 abnormalities in CBF leukemias are virtually restricted to atypical cases.
FLT3-ITDs were seen in more than 30% of patients with MLL duplication or topoII breakage, representing the highest incidence after PML-RARA and DEK-CAN, equivalent to that seen in cases with a normal karyotype, to which category most of these cases belong. Schnittger et al25 identified FLT3-ITD in only 1 of 33 karyotypic 11q23 aberrations but found MLL duplication in approximately 10% of their FLT3-ITD group, in keeping with our data. AML cases with MLL duplication and topoII breakage also showed D835 abnormalities in more than 20% of cases. No subgroup with such a high incidence has been described, although D835 are more common in monocytic/M5 AML and in cases with normal karyotype.5,23 We found only 3% in the latter, reflecting classification of cases with MLL duplication and topoII breakage elsewhere. Mutations around D835 in AML cases with MLL translocation may be relatively frequent in children, since we found 2 of 28 (14%) compared with only 2.5% in pediatric AML cases in general.31 These preferentially involve microdeletion, as recently described for I836 in MLL+ infant ALL.29 D835 mutations are found in 20% of MLL+ infant ALL compared with 6% of cases without MLL translocation.31 We found neither FLT3-ITD nor D835 in 12 B-lineage ALL cases with MLL-AF4, but 8 of our 12 MLL-AF4 were adults. Thiede et al identified 13 I836 deletions and 2 insertions among 87 mutations in adult AML,5 but their MLL status was not specified. Taken together, FLT3 mutation, and particularly microdeletion, appears to be particularly common in pediatric AML and ALL with MLL translocation, whereas FLT3-ITD is relatively rare.
The correlation between FLT3-ITD/D835 with MLL topoII breakage suggests that they may be initiated by double-strand breaks induced by topoII inhibition. Sequence analysis identified 2 overlapping topoII sites within the EcoRV/D835 site in FLT3 exon 20 and also within the exon 14 ITD hot spot (Figure 4). TopoII breakages are repaired by both nonhomologous end joining (NHEJ) and homologous recombination (HR).32 NHEJ can result in excision of several bases. AML cases demonstrate increased activity of a particularly error-prone NHEJ system.33 Such a pathway may lead to FLT3 microdeletions, reinforced by the fact that our FLT3 microdeletions were restricted to MLL+ cases, known to be linked to topoII inhibition.34 The environmental agents responsible for topoII inhibition, other than chemotherapeutic agents, include dietary flavonoids,18 probably potentiated by genetic heterogeneity in the capacity to detoxify such substances.35 MLL duplications, which also correlated with FLT3-ITD and D835, are thought to result from defective DNA repair by HR, often involving Alu repeats.36 Alu repeats are unlikely to be involved in FLT3-ITD since the closest such repeats are situated 200 bp downstream to exon 14. FLT3-ITDs also have been reported to be due to HR errors, following loop formation within a palindromic hot spot.37
Potential topoII sites in FLT3 exons 14 and 20. Top: consensus topoII recognition sequence. Bottom: the entire exons 14 and 20 were searched, but only sequences flanking potential sites are shown. Percentages under potential sites refer to homology on the sense/antisense DNA strands, respectively. Homologous nucleotides are underlined and tyrosine residues identified as vertical arrows. The EcoRV site that covers D835 and I836 in exon 20 is underlined.
Potential topoII sites in FLT3 exons 14 and 20. Top: consensus topoII recognition sequence. Bottom: the entire exons 14 and 20 were searched, but only sequences flanking potential sites are shown. Percentages under potential sites refer to homology on the sense/antisense DNA strands, respectively. Homologous nucleotides are underlined and tyrosine residues identified as vertical arrows. The EcoRV site that covers D835 and I836 in exon 20 is underlined.
To our knowledge, there is no evidence that FLT3-ITD or D835 modifies FLT3 transcription level or message stability. FLT3 D835 were clearly associated with higher levels of FLT3 expression, in keeping with the higher incidence of D835 in monocytic M4/M5 AML.5 It suggests that high-level transcription and/or prolonged accessibility predisposes to D835 abnormalities. FLT3-ITD in the context of high-level FLT3 expression also may correspond to this mechanism. In contrast, frequent FLT3-ITD but relatively low-level FLT3 expression was found within PML-RARA, DEK-CAN, and/or a proportion of AML M1 and cases with normal cytogenetics. The DEK gene on chromosome 6p23 codes for a protein, which modifies DNA topology by interacting with topoII, which alters superhelical density of DNA in chromatin, in a histone-dependent manner. DEK also substantially reduces the replication efficiency of chromatin, but not of naked, DNA templates.38 The identification of FLT3-ITD in 60%-90% of cases of DEK-CAN AML5 suggests that DEK-CAN also modifies DNA topology and/or replication efficiency. The PML protein also plays a role in genomic stability, since it colocalizes in nuclear bodies with the Bloom syndrome gene protein, BLM, which is required for normal PML function. BLM encodes a RecQ DNA helicase, whose absence leads to genomic instability, characterized by high levels of sister chromatid exchange (SCE) and a predisposition to cancer, including myeloid leukemia.39 PML-RARA delocalizes PML and BLM, leading to functional inactivation and increases in SCE.40 It is possible that this is the mechanism responsible for FLT3-ITD in AML M3, although PML-RARA can also modify histone acetylation.41 Unequal SCE during HR also could explain loss of the wild-type FLT3 allele in the absence of allelic deletion by FISH.5 It is therefore likely that FLT3-ITD in PML-RARA and DEK-CAN AML result from altered DNA/chromatin configuration with consequent replication defects.
Not all acute leukemias with high-level FLT3 expression are at risk of FLT3-ITD, as exemplified by AML M0 in the present series. Other AML with rare or absent FLT3-ITD include most MLL translocations and typical CBF AML. FLT3 is unlikely to play a leukemogenic role in these cases. MLL translocated AML have been divided into those demonstrating a short latency, including MLL-AF9 and MLL-AF1012 and those with longer latency and a probable requirement for a second event, including MLL-CBP and MLL-ELL.42,43 We would expect that the rare MLL translocated cases with FLT3-ITD would be found in the latter category.
All 5 CBF leukemias demonstrating FLT3 abnormalities were atypical, either with regard to their stage of maturation arrest for AML1-ETO AMLs or the type of CBFβ-MYH11 fusion transcript. Typical CBF leukemias are more frequently associated with c-KIT or RAS mutations.44 Variant CBFβ-MYH11 and t(15;17) have been described in AML secondary to treatment with topoII inhibitors.27,45,46 The coexistence of a variant CBFβ-MYH11 and an MLL duplication in one of our, apparently de novo, AML cases is in keeping with a possible common etiological mechanism. Since FLT3-ITD is particularly common in PML-RARA AML with bcr3 breakpoints,4,25 it is possible that the mechanism(s) inducing bcr3 breakage might be similar to that of FLT3-ITD.
Genetic abnormalities in genes situated on trisomic chromosomes are increasingly recognized in AML. AML1 mutations are found in 19% of cases of AML with trisomy 21,47 and MLL duplication is frequent in AML with trisomy 11,14 when it can coincide with FLT3-ITD.48 We found a probable MLL point mutation, identified as a BamHI polymorphism, in an AML with trisomy 11. Similarly, the only pediatric case of FLT3 occurred in an AML patient with trisomy 13. Trisomy 13 was seen in fewer than 2% of more than 1000 successful AML10 karyotypes. While it is possible either that mismatch repair is less efficient for aneuploid chromosomes or that mutation predisposes to aneuploidy, it is more likely that both aneuploidy and mutation result from related DNA replication defects.
Prepublished online as Blood First Edition Paper, June 5, 2003; DOI 10.1182/blood-2003-01-0162.
Supported by the European Organisation for the Research and Treatment of Cancer (EORTC) Leukemia Group, the Fondation contre la leucémie of the Fondation de France, l'Association de Recherche sur le Cancer (ARC), the Direction de Recherche Clinique de l'Assistance Publique-Hopitaux de Paris (PHRC 97-106), and the Institut National de la Santé et de la Recherche Médicale (INSERM).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Isabelle Radford-Weiss, Alain Bernheim, Frank Viguier, Sylvie Ramon, Christian Bastard, and Françine Mugneret for cytogenetic data; André Baruchel, Claire Berger, and all the clinicians from the French EORTC AML10 centers for contributing patient material; and the members of the EORTC leukemia group. Marta Libura is a recipient of a scholarship from the Postgraduate School of Molecular Medicine at the Medical University of Warsaw and is currently working in the Department of Hematology of the Jagellonian University in Cracow.

![Figure 1. Fluorescent genescan detection of FLT3-ITD. (A) Sensitivity of detection from cDNA (left column) versus DNA (right column). PCR products from a representative case (unique patient number [UPN] 529), after dilution into PBL DNA/RNA. The size of the ITD is given relative to the wild-type allele and the fluorescent scale indicated. (B) Subclone detection: minor subclones were more frequently detected from cDNA, as demonstrated for 2 representative patients. (C) Differences in relative intensity of the wild-type compared with FLT3-ITD alleles for cDNA versus DNA in a representative AML.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/6/10.1182_blood-2003-01-0162/6/m_h81834923001.jpeg?Expires=1765901743&Signature=5MuJWubcjjW1~wZ1SF~beX-TxGNvY~-YIbXRXzqERq0ReVm6GqCxB5E7au4RhJa2u2Uzbni9mgRezK6ujm~tmTb-8V7t1DBM80MCxLoJbTiabI8envcPVaQth5heJn6P3OTgxRf22Tgu3rrSbRJpHeyTACPAjKFRRoFchDf57sTwkKQ0rO9GfYC5j-FdYN8L9YaRSMjxAOXpr8AX~8tAfxtzc9kjjfEoYvG4ibn6bj7WWSRapV2QIsc1w0NfggnqVT9xCapRmShfUHXE0~HtKzb8hFcGJjedcFtsrSoh5Z372nuzLzkrGF2gtNcXEmX9XP5Ji7s9YoCFdB-sIbfyKQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal