Abstract
To elucidate the biologic and clinical heterogeneity of adult pro-B acute lymphoblastic leukemia (ALL) (ie, terminal deoxynucletidyl-transferase–positive[TdT+], CD19+, CD10–, surface immunoglobulin–negative [SIg–]), we evaluated 66 patients enrolled in the Italian multicentric Gruppo Italiano Malattie Ematologiche dell'Adulto (GIMEMA) 0496 study between October 1996 and December 1999. The ALL1/AF4 fusion transcript, originating from the t(4;11) translocation, was detected in 24 patients (36.4%), and the BCR/ABL chimeric product was found in 6 patients (9%), while the remaining 36 cases (54.6%) were ALL1/AF4-BCR/ABL–negative. A white blood cell (WBC) count higher than 50 × 109/L was found in 13 of 24, 2 of 6, and 6 of 36 of the ALL1/AF4-positive, BCR/ABL-positive, and ALL1/AF4-BCR/AB–negative patients, respectively (P = .007). None of the 24 ALL1/AF4-positive patients coexpressed the CD13 and/or CD33 myeloid antigens. By contrast, CD13 and CD33 molecules were detected, respectively, in 3 of 6 and in 14 of 33 cases of the BCR/ABL-positive patient group, and in 2 of 6 and 9 of 35 cases of the ALL1/AF4-BCR/ABL–negative patient group. These differences still remained statistically significant even if the BCR/ABL-positive patients were excluded from the analysis. A complete remission (CR) was achieved in 52 (83.4%) of the 62 patients with ALL evaluable for response to treatment. CR rates were similar in the 3 genotypic groups. By contrast, comparing patients with or without the ALL1/AF4 gene the probability of remaining in continuous complete remission (CCR) at 3.5 years was 16% and 49.8%, respectively (P = .005). Our data demonstrate that in adult pro-B-ALL a distinction should be made between pro-B-ALL cases with and without the ALL1/AF4 or the BCR/ABL chimeric genes, since the absence of both of these fusion genes correlates with a significantly better clinical outcome after intensive polychemotherapy treatment without hematopoietic stem cell transplantation.
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous entity that includes distinct clinico-biologic and genotypic forms, as well as variable therapeutic response rates and outcome. Therefore, several classification systems have been proposed over the years in an attempt to better identify ALL subtypes and to stratify patients into prognostic subgroups. Immunophenotypic-based profiles have represented until recently the most widely used categorization system.1,2 Over the last few years, an increasingly relevant significance has been attributed to genetic lesions in acute leukemia (AL).3,4 The cloning and characterization of genes involved in the most frequent translocations associated with AL has allowed the identification of genetic markers in approximately 50% of patients with either ALL or acute myeloid leukemia (AML). A number of clinical studies have shown that these genetic alterations represent better prognostic markers compared with other previously used indicators.4,5
Pro-B-ALL, one of the most immature ALL subtypes, is characterized by the expression of cytoplasmic (cy) or membrane CD22, CD19, CD24, and CD79a, and by the absence of CD10 and of cy or surface immunoglobulins. A relevant fraction (∼ 30%) of pro-B-ALL cases is associated with the t(4;11)(q22;q23) translocation that fuses the ALL1 (MLL/HRX/Hrtx1) gene at 11q23 to the AF4 (FEL) gene at 4q22.6-10 This abnormality is one of the most frequent genetic alterations in ALL, and it characterizes a subset of ALL with aggressive clinical features, such as hyperleukocytosis, organomegaly, frequent central nervous system (CNS) involvement, and poor prognostic outcome. The prevalence of this aberration in ALL varies in the different age groups: it is extremely frequent in infants younger than 12 months (up to 60%-70%) and less common in children and adults (5% and 10% of cases, respectively).11,12
The close association between pro-B immunophenotype, neonatal leukemia, and t(4;11) translocation accounts for the well-known poor prognostic significance usually attributed to this immunophenotypic subtype in childhood ALL. By contrast, little is known about the correlation between pro-B-ALL, genetic markers, and clinical characteristics in adults. In the experience of the German Multicentric Group, patients with pro-B-ALL were considered at high prognosis risk, regardless of the presence or absence of the t(4;11) alteration, and, as a consequence, they were considered as candidates for stem cell allotransplantation (SCT) in first complete remission (CR).13
To better elucidate the biologic and clinical heterogeneity of this immunophenotypic subset, in the present study we characterized by a multiparametric approach, including genotypic analysis, 66 adult patients with pro-B-ALL enrolled in the Gruppo Italiano Malattie Ematologiche dell'Adulto (GIMEMA) 0496 multicenter protocol. We show that the absence of both the ALL1/AF4 or BCR/ABL fusion genes correlates with a significantly better clinical outcome for patients receiving intensive consolidation therapy that does not include SCT.
Patients, materials, and methods
Patients
Between October 1996 and December 1999, 403 adult patients aged 15 to 60 years with newly diagnosed ALL with the exclusion of mature B-ALL were enrolled in the GIMEMA 0496 protocol. Of these, 66 patients (16.3%) exhibited a pro-B-ALL immunophenotype and were included in this study. The GIMEMA 0496 study contemplates a central handling of cell samples at diagnosis and at predetermined time intervals during the clinical follow-up.14 Since the availability of patients' cell samples was regarded as an essential prerequisite to enter the study, evaluable cytogenetic and molecular data were obtained from 302 (75%) of 403 and in 395 (98%) of 403 patients, respectively, of the GIMEMA 0496 trial. As a consequence, evaluable karyotypes and molecular results were available for 56 (84.8%) of 66 and 66 (100%) of 66 of pro-B-ALL cases, respectively. Cytogenetic and molecular studies on all cases were performed at presentation in the laboratories of the Department of Cellular Biotechnologies and Hematology of the University “La Sapienza” of Rome, Department of Biomedical Sciences of the University of Turin, Department of Hematology of the University of Perugia, Department of Biomedical Sciences, Hematology Unit, of the University of Ferrara, and Unity of Molecular Onco-Hematology, faculty of Medicine of the University “Federico II” of Napoli. The diagnosis of ALL was based on standard morphologic and cytochemical evaluation,15 and on immunophenotypic criteria. Blast cells from all cases were negative for myeloperoxidase (MPO) and nonspecific esterase using conventional cytochemistry. Bright uniform expression of CD19 and absence of CD10 surface antigen were required for the diagnosis of pro-B-ALL.
Mononuclear cells were obtained from bone marrow (BM) samples after centrifugation on a Ficoll-Hypaque gradient, washed, and cryopreserved in 4 M guanidium isothiocyanate at –20°C for total RNA extraction or as dry pellet for DNA extraction.
Informed consent for both treatment protocol and diagnostic procedures was obtained from all patients. The present study was approved by the institutional review board of the University “La Sapienza” of Rome, Italy.
Immunophenotyping
Immunophenotyping was not carried out at central reference laboratories, but it was performed at the single peripheral hematologic center. However, a uniformly agreed to immunophenotypic work-up was performed in all cases and the results were centrally reviewed. The following panel of monoclonal antibodies directed against lymphoid and myeloid antigens: terminal deoxynucletidyl-transferase (TdT), CD22, CD3, CD7, CD10, CD19, CD22, CD13, CD33, CD34, HLA-DR, intracytoplasmic immunoglobulin M (IgM), and surface Ig.
A sample was considered positive for surface antigens if more than 20% of leukemic cells expressed fluorescence intensity. Positivity for TdT and cytoplasmic antigens was arbitrarily defined as more than 10% of cells exhibiting nuclear or intracytoplasmic fluorescence compared with negative control.
Cytogenetic analysis
Cytogenetic analyses were performed on BM cells after 24 hours of unstimulated culture. GTG bands with trypsin were obtained. Karyotypes were reviewed and defined according to the International System for Human Cytogenetic Nomenclature (ISCN) criteria.
DNA analysis
High-molecular-weight DNA was obtained from cell pellets following proteinase-K digestion and phenol-chloroform extraction, digested to completion with BamHI and BglII, size fractionated by electrophoresis through a 0.8% agarose gel, and transferred to nitrocellulose membranes. Filters were prehybridized for 16 to 24 hours at 42°C in a solution containing 50% deionized salmon sperm, 5 × SSC, 5 × Denhardt solution, and 100 μg/mL denatured salmon sperm DNA, and then hybridized in the same solution with the addition of the denatured B859 probe, previously labeled with the 32P random priming technique. Filters were washed twice for 30 minutes in 0.2 × SSC and 0.1% sodium dodecyl sulfate (SDS) at 65°C and exposed with an intensifying screen at –70°C for 24 to 72 hours. The B859 probe is a cDNA insert that explores the entire ALL1 breakpoint cluster region, as previously reported.6
RNA preparation
Total RNA was extracted from cells cryopreserved in guanidium isothiocyanate according to the method of Chomczynsky and Sacchi.16 The quality of RNA was assessed on an ethidium bromide–stained 1% agarose gel containing 2.2 M formaldehyde.
Reverse transcription–polymerase chain reaction (RT-PCR) amplification
In vitro reverse transcription of 1 μg total RNA to cDNA was performed at 42°C for 20 minutes in a 20-μL reaction volume containing 2.5 U cloned Moloney murine leukemia virus (Mo-MLV), reverse transcriptase, and random examers as primers, using a commercial kit (Gene Amp RNA PCR kit; Applied Biosystems, Foster City, CA). RT-PCR amplification was performed following the procedures previously reported.17 Briefly, a volume of 5 μL was then diluted with 95 μL of a PCR mixture containing 1.5 mM Mg2Cl, 50 mM KCl, 10 mM Tris (tris(hydroxymethyl)aminomethane) HCl (pH 8.3), 200 μM deoxynucleoside triphosphates (dNTPs), 2.5 U TAQ DNA polymerase (Applied Biosystems), and 15 pmol of primers Ex5 and AF4.1. After initial denaturation at 94°C for 2 minutes, 30 cycles of amplification were performed on a DNA Thermal Cycler (Applied Biosystems). One cycle of denaturation, annealing, and extension consisted of 94°C for one minute, 56°C for one minute, and 72°C for one minute, respectively. At the end, 1 μL of a 1:10 dilution of the first PCR product was used for a second round of amplification for 30 further cycles using the primers AF4.1 and EX6 (half-nested PCR). Finally, one tenth of PCR products was run on a 2% agarose gel stained with ethidium bromide and visualized under an ultraviolet lamp. The sequences of the primers used were as follows: Ex 5, 5′-GAGGATCCTGCCCCAAAGAAAAG-3′ (sense); Ex 6, 5′-CGCCCAAGTATCCCTGTAAAAC-3′ (sense); and AF4.1, 5′-TGAGCTGAAGGTCGTCTTCGAGCAT-3′ (antisense).
Amplification of the normal ALL1 gene mRNA was performed with the same cDNA preparation and the same conditions used to identify the ALL1/AF4 junctions, using the following primers: Ex 5 and Ex6 as sense primers, and the Ex 9 antisense primer (Ex9, 5′-TTGTAGCCTGATGTTGCCTTCCACA-3′).
Negative controls (all reagents plus water) were included in all PCR experiments. To rule out the possibility of a cDNA contamination of RNA samples, all positive tests were repeated mixing all reagents without RNA.
Treatment strategy
Patients were treated according to the GIMEMA 0496 protocol that was derived from the ALLVR589 scheme reported by Todeschini et al18 that consists of induction, consolidation, CNS prophylaxis, and maintenance/reinduction phases, as detailed in Figure 1. This program includes a 4-drug (prednisone [PDN], vincristine [VCR], danorubicin [DNR], and asparaginase [ASP]) induction with high-dose danorubicin (90 mg/m2), followed by 2 courses of high-dose cytosine arabinoside (Ara-C; 2 g/m2 every 12 hours as 3-hour infusion on days 1 and 2) with etoposide as consolidation, and by 3 years of maintenance treatment. In patients with a white blood cell (WBC) count higher than 100 × 109/L at diagnosis, cyclophosphamide (300 mg/mq × 6 doses) was added during the consolidation phase. Philadelphia (Ph)–positive ALL patients received a different consolidation treatment that consisted of high-dose Ara-C and mitoxantrone (HAM) followed by autologous or allogeneic stem cell transplantation in first CR, based on the availability of an HLA-identical sibling and according to the EIGLE study.19
A schematic representation of the GIMEMA 0496 treatment plan. The figure was derived from Todeschini et al18 with permission.
A schematic representation of the GIMEMA 0496 treatment plan. The figure was derived from Todeschini et al18 with permission.
Patients were considered in CR if, after completing the induction treatment, they had normal peripheral blood counts (polymorphonuclear leukocyte [PMN] count > 1.5 × 109/L; hemoglobin [Hb] level > 100 g/L [10 g/dL]; platelet count > 100 × 109/L with no circulating blast cells) and less than 5% blast cells in the BM with a normal cellularity. Patients who did not obtain CR after the induction course were considered resistant.
Statistical analysis
The primary study end points were achievement of CR, duration of first CR (continuous complete remission [CCR]), and overall survival (OS). We compared the prevalence of risk factors and the incidence of end points among patient groups using χ2 or the Fisher exact test. Survival was defined as the time from diagnosis to death or date of the last follow-up. CCR was calculated from the time of achieving CR to relapse or date of last follow-up. For the calculation of the actuarial probability of CCR, patients who died in CR were censored at the time of death.
The probability of OS and CCR were estimated using the Kaplan-Meier method.20 The log-rank test21 was used to compare treatment effect and risk factor categories. Using the Simon and Lee22 method, 95% confidence intervals (95% CIs) for these probabilities and the median survival times were obtained. All tests were 2-sided, accepting P < .05 as indicating a statistically significant difference. Median follow-up time was estimated by reversing the codes for the censoring indicator in a Kaplan-Meier analysis. Logistic regression and Cox proportional hazard regression models23,24 were performed to examine and check for treatment results and the risk factors affecting CR rate and time to event. The logistic and Cox models were performed using the SAS procedures LOGISTIC and PHREG, respectively (SAS Institute, Cary, NC).24 Analysis of the full trial population was followed by analysis of relevant subgroups.
Results
Enrolled into the 0496 protocol between October 1996 and December 1999 were 403 patients with ALL from 49 Italian centers participating in the GIMEMA studies. Of these, 66 (16.3%) were diagnosed as having a pro-B-ALL. Considering only B-lineage ALL, pro-B cases accounted for 21.4% of cases. As stated in the patients' enrollment criteria, the availability of diagnostic cell samples allowed for a successful molecular evaluation in all 66 pro-B-ALL cases. At Southern blot analysis, an ALL1 genomic rearrangement resulting in the ALL1/AF4 fusion transcript, originating from the t(4;11) translocation, was detected in 24 patients (36.4%). The BCR/ABL chimeric product was found in 6 patients (9%), while the remaining 36 cases (54.6%) were negative for both fusion transcripts. The clinico-biologic characteristics of the 66 patients grouped according to the presence/absence of ALL1/AF4 or BCR/ABL fusion transcripts are reported in Table 1. The median age of the entire patient population was 32.5 years, ranging from 15 to 59.4 years, whereas the median age in the ALL1/AF4-positive, BCR/ABL-positive, and ALL1/AF4-BCR/ABL–negative groups were 32.4 years (range, 15-55.6 years), 34.1 years (range, 19.6-59.4 years), and 31.3 years (range, 18.4-55.8 years), respectively. As shown in Tables 1 and 3, the statistical analysis of the prevalence of risk factors and of the incidence of end points among patient groups was performed either considering the 3 groups of patients (ie, ALL1/AF4-positive, BCR/ABL-positive, and ALL1/AF4-BCR/ABL–negative) or limiting the comparison to the ALL1/AF4-positive and ALL1/AF4-BCR/ABL–negative groups. In both cases, we did not observe any statistical differences with regard to sex and age distribution. By contrast, a statistically significant greater percentage of patients with WBC counts higher than 50 × 109/L was present in the ALL1/AF4-positive patients compared with ALL1/AF4-BCR/ABL–negative and BCR/ABL-positive patients (P = .001 and P = .006).
Clinico-biologic characteristics of pro-B-ALL patients grouped according to the 3 genotypic subtypes
. | . | 1 . | 2 . | . | 3 . | . |
|---|---|---|---|---|---|---|
| Features . | Total no. of patients (%), n = 66 . | ALL1/AF4-positive (%), n = 24 . | ALL1/AF4-BCR/ABL-negative (%), n = 36 . | P, 1 vs 2 . | BCR/ABL-positive (%), n = 6 . | P, 1 vs 2 vs 3 . |
| Sex | NS | NS | ||||
| M | 32 (48) | 13 (54) | 15 (42) | 4 (67) | ||
| F | 34 (52) | 11 (46) | 21 (58) | 2 (33) | ||
| Age | NS | NS | ||||
| 16 y to 30 y | 29 (44) | 7 (29) | 20 (56) | 2 (33) | ||
| 31 y to 50 y | 30 (45) | 14 (58) | 14 (39) | 2 (33) | ||
| 51 y to 60 y | 7 (11) | 3 (13) | 2 (5) | 2 (33) | ||
| WBC count | .001 | .001 | ||||
| 50 × 109/L or lower | 45 (68) | 11 (46) | 30 (83) | 4 (67) | ||
| Higher than 50 × 109/L | 21 (32) | 13 (54) | 6 (17) | 2 (33) | ||
| CD13 | .0001 | .0001 | ||||
| Positive | 17 (27) | 0 (0) | 14 (42) | 3 (50) | ||
| Negative | 46 (73) | 24 (100) | 19 (58) | 3 (50) | ||
| CD33 | .007 | .007 | ||||
| Positive | 11 (17) | 0 (0) | 9 (26) | 2 (33) | ||
| Negative | 53 (83) | 23 (100) | 26 (74) | 4 (67) | ||
| CD34 | .00006 | .00006 | ||||
| Positive | 44 (64) | 10 (42) | 33 (92) | 5 (83) | ||
| Negative | 22 (33) | 14 (58) | 3 (8) | 1 (17) |
. | . | 1 . | 2 . | . | 3 . | . |
|---|---|---|---|---|---|---|
| Features . | Total no. of patients (%), n = 66 . | ALL1/AF4-positive (%), n = 24 . | ALL1/AF4-BCR/ABL-negative (%), n = 36 . | P, 1 vs 2 . | BCR/ABL-positive (%), n = 6 . | P, 1 vs 2 vs 3 . |
| Sex | NS | NS | ||||
| M | 32 (48) | 13 (54) | 15 (42) | 4 (67) | ||
| F | 34 (52) | 11 (46) | 21 (58) | 2 (33) | ||
| Age | NS | NS | ||||
| 16 y to 30 y | 29 (44) | 7 (29) | 20 (56) | 2 (33) | ||
| 31 y to 50 y | 30 (45) | 14 (58) | 14 (39) | 2 (33) | ||
| 51 y to 60 y | 7 (11) | 3 (13) | 2 (5) | 2 (33) | ||
| WBC count | .001 | .001 | ||||
| 50 × 109/L or lower | 45 (68) | 11 (46) | 30 (83) | 4 (67) | ||
| Higher than 50 × 109/L | 21 (32) | 13 (54) | 6 (17) | 2 (33) | ||
| CD13 | .0001 | .0001 | ||||
| Positive | 17 (27) | 0 (0) | 14 (42) | 3 (50) | ||
| Negative | 46 (73) | 24 (100) | 19 (58) | 3 (50) | ||
| CD33 | .007 | .007 | ||||
| Positive | 11 (17) | 0 (0) | 9 (26) | 2 (33) | ||
| Negative | 53 (83) | 23 (100) | 26 (74) | 4 (67) | ||
| CD34 | .00006 | .00006 | ||||
| Positive | 44 (64) | 10 (42) | 33 (92) | 5 (83) | ||
| Negative | 22 (33) | 14 (58) | 3 (8) | 1 (17) |
NS indicates not significant.
Therapeutic results of the GIMEMA 0496 trial according to the 3 genotypic subgroups
. | . | 1 . | 2 . | . | 3 . | . |
|---|---|---|---|---|---|---|
| Therapeutic response . | Total no. of patients (%), n = 62 . | ALL1/AF4-positive (%), n = 23 . | ALL1/AF4-BCR/ABL-negative (%), n = 33 . | P, 1 vs 2 . | BCR/ABL-positive (%), n = 6 . | P, 1 vs 2 vs 3 . |
| CR | 52 (84) | 21 (91) | 28 (85) | 3 (50) | ||
| Resistant | 4 (6) | 0 (0) | 3 (9) | NS | 1 (17) | NS |
| DI | 6 (10) | 2 (9) | 2 (6) | 2 (33) | ||
| Relapses | .003 | .003 | ||||
| Yes | 31 (60) | 17 (81) | 11 (39) | 3 (100) | ||
| No | 21 (40) | 4 (19) | 17 (61) | 0 (0) | ||
| Deaths | .01 | .002 | ||||
| Yes | 36 (58) | 17 (74) | 13 (39) | 6 (100) | ||
| No | 26 (42) | 6 (26) | 20 (61) | 0 (0) |
. | . | 1 . | 2 . | . | 3 . | . |
|---|---|---|---|---|---|---|
| Therapeutic response . | Total no. of patients (%), n = 62 . | ALL1/AF4-positive (%), n = 23 . | ALL1/AF4-BCR/ABL-negative (%), n = 33 . | P, 1 vs 2 . | BCR/ABL-positive (%), n = 6 . | P, 1 vs 2 vs 3 . |
| CR | 52 (84) | 21 (91) | 28 (85) | 3 (50) | ||
| Resistant | 4 (6) | 0 (0) | 3 (9) | NS | 1 (17) | NS |
| DI | 6 (10) | 2 (9) | 2 (6) | 2 (33) | ||
| Relapses | .003 | .003 | ||||
| Yes | 31 (60) | 17 (81) | 11 (39) | 3 (100) | ||
| No | 21 (40) | 4 (19) | 17 (61) | 0 (0) | ||
| Deaths | .01 | .002 | ||||
| Yes | 36 (58) | 17 (74) | 13 (39) | 6 (100) | ||
| No | 26 (42) | 6 (26) | 20 (61) | 0 (0) |
DI indicates deaths during induction treatment.
By definition, all 66 patients with pro-B-ALL displayed a bright CD19 expression and absence of the CD10 surface antigen. In particular, the CD10 antigen was detected in less than 10% of bone marrow cells in 62 of the 66 cases, and between 10% to 20% of cells in the remaining 4 cases. A coexpression of CD13 or CD33 myeloid antigens was detected in 27% and 17% of cases, respectively. However, as listed in Table 1, none of the ALL1/AF4-positive patients expressed CD13 or CD33, whereas these antigens were detected in 3 (50%) of 6 and 14 (42%) of 33 of the BCR/ABL-positive cases and in 2 (33%) of 6 and 9 (26%) of 35 of the ALL1/AF4-BCR/ABL–negative patients, respectively. The CD34 antigen was expressed in 44 (66%) of 66 patients with a different distribution among the 3 genotypic groups. Again, as reported in Table 1, these differences were statistically significant both when the 3 groups of patients were considered together and when the comparison was limited to the ALL1/AF4-positive and ALL1/AF4-BCR/ABL–negative groups only.
Of the 66 patients, 56 (84.8%) also had an evaluable karyotype. As illustrated in Table 2, the comparison between cytogenetic and molecular analyses showed that of the 23 ALL1/AF4-positive cases with successful cytogenetic analysis 18 presented the corresponding t(4;11) translocation, while 3 cases had a normal and 2 a pseudodiploid karyotype, with alterations different from the t(4;11) translocation. The corresponding t(9;22) alteration was detected in 3 of the 4 BCR/ABL-positive patients with an evaluable cytogenetic analysis, while the fourth patient had a normal karyotype. Finally, among the 29 ALL1/AF4-BCR/ABL–negative cases with an available cytogenetic study, 13 presented a normal karyotype, whereas a pseudodiploid and an hyperdiploid karyotype were detected in 8 and 8 cases, respectively.
Cytogenetic analysis according to the 3 genotypic groups
Karyotype . | Total no. of patients, n = 66 . | ALL1/AF4-positive, n = 24 . | BCR/ABL-positive, n = 6 . | ALL1/AF4-BCR/ABL-negative, n = 36 . |
|---|---|---|---|---|
| Normal | 17 | 3 | 1 | 13 |
| t(4;11) | 18 | 18 | — | — |
| t(9;22) | 3 | — | 3 | — |
| Pseudodiploid | 10 | 2 | — | 8 |
| Hyperdiploid | 8 | — | — | 8 |
| Failure | 5 | 1 | — | 4 |
| Not done | 5 | — | 2 | 3 |
Karyotype . | Total no. of patients, n = 66 . | ALL1/AF4-positive, n = 24 . | BCR/ABL-positive, n = 6 . | ALL1/AF4-BCR/ABL-negative, n = 36 . |
|---|---|---|---|---|
| Normal | 17 | 3 | 1 | 13 |
| t(4;11) | 18 | 18 | — | — |
| t(9;22) | 3 | — | 3 | — |
| Pseudodiploid | 10 | 2 | — | 8 |
| Hyperdiploid | 8 | — | — | 8 |
| Failure | 5 | 1 | — | 4 |
| Not done | 5 | — | 2 | 3 |
— indicates no cases.
Table 3 summarizes the overall results of induction therapy and clinical outcome of adult patients with pro-B-ALL according to genotypic groups. Of 66 patients, 4 were not evaluable for therapeutic response (ALL1/AF4-positive, 1 case; ALL1/AF4-BCR/ABL–negative, 3 cases) due to major protocol violation, treatment refusal, or psychotic disorder. Thus, treatment results were evaluated considering the remaining 62 patients. No statistically significant differences in CR rate were observed either when the 3 genotypic groups were considered all together or when the ALL1/AF4-positive and the ALL1/AF4-BCR/ABL–negative groups were considered alone. Of 62 patients, 52 (83.8%) achieved CR after induction treatment. Due to treatment failure, 4 patients (BCR/ABL-positive, 1 case; ALL1/AF4-BCR/ABL–negative, 3 cases) failed to achieve a CR, and 6 patients died during induction therapy due to infection (5 cases) and myocardial infarction (1 case). Of the 52 complete responders, 31 relapsed (59%). In particular, as shown in Table 3, disease recurrence occurred in 17 (81%) of 21, 3 (100%) of 3, and 11 (39%) of 28 patients in the ALL1/AF4-positive, BCR/ABL-positive, and ALL1/AF4-BCR/ABL–negative groups, respectively. Statistically significant differences in the distribution of relapses were observed considering the 3 subgroups of patients all together or the ALL1/AF4-positive and the ALL1/AF4-BCR/ABL–negative groups separately (Table 3). Among the entire group of patients we observed 36 deaths: 6 died during induction treatment, 3 patients died due to resistant disease, and 26 patients died due to relapse of their disease. The remaining ALL1/AF4-BCR/ABL–negative patient died in CCR. Similar to what was observed for relapses, deaths were differently distributed among the 3 patient groups.
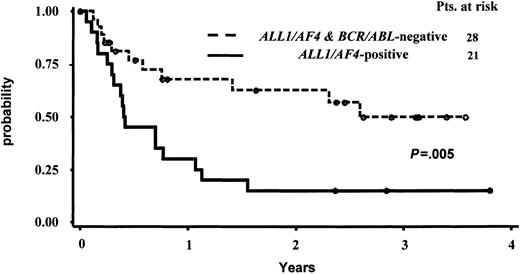
Due to the limited number of cases with the BCR/ABL alteration, which are much more frequent in CD10+ cases, and to the different postinduction treatment received by these latter 6 patients, the statistical analysis on OS and CCR was calculated comparing the ALL1/AF4-positive and ALL1/AF4-BCR/ABL–negative groups only. The median survival follow-up for the entire patient series and for the 26 alive patients was 376 days and 921 days, respectively. The median duration CCR time was 223 days for the responder patients and 876 days for the 21 patients persisting in CCR at the time of analysis. The actuarial probabilities of CCR and OS are shown in Figures 2 and 3. For the 28 CR pro-B-ALL patients without ALL1/AF4 or BCR/ABL alterations, the median remission duration was 949 days and the probability of being in CCR at 3.5 years was 49.8% (95% CI, 35%-73%) By contrast, for the 21 CR patients with ALL1/AF4 the median remission duration was 152 days and the probability of being in CCR at 3.5 years was 15% (95% CI, 0%-31%). These differences were statistically significant (P = .005). The estimated median survival was 370 versus 1360 days in patients with the ALL1/AF-4 or without the ALL1/AF-4 or BCR/ABL alterations (P = .03), while the probability of OS was 23% (95% CI, 6%-41%) and 53% (95% CI, 33%-73%), respectively. With respect to the clinical outcome of the 6 BCR/ABL-positive patients, they all died within 4.5 years from diagnosis; the 3 responders relapsed within 2 years from the achievement of CR.
Actuarial probability of survival in continuous complete remission (CCR) of responder pro-B-ALL patients with and without the ALL1/AF4 fusion transcript.
Actuarial probability of survival in continuous complete remission (CCR) of responder pro-B-ALL patients with and without the ALL1/AF4 fusion transcript.
Actuarial probability of survival of pro-B-ALL patients with and without the ALL1/AF4 fusion transcript.
Actuarial probability of survival of pro-B-ALL patients with and without the ALL1/AF4 fusion transcript.
We also evaluated, by univariate analysis, the influence of age, sex, WBC count, and CD34, CD13, and CD33 expression with respect to CCR and OS rates. Only CD33 expression emerged as another risk factor that significantly affected the actuarial probability of CCR (P = .01). Therefore, to further define the potential prognostic value of CD33 expression, and considering that ALL1/AF4-positive patients did not express this antigen, we focused our analysis on the 35 ALL1/AF4-BCR/ABL–negative pro-B-ALL cases. In this group, CD33 was detected in 9 of the 35 analyzed cases. CR was achieved in 9 of 9 CD33+ and in 18 (69%) of the 26 patients without CD33, whereas a hematologic relapse occurred in 1 of 9 and in 9 of 18 patients with and without CD33 antigen expression, respectively. Therefore, the estimated median duration of CR was not reached in patients in whom the blasts expressed the CD33 molecule, while it was 2.3 years in patients without CD33 coexpression (P = .05).
The results of multivariate analysis showed that among the variables analyzed (ie, age, WBC count, CD34 antigen positivity, and absence or presence of ALL1/AF4 fusion transcript), the presence of the ALL1/AF4 fusion transcript was the only powerful independent prognostic factor influencing CCR (relative risk, 1.7; 95% CI, 1.15-2.49; P = .007).
Discussion
The present study describes the clinico-biologic characterization of the largest series of adult pro-B-ALL reported to date, which includes patients who received a homogeneous treatment program. Our results indicate that this leukemic subset, although immunophenotypically homogeneous, includes genotypically distinct entities that are characterized by a variable response to therapy and outcome.
In our study, the frequency of pro-B-ALL cases, calculated within the entire ALL population, was 16%. This incidence is slightly superior to the percentage reported in other recent studies on adult ALL,13,25,26 but it is close to the percentage 13.4% reported in a previous GIMEMA study carried out in 304 adult patients with ALL immunophenotyped in 43 Italian centers between 1988 and 1991.27 Within B-lineage ALL, the prevalence of pro-B-ALL was 21.4%. With regard to prevalence of genotypic subtypes in pro-B-ALL, we confirm that a consistent group of these patients (36%) expressed the ALL1/AF4 fusion transcripts, while an additional 9% of patients harbored the BCR/ABL gene in their leukemic cells. The ALL1/AF4 fusion gene was never detected in other immunophenotypic ALL subtypes, and its overall frequency was 5.9% among the entire population of 403 ALL patients enrolled in the GIMEMA 0496 study. This incidence is similar to the 4% to 7% reported in other studies, in which, however, the frequency of t(4;11) was calculated by cytogenetic analysis only.28,29 We believe, therefore, that the present data, achieved by combining cytogenetic, Southern blot, and RT-PCR analyses, are likely to closely represent the true incidence of the ALL1/AF4 gene within the overall adult ALL patient population, as well as within the pro-B-ALL subtype.
Our findings confirm that hyperleukocytosis is significantly associated with ALL1/AF4-positive pro-B-ALL and that leukemic cells of adult patients with the ALL1/AF4 fusion never express the myeloid antigens CD13 and CD33.13 This latter feature distinguishes adult ALL1/AF4 pro-B-ALL from infant ALL with t(4;11) translocation, in which the CD13/CD33 antigens are very frequently coexpressed.30,31 We hypothesize that such biologic differences may reflect variable functional states of the hematopoietic precursor during its lifetime. Compared with CD34+ cord blood stem cells, BM CD34+ precursors, for example, show different features, such as the expression of either MPO mRNA and/or protein, and of other myeloid features, and so forth.32-34 However, other panmyeloid markers, such as CD65s and the CD15 antigen or the MPO mRNA, not tested in the present study, are very frequently detected in blasts from both infants and adults with the ALL1/AF4 gene, regardless of the coexpression of the CD13 and/or CD33 antigens.13,35-37
With regard to the clinical outcome of pro-B-ALL, it is well known that in childhood ALL this leukemic subtype is associated with a poor prognosis, while in adults this issue remains controversial. Data from the large French multicentric trial LALA87 did not show a significantly worse prognosis in the group of 45 ALL patients with a pro-B phenotype.25 On the contrary, a German study on the use of a conventional chemotherapy program including neither consolidation courses with high-dose Ara-C and/or methotrexate nor allogeneic SCT in first CR showed that pro-B-ALL patients fared poorer than patients with the other ALL subtypes.38 Therefore, in the German multicentric 03/87 and 04/89 studies, patients with pro-B-ALL were assigned to receive an intensified postremission treatment, consisting of high-dose Ara-C and/or methotrexate followed by SCT, irrespective of the presence of the t(4;11) aberration. This strategy resulted in a significant improvement in remission duration. Moreover, in these latter studies the presence of the t(4;11) translocation did not adversely affect the clinical outcome of pro-B-ALL patients, as the actuarial CCR rates were similar in patients with and without the t(4;11) translocation (ie, 40% vs 45%, at 7 years).13 By contrast, our results demonstrate that, even if BCR/ABL-positive ALL patients were excluded, the presence of the ALL1/AF4 fusion gene is the only powerful independent unfavorable prognostic factor for the clinical outcome of pro-B-ALL patients. In fact, while the therapeutic response rate to induction treatment was similar in the 3 genotypic subgroups, the number of relapses and deaths was significantly higher in ALL1/AF4-positive cases. Thus, the 4-year actuarial CCR and OS rates of our patients were significantly higher in ALL1/AF4-BCR/ABL–negative patients compared with ALL1/AF4-positive patients. In addition, we showed that, after an intensive chemotherapeutic protocol not including SCT, ALL1/AF4-BCR/ABL–negative patients had a relatively favorable clinical outcome with an actuarial CCR rate of 49.8% at 4 years. This observation suggests that ALL1/AF4-BCR/ABL–negative pro-B-ALL patients may be very sensitive to treatments including high doses of antracycline and cytarabine, as scheduled in the GIMEMA 0496 protocol. Moreover, these observations may parallel some in vivo and in vitro data demonstrating the high sensibility of childhood pro-B-ALL to cytarabine.39,40
Compared with the data recently reported by the German multicentric 03/87 and 04/89 trials, which, however, included SCT in first CR, our results indicate that a distinction should be made between pro-B-ALL cases with and without ALL1/AF4-BCR/ABL alterations. In fact, we show that ALL1/AF4-BCR/ABL–negative cases could be successfully managed without hematopoietic SCT in first CR. By contrast, transplantation procedures in first CR should be attempted in ALL1/AF4-positive cases, as they seem to improve the clinical outcome of these patients.
An additional observation that derives from our study is that within the 35 ALL1/AF4-BCR/ABL–negative pro-B-ALL cases, the 9 CD33+ patients fared significantly better than the remaining 26 CD33– cases. The significance of myeloid antigen expression in ALL is controversial, and the antigens may not have an adverse prognostic value.41-43 However, due to the fact that our findings regard a small number of patients referring to a specific ALL subset (namely, ALL1/AF4-BCR/ABL–negative pro-B-ALL) and due to the present observation showing that CD33 coexpression occurred only in the prognostically favorable group of ALL1/AF4-BCR/ABL–negative cases, further data are needed to draw final conclusions on the potential prognostic significance of CD33 expression in ALL patients with the pro-B-ALL immunophenotype.
In conclusion, the present findings demonstrate that a multiparametric characterization of adult ALL that contemplated a uniform immunophenotypic, cytogenetic, and molecular work-up allowed patients with a different prognostic likelihood to be distinguished within pro-B-ALL cases, identifying those patients who may be successfully treated with intensive conventional chemotherapy alone versus those who require SCT in first CR.
Appendix
Participating centers and physicians in the GIMEMA study were: Alessandro Levis, Ospedale SS Antonio e Biagio, Alessandria; Pietro Leoni, Nuovo Ospedale Torrette, Ancona; Ettore Volpe, A. O. S. G. Moscati, Avellino; Umberto Tirelli, CRO Unità Operativa Leucemia, Aviano; Vincenzo Liso, Ematologia-Università degli Studi, Bari; Michele Baccarani, Istituto di Ematologia L. A. Seragnoli, Bologna; Giovanni Quarta, Azienda Ospedaliera “A: Di Summa”, Brindisi; Rosario Giustolisi, Ospedale Ferrarotto, Catania; Antonio Peta, Ospedale A. Pugliese, Catanzaro; Andrea Gallamini, Ospedale S. Croce, Cuneo; Gianluigi Castoldi, Arcispedale S. Anna, Ferrara; Alberto Bosi, Ematologia-Policlinico di Careggi, Florence; Ruggerro Mozzana, Ospedale S. Antonio Abate, Gallarate, Gino Santini, Ospedale S. Martino, Genoa; Franco Patrone, Ematologia-Università degli Studi, Genoa; Angelo De Blasio, Ospedale S. Maria Goretti, Latina; Giulio Nalli, Ospedale Maggiore, Lodi; Marco Bregni, Istututo di Ricerca e Cura a Carattere Scientifico San Raffaele, Milan; Giuseppe Torelli, Università degli Studi, Modena; Marco Montanaro, Ospedale Civile, Montefiascone; Eustachio Miraglia, Ospedale S. Giovanni Bosco, Napoli; Felicetto Ferrara, Ospedale A. Cardarelli, Napoli; Bruno Rotoli, Università Federico II, Napoli; Vincenzo Mettiver, Divisione TERE-Ospedale A. Carderelli, Napoli; Enrica Morra, Ospedale Niguarda “Cà Grande”, Milan; Giancarlo Avanzi, Ospedale Maggiore della Carità, Novara; Attilio Gabbas, Ospedale S. Francesco, Nuoro; Giuseppe Saglio, A. O. S. Luigi Gonzaga, Orbassano-Turin; Salvatore Mirto, Ospedale Cervello, Palermo; Guglielmo Mariani, Università degli Studi, Palermo; Pietro Citarella, Università degli Studi, Palermo; Edoardo Ascari, IRCCS S. Matteo, Pavia; Massimo Martelli, Policlinico Monteluce; Perugia, Giuseppe Visani, Ospedale S. Salvatore, Pesaro; Giuseppe Fioritoni, Ospedale Civile, Pescara; Francesco Ricciuti, Ospedale S. Carlo, Potenza; Francesco Nobile, A. O. Bianchi-Melacrino-Morelli, Reggio Calabria; Luigi Gugliotta, Arcispedale S. Maria Nuova, Reggio Emilia; Franco Mandelli, Università “La Sapienza”, Rome; Giuseppe Leone, Università Cattolica del Sacro Cuore, Rome; Sergio Amadori, Ospedale S. Eugenio, Rome; A. Michele Carella, Ospedale Casa Sollievo Della Sofferenza, San Giovanni Rotondo; Maurizio Longinotti, Università degli Studi, Sassari; Francesco Lauria, A. O. A. Sclavo, Siena; Dr, Patrizio Mazza, Ospedale S. S. Annunziata, Taranto; Mario Boccadoro, Ospedale S. Giovanni, Turin; Eugenio Gallo, Opsedale “le Molinette,” Turin and Giovanni Pizzolo, Policlinico G. B. Rossi, Verona; Italy.
Prepublished online as Blood First Edition Paper, June 5, 2003; DOI 10.1182/blood-2002-12-3822.
Supported by Associazione Italiana contro la Leucemia–Sezione di Roma (ROMAIL), and Consiglio Nazionale delle Ricerche (CNR), legge 27 dicembre 1997 no. 449 (G.C.); by Associazione Italiana per la Ricerca sul Cancro (AIRC), CNR, and Ministero dell'Istruzione e Ricerca Scientifica (MIUR) (C.M.); and by MIUR, Istituto Superiore di Sanità, and AIRC (R.F.).
A complete list of the members of the Gruppo Italiano Malattie Ematologiche dell'Adulto (GIMEMA) appears in the “Appendix.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal