Abstract
The pathogenesis of AIDS-related non-Hodgkin lymphomas (AIDS-NHLs) is associated with chromosomal translocations that deregulate the expression of various oncogenes. Recently, a novel mechanism of genetic lesion, termed aberrant hypermutation, has been identified in diffuse large B-cell lymphoma (DLBCL) of immunocompetent hosts. In these tumors, the somatic hypermutation (SHM) process that normally targets immunoglobulin V (IgV) genes in B cells appears to misfire and causes mutations in the 5′ sequences of multiple proto-oncogenes, including PIM-1, PAX-5, RhoH/TTF, and c-MYC. To investigate whether aberrant hypermutation occurs also in AIDS-NHL, we studied the mutation profile of these 4 genes in various histologic subtypes. Mutations in 1 gene or more were detected in 19 of 39 (48.7%) AIDS-NHL cases (10 of 18 AIDS-diffuse large B-cell lymphoma; 4 of 11 AIDS-Burkitt lymphoma; 4 of 6 AIDS-primary effusion lymphoma; 1 of 4 AIDS-primary central nervous system lymphoma), with 9 of 39 (23.1%) cases carrying mutations in 2 or more genes. Overall, PIM-1 was mutated in 5 of 39 (12.8%), PAX-5 in 8 of 39 (20.5%), RhoH/TTF in 9 of 39 (23.1%), and c-MYC in 7 of 27 (25.9%) AIDS-NHL cases. Mutations were represented mainly by single base pair substitutions (n = 63) with rare deletions/insertions (n = 5) and displayed features typical of the IgV-associated SHM process. In addition, a number of mutations in PIM-1 and c-MYC were found to affect coding exons, leading to amino acid substitutions with likely functional consequences. Analysis of intraclonal heterogeneity documented that the aberrant hypermutation activity may be ongoing in at least some cases. These data indicate that aberrant hypermutation is associated with various subtypes of AIDS-NHL and may represent a major contributor to their pathogenesis.
Introduction
Acquired immunodeficiency syndrome–related non-Hodgkin lymphomas (AIDS-NHLs) represent a significant source of morbidity and mortality among HIV-infected individuals.1,2 According to body location and histologic criteria, the pathologic spectrum of AIDS-NHL includes systemic AIDS-NHL, primary central nervous system lymphoma (AIDS-PCNSL), and primary effusion lymphoma (AIDS-PEL).1-3 Systemic AIDS-NHLs are histologically classified into AIDS-related Burkitt lymphoma (AIDS-BL) and AIDS-related diffuse large B-cell lymphoma (AIDS-DLBCL).3 All AIDS-PCNSLs are histologically represented by DLBCL.3 Despite their clinicopathologic heterogeneity, most AIDS-NHLs derive from B cells that have experienced the germinal center (GC) reaction4 and, therefore, have been exposed to the mechanism of somatic hypermutation (SHM) that normally targets the immunoglobulin variable region (IgV), BCL-6, and FAS genes.5-10 In particular, AIDS-BL and a fraction of AIDS-DLBCLs and AIDS-PCNSLs express markers typical of GC B cells, whereas AIDS-PEL and the remaining fraction of AIDS-DLBCLs and AIDS-PCNSLs are composed of post-GC B cells maturing toward the plasma cell pathway of differentiation.4
The molecular pathogenesis of AIDS-NHL is associated with heterogeneous genetic lesions, including mainly chromosomal translocations that deregulate the expression of various proto-oncogenes.2 In fact, all AIDS-BLs harbor a reciprocal chromosomal translocation between band 8q24, the site of the c-MYC proto-oncogene, and one of the immunoglobulin gene loci, causing transcriptional deregulation of the proto-oncogene.2 A significant fraction of AIDS-DLBCLs associates with chromosomal translocations of BCL-6, a proto-oncogene initially cloned from 3q27 breakpoints in DLBCL of the immunocompetent host and found to encode a zinc-finger transcriptional repressor.2 Chromosomal rearrangements lead to substitution of the BCL-6 promoter with heterologous regulatory elements that derive from different chromosomal sites, including, although not restricted to, the immunoglobulin loci.2 In addition, a significant fraction of AIDS-BLs displays deletions and mutations of the p53 tumor suppressor gene.2
Recently, a second mechanism of genetic lesion, termed aberrant somatic hypermutation, has been identified in DLBCL of the immunocompetent host.11 Normally, the SHM process introduces mainly single nucleotide substitutions into the IgV genes of GC B cells, to produce antibodies with high affinity for the antigen.5,6 SHM requires transcription, targets sequences within 2 kb downstream from the transcription initiation site, and is not restricted to the IgV sequences.7-14 In fact, at least 2 nonimmunoglobulin genes, BCL-6 and Fas/CD95, accumulate mutations in normal GC B cells, suggesting that this mechanism may target more genes than originally suspected.7-10 Although the molecular basis of SHM remains largely unknown, studies from the past 2 years have identified the AID gene, encoding for the activation-induced cytidine deaminase, as an absolute requirement for SHM in humans and mice.15-17 In more than half of the patients with DLBCL, the SHM process appears to misfire and aberrantly targets multiple loci, including well-known proto-oncogenes already implicated in the pathogenesis of lymphoid malignancies (PIM-1, PAX-5, RhoH/TTF, and c-MYC).11 In these genes, the mutations are distributed in the 5′ region, including regulatory as well as coding sequences, occur independently of chromosomal translocations to the immunoglobulin genes, and display similar features to those observed in the IgV genes, although at lower rates.11 In contrast to IgV and BCL-6, however, mutations of PIM-1, PAX-5, RhoH/TTF, and c-MYC do not occur at a significant level in normal GC B cells or in other B-cell malignancies, suggesting a malfunction of SHM associated to DLBCL.11
The present study was aimed at investigating the role of aberrant somatic hypermutation in AIDS-NHL. We have performed a comprehensive analysis exploring the mutation profile of PIM-1, PAX-5, RhoH/TTF, and c-MYC, along with IgV and BCL-6, throughout the clinicopathologic spectrum of AIDS-NHL. We report that aberrant hypermutation is a frequent event in AIDS-NHL, can be detected in lymphomas displaying a GC or post-GC profile, and is ongoing at least in some cases. Overall, these findings indicate that aberrant hypermutation may represent a major contributor to lymphomaganesis in AIDS-NHL.
Materials and methods
Characteristics of AIDS-NHL panel
This study was based on 39 samples of AIDS-NHL, including 29 systemic AIDS-NHL (all primary samples), 4 AIDS-PCNSL (all primary samples), and 6 AIDS-PEL cell lines. According to the Revised European-American classification of lymphoid neoplasms (REAL) and the World Health Organization classification of hematopoietic tumors,3,18 systemic AIDS-NHLs were classified into AIDS-DLBCL (n = 18) and AIDS-BL (n = 11). AIDS-PCNSLs were histologically classified as DLBCL. Primary samples of AIDS-NHL were derived from lymph nodes, bone marrow, peripheral blood, body cavity effusions, or other involved organs and were obtained in the course of routine diagnostic procedures. In all instances, the specimen was collected at diagnosis before specific therapy. According to morphologic and immunophenotypic analysis, the fraction of malignant cells in the pathologic specimen was 95% or more in the case of AIDS-BL and AIDS-PEL, and 70% or more in the case of AIDS-DLBCL. All AIDS-NHL primary samples displayed a major monoclonal B-cell population, as determined by immunophenotypic or immunogenotypic studies. The PEL cell lines included in this study were HBL-6, BC-2, CRO-AP/3, CRO-AP/5, CRO-AP/6, and BCBL-1.19-23 BCBL-1 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH, Bethesda, MD). Gross rearrangements of the c-MYC gene, tested by conventional strategies,24 were detected in all AIDS-BL cases and in 1 of 18 AIDS-DLBCL cases (case 1905).
DNA extraction
Genomic DNA from primary samples and cell lines was isolated by cell lysis followed by digestion with proteinase K and purified by “salting out” extraction and ethanol precipitation.25 In selected cases of AIDS-NHL, paired germ line DNA was also available and was included in the analysis.
Oligonucleotides
The sequences of the oligonucleotides used for mutational analysis of PIM-1, PAX-5, RhoH/TTF, and c-MYC were as follows: PIM1-F, 5′-CCA CAG CCA CAG CCA CAG CC-3′, and PIM1-R, 5′-CGC AGT TGT GGC AGT GCC GC-3′ (for PIM-1); PAX5-F, 5′-CCC AGA GAC TCG GAG AAG CA-3′, and PAX5-R, 5′-AAG AGC TGA AAT GTC GCC GCC G-3′ (for PAX-5); TTF-F, 5′-CCT TAA AAG TAT TTC TTT GGT GTC-3′, and TTF-R, 5′-AAC TCT TCA AGC CTG TGC TG-3′ (for RhoH/TTF); MYC1-F, 5′-CAC CGG CCC TTT ATA ATG CG-3′, and MYC-1R, 5′-CGA TTC CAG GAG AAT CGG AC-3′ (for c-MYC exon 1 fragment); and MYC2-F, 5′-CTT TGT GTG CCC CGC TCC AG-3′, and MYC-2R, 5′-GCG CTC AGA TCC TGC AGG TA-3′ (for c-MYC exon 2 fragment). Where needed, additional primers internal to each amplicon were used, and their sequence is available on request. The oligonucleotides used for mutational analysis of BCL-6 have been previously reported.26 For analysis of the rearranged IgVH genes, a set of 6 VH gene family–specific primers that hybridize to sequences in the VH leader region and an antisense-degenerated JH primer (5′-CTY ACC TGA RGA GAC RGT GAC C-3′) were used.27,28
Sequencing analysis of PIM-1, PAX-5, RhoH/TTF, and c-MYC
Mutational analysis of the 4 proto-oncogenes was performed on selected regions previously shown to contain more than 90% of the mutations found in DLBCL of immunocompetent hosts.11 Mutational analysis of PIM-1 (GenBank accession no. AF386792) was performed by DNA sequencing of a unique polymerase chain reaction (PCR) product of 901 bp encompassing exon 1 to the 3′ region of exon 4 (nucleotides +1062 to +1963). Mutational analysis of PAX-5 (GenBank accession no. AF386791) was performed by direct sequencing of a unique PCR product of 859 bp encompassing the 3′ region of exon 1B and part of intron 1 (nucleotides +630 to +1489). Mutational analysis of RhoH/TTF (GenBank accession no. AF386789) was performed by direct sequencing of a unique PCR product of 844 bp encompassing the 5′ region of intron 1 (nucleotides +265 to +1109). Mutational analysis of c-MYC (GenBank accession no. X00364) was performed by direct sequencing of 2 PCR products of 1300 bp and 582 bp encompassing, respectively, the region downstream to the major P1/P2 promoter (c-MYC exon 1; nucleotides +2289 to +3589) and the region downstream to the minor P3 promoter (c-MYC exon 2, nucleotides +4486 to +5068). PCR products were purified and directly sequenced using a commercially available kit (Thermosequenase; Amersham Life Sciences, Oxford, United Kingdom). [α-33P]-labeled terminator dideoxynucleotides (Amersham Life Sciences) were included in the sequencing mixture. For each DNA fragment analyzed, sequencing of both strands was performed on independent PCR reactions. Sequences obtained were compared with the corresponding germ line sequences, and the first nucleotide of the reference sequence was arbitrarily defined as position +1. Nucleotide changes due to previously reported polymorphisms or occurring more than once in separate cases were considered as polymorphic variants, unless their somatic origin could be proven.11
Sequencing analysis of the BCL-6 gene
Mutations of the BCL-6 5′ noncoding region were assessed by PCR amplification and direct sequencing of a 739-bp genomic region located in the gene intron 1 and previously shown to harbor more than 95% of the mutations detected in B-cell lymphoma (GenBank AY189709; note that the numbering has been corrected [+1 bp] compared with the one reported in our previous publications).8,26,29 PCR conditions have been reported in detail previously.26
Sequencing analysis of IgVH genes
The rearranged IgVH genes were amplified using a set of 6 VH gene family–specific primers that hybridize to sequences in the VH leader region and a JH degenerated primer in separate reactions for each VH primer.26 Samples for which no clonal IgVH gene rearrangement was obtained with leader primers were amplified with a set of 6 VH gene family–specific primers that hybridize to sequences in framework region (FR)I.28 PCR products were purified and subjected to DNA direct sequencing. The rearranged IgVH sequences were then compared with the most homologous germ line sequences using the V-BASE sequence directory (MRC Centre for Protein Engineering, Cambridge, United Kingdom) and MacVector 6.0.1 software (Oxford Molecular Group PLC, Oxford, United Kingdom).
Clonal analysis of PIM-1, PAX-5, and RhoH/TTF genes
The presence of intraclonal heterogeneity, indicative of ongoing hypermutation, was assessed in 7 AIDS-NHL cases (4 AIDS-BL, 2 AIDS-DLBCL, and 1 AIDS-PCNSL) previously shown to harbor mutations in PIM-1, PAX-5, and/or RhoH/TTF by DNA direct sequencing. Genomic DNA was subjected to PCR amplification using the Pyrococcus furiosus (Pfu) Turbo polymerase (Stratagene, La Jolla, CA), as described previously.11 Sizes of PCR products were 715 bp for PIM-1, 932 bp for PAX-5, 875 bp for RhoH/TTF, and approximately 230 bp for the rearranged IgVH genes.11 After 30 cycles (35 for IgVH), PCR products were gel purified, incubated with 0.2 mM dATP (deoxyadenosine triphosphate) and Taq polymerase (GIBCO BRL, Karlsruhe, Germany) for 15 minutes at 72°C, and cloned into the pGEM-T vector (Promega, Madison, WI) as recommended by the manufacturer. For each gene, 25 to 65 clones derived from independent PCR reactions were sequenced and analyzed. As a control for Pfu-introduced background error, a fibroblast cell line (IMR91) and purified tonsillar naive B cells were analyzed using the same approach.11
Analysis of viral infection
Infection by Epstein-Barr virus (EBV) was investigated by PCR with primers SL-1 and SL-3 representative of the EBV nuclear antigen 1 (EBNA-1) gene, as previously reported.29
Analysis of AIDS-NHL histogenetic markers
Histogenetic markers of AIDS-NHL included mutations of IgVH and BCL-6 genes (“Sequencing analysis of IgVH genes”) as well as protein expression of the BCL-6 and CD138 antigens that distinguish the GC phenotype from the post-GC phenotype of AIDS-NHL.4,30 On the basis of previously reported protocols, CD138 and BCL-6 expression were revealed using the B-B4 (Serotec, Oxford, England) and the PG-B6 (Dakopatts A/S, Glostrup, Denmark) monoclonal antibodies, respectively.4
Results
Frequency of aberrant hypermutation in AIDS-NHL subtypes
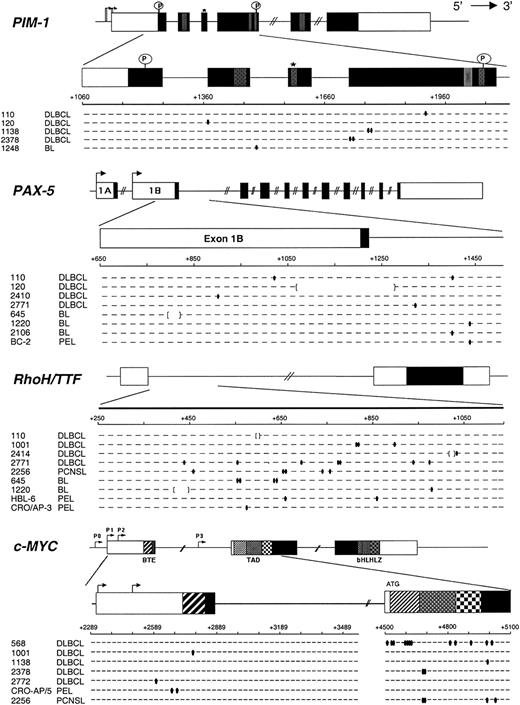
All 39 samples of AIDS-NHL were subjected to sequence analysis of PIM-1, PAX-5, RhoH/TTF, and c-MYC. Mutations of c-MYC were not considered when detected in cases harboring gross rearrangements of the gene with the immunoglobulin loci (all AIDS-BL cases and AIDS-DLBCL 1905), because these mutations may be related to the juxtaposed immunoglobulin sequences, as described for translocated c-MYC alleles in BL.31-33
Results are summarized in Table 1 and are graphically represented in Figure 1. Overall, mutations in at least one of the 4 genes were detected in 19 of 39 (48.7%) AIDS-NHL cases, including 10 of 18 (55.5%) AIDS-DLBCL, 4 of 11 (36.3%) AIDS-BL, 4 of 6 (66.6%) AIDS-PEL, and 1 of 4 (25%) AIDS-PCNSL. Mutations in more than one gene were found in 9 of 39 (23.1%) AIDS-NHL cases, namely 6 of 18 (33.3%) AIDS-DLBCL, 2 of 11 (18.1%) AIDS-BL, and 1 of 4 (25.0%) AIDS-PCNSL. Each of the 4 genes analyzed was altered in a significant fraction of AIDS-NHL, because PIM-1 was mutated in 5 of 39 (12.8%), PAX-5 in 8 of 39 (20.5%), RhoH/TTF in 9 of 39 (23.1%), and c-MYC in 7 of 27 (25.9%) cases. Analysis of the corresponding germ line DNA, performed in selected cases, formally confirmed the somatic nature of the mutations, as previously reported in DLBCL of the immunocompetent host. The observed changes clearly belong to the neoplastic clone and not to nonlymphomatous surrounding B cells or non-B cells according to the following lines of evidence. (1) Previous studies performed on purified naive and GC B cells obtained from reactive tonsils unequivocally demonstrated that normal B cells and fibroblasts do not undergo SHM in these loci at a detectable level.11 (2) Because the analysis was performed by PCR amplification and direct sequencing, only changes present in a significant fraction (≥ 10%) of the template DNA, ie, only clonal mutations, would be detected in the electropherograms. Therefore, the mutations seen can only reflect the status of the tumor clone, as also documented by the amplification of clonal VH gene rearrangements from the same samples. (3) The fraction of malignant cells in the biopsies was 95% or more in AIDS-BL and AIDS-PEL, and 70% or more in AIDS-DLBCL.
Distribution of PIM-1, PAX-5, RhoH/TTF, c-MYC, and BCL-6 mutations throughout the clinicopathologic spectrum of AIDS-NHL
. | Mutated/tested (%) . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | c-MYC . | c-MYC . | . | |||||
| Histology . | PIM-1 . | PAX-5 . | RhoH/TTF . | exon 1 . | exon 2 . | BCL-6 . | |||||
| AIDS-NHL (all) | 5/39 (12.8) | 8/39 (20.5) | 9/39 (23.1) | 3/27 (11.1) | 4/27 (14.8) | 24/39 (61.5) | |||||
| Systemic AIDS-NHL | |||||||||||
| Burkitt lymphoma | 1/11 (9.1) | 3/11 (27.3) | 2/11 (18.2) | NA | NA | 5/11 (45.4) | |||||
| Diffuse large B-cell lymphoma | 4/18 (22.2) | 4/18 (22.2) | 4/18 (22.2) | 2/17 (11.8) | 3/17 (17.6) | 11/18 (61.1) | |||||
| AIDS-PCNSL | 0/4 | 0/4 | 1/4 (25) | 0/4 | 1/4 (25) | 3/4 (75) | |||||
| AIDS-PEL | 0/6 | 1/6 (16.7) | 2/6 (33.3) | 1/6 (16.7) | 0/6 | 5/6 (83.3) | |||||
. | Mutated/tested (%) . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | c-MYC . | c-MYC . | . | |||||
| Histology . | PIM-1 . | PAX-5 . | RhoH/TTF . | exon 1 . | exon 2 . | BCL-6 . | |||||
| AIDS-NHL (all) | 5/39 (12.8) | 8/39 (20.5) | 9/39 (23.1) | 3/27 (11.1) | 4/27 (14.8) | 24/39 (61.5) | |||||
| Systemic AIDS-NHL | |||||||||||
| Burkitt lymphoma | 1/11 (9.1) | 3/11 (27.3) | 2/11 (18.2) | NA | NA | 5/11 (45.4) | |||||
| Diffuse large B-cell lymphoma | 4/18 (22.2) | 4/18 (22.2) | 4/18 (22.2) | 2/17 (11.8) | 3/17 (17.6) | 11/18 (61.1) | |||||
| AIDS-PCNSL | 0/4 | 0/4 | 1/4 (25) | 0/4 | 1/4 (25) | 3/4 (75) | |||||
| AIDS-PEL | 0/6 | 1/6 (16.7) | 2/6 (33.3) | 1/6 (16.7) | 0/6 | 5/6 (83.3) | |||||
NA indicates not applicable; in these cases mutations of c-MYC were not considered because of the presence of gross rearrangements of the gene to the immunoglobulin loci.
Mutational analysis of PIM-1, PAX-5, RhoH/TTF, and c-MYC in AIDS-NHL. The 4 genomic loci are shown, with untranslated and translated sequences (open and filled boxes, respectively); hatched boxes represent relevant protein functional domains. Arrows indicate transcriptional start sites. For each gene, the region amplified for analysis is expanded below and aligned to sequences of mutated AIDS-NHL cases (1 line = 2 alleles). Each small segment represents a 20-bp (PIM-1, PAX-5, and RhoH/TTF) or 25-bp (c-MYC) interval (numbered according to the relative GenBank accession numbers). Ovals indicate single bp substitutions; brackets indicate deletions. *Indicates site of ATP binding in the Pim-1 protein; P, putative phosphorylation sites; BTE, segment controlling block of transcriptional elongation; TAD, transactivation domain; bHLHLZ, basic Helix Loop Helix Leucine Zipper domain.
Mutational analysis of PIM-1, PAX-5, RhoH/TTF, and c-MYC in AIDS-NHL. The 4 genomic loci are shown, with untranslated and translated sequences (open and filled boxes, respectively); hatched boxes represent relevant protein functional domains. Arrows indicate transcriptional start sites. For each gene, the region amplified for analysis is expanded below and aligned to sequences of mutated AIDS-NHL cases (1 line = 2 alleles). Each small segment represents a 20-bp (PIM-1, PAX-5, and RhoH/TTF) or 25-bp (c-MYC) interval (numbered according to the relative GenBank accession numbers). Ovals indicate single bp substitutions; brackets indicate deletions. *Indicates site of ATP binding in the Pim-1 protein; P, putative phosphorylation sites; BTE, segment controlling block of transcriptional elongation; TAD, transactivation domain; bHLHLZ, basic Helix Loop Helix Leucine Zipper domain.
The presence of aberrant SHM was independent of EBV infection, because mutations of one or more of the tested proto-oncogenes were detected in 10 of 23 (43.5%) EBV-positive AIDS-NHL and in 9 of 16 (56.2%) EBV-negative cases.
Detailed characterization of phenotypic markers of histogenesis (expression of BCL-6 and CD138 antigens) was available in 23 cases of AIDS-NHL. All AIDS-BL cases displayed the BCL-6+/CD138– phenotype typical of GC B cells, and all AIDS-PEL cases displayed the BCL-6–/CD138+ post-GC phenotype. Among systemic AIDS-DLBCL and AIDS-PCNSL, 9 cases expressed the BCL-6–/CD138+ phenotype, and 1 case expressed the BCL-6+/CD138– phenotype. Comparison of histogenetic profiles with the occurrence of mutations in PIM-1, PAX-5, RhoH/TTF, and c-MYC showed the presence of aberrant hypermutation in 8 of 15 (53.3%) BCL-6–/CD138+ cases and in 3 of 8 (37.5%) BCL-6+/CD138– cases (not shown).
Mutational spectrum of PIM-1, PAX-5, RhoH/TTF, and c-MYC in AIDS-NHL
The detailed characterization of PIM-1, PAX-5, RhoH/TTF, and c-MYC mutations in AIDS-NHL is reported in Table 2, and their general features are summarized in Table 3.
Mutational analysis of IgVH, BCL-6, PIM-1, PAX-5, RhoH/TTF, and c-MYC in AIDS-NHL
. | . | IgV gene analysis* . | . | Mutations (position)† . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case . | Histology . | VH family (gene) . | % homology . | BCL-6 . | PIM-1 . | PAX-5 . | RhoH/TTF . | c-MYC . | |||||
| 110 | AIDS-DLBCL | VH4 (DP-63) | 77.4 | 440T>G, 476C>G, 480C>T, 822A>C | 1916G>A | 1018G>C, 1400G>A | ΔGTTG (604-607) | — | |||||
| 112 | AIDS-DLBCL | VH4 (DP-63) | 90.0 | 480C>G | — | — | — | — | |||||
| 120 | AIDS-DLBCL | NA | NA | 528A>G, ΔTTCT (784-787) | 1363C>A | Δ199 bp (1069-1267) | — | — | |||||
| 568 | AIDS-DLBCL | NA | NA | — | — | — | — | 4524C>A, 4537G>T, 4547C>G, 4595C>G, 4600A>C, 4606A>G, 4619G>T, 4623C>G, 4814G>A, 4832G>C, 4902A>G, 4975A>T, 5009C>A | |||||
| 1001 | AIDS-DLBCL | NA | NA | — | — | — | 819A>T, 820C>T, 898A>C | 2778C>T | |||||
| 1138 | AIDS-DLBCL | VH2 (S12-9) | 97.8 | — | 1764G>A, 1765G>C | — | — | 4996G>T | |||||
| 1139 | AIDS-DLBCL | NA | NA | 419T>A, 426C>A, 504G>A, 602C>A, 679G>A | — | — | — | — | |||||
| 1902 | AIDS-DLBCL | VH1 (2M27) | 100.0 | — | — | — | — | — | |||||
| 1904 | AIDS-DLBCL | NA | NA | — | — | — | — | — | |||||
| 1905 | AIDS-DLBCL | NA | NA | 480C>G, 610A>T | — | — | — | ‡ | |||||
| 2378 | AIDS-DLBCL | VH3 (DP-48) | 97.3 | 886C>T | 1720C>T, 1721C>G | — | — | 4686C>T, 4689C>T | |||||
| 2410 | AIDS-DLBCL | VH4 (DP-63) | 84.3 | 480C>G, 534G>A, 538A>T, 789C>G | — | 904G>A | — | — | |||||
| 2413 | AIDS-DLBCL | VH1 (DP-75) | 95.3 | 734G>T, 745C>T | — | — | — | — | |||||
| 2414 | AIDS-DLBCL | NA | NA | ΔCGCGC (764-768) | — | — | ΔGTA (1026-1028), 1034G>C | — | |||||
| 2768 | AIDS-DLBCL | VH4 (V4-34) | 93.7 | — | — | — | — | — | |||||
| 2771 | AIDS-DLBCL | VH4 (DP64) | 85.6 | 602C>T, 686C>T, 759G>C, 781T>C, ΔT837, 876C>G, 921C>G, 955T>C, 976G>A, 1024T>C | — | 1324A>T | 448C>T, 558T>C, 694G>A, 783T>C, 784C>T, 945C>A, 987T>C | — | |||||
| 2772 | AIDS-DLBCL | VH4 (DP63) | 86.8 | 780C>T | — | — | — | 2594G>A | |||||
| 2790 | AIDS-DLBCL | NA | NA | — | — | — | — | — | |||||
| 642 | AIDS-BL | VH2 (VII-5) | 94.7 | 480C>T, 518C>A | — | — | — | ‡ | |||||
| 645 | AIDS-BL | NA | NA | — | — | Δ25 bp (805-829) | 557T>G, 563A>G, 638C>T, 643A>G | ‡ | |||||
| 1142 | AIDS-BL | VH4 (DP-79) | 85.1 | +G707, ΔT1099 | — | — | — | ‡ | |||||
| 1143 | AIDS-BL | VH3 (DP-47) | 85.4 | 706T>C, 718T>A, 757G>C | — | — | — | ‡ | |||||
| 1220 | AIDS-BL | VH4 (V4-34) | 96.7 | 563T>G, 836G>A | — | 1436C>G | Δ31 bp (418-448), 987T>C | ‡ | |||||
| 1246 | AIDS-BL | NA | NA | — | — | — | — | ‡ | |||||
| 1248 | AIDS-BL | VH4 (DP-66) | 91.4 | — | 1496G>A | — | — | ‡ | |||||
| 1249 | AIDS-BL | NA | NA | — | — | — | — | ‡ | |||||
| 1907 | AIDS-BL | VH3 (DP-33) | 93.0 | — | — | — | — | ‡ | |||||
| 2106 | AIDS-BL | NA | NA | — | — | 1394C>T | — | ‡ | |||||
| 2415 | AIDS-BL | VH4 (DP-63) | 87.8 | 563T>C | — | — | — | ‡ | |||||
| HBL-6 | AIDS-PEL | VH5 (DP-73) | 94.3 | 821C>G, 1085T>G | — | — | 658A>G, 854A>G | — | |||||
| BC-2 | AIDS-PEL | VH3 (3-73) | 94.0 | 605T>C | — | 1436C>T | — | — | |||||
| BCBL-1 | AIDS-PEL | NA | NA | 843C>G, 850T>A | — | — | — | — | |||||
| CRO-AP/3 | AIDS-PEL | VH3 (DP-47) | 93.5 | 447T>C, 480C>T, 522T>C, 539C>G, 564T>C, 794G>A | — | — | 573G>A | — | |||||
| CRO-AP/5 | AIDS-PEL | NA | NA | 543T>C, 549T>A | — | — | — | 2678T>A, 2691G>A | |||||
| CRO-AP/6 | AIDS-PEL | VH4 (DP-79) | 94.3 | — | — | — | — | — | |||||
| 2254 | AIDS-PCNSL | VH3 (DP-47) | 93.9 | 480C>T, 538A>G | — | — | — | — | |||||
| 2255 | AIDS-PCNSL | VH6 (DP-74) | 91.3 | — | — | — | — | — | |||||
| 2256 | AIDS-PCNSL | VH4 (DP-63) | 78.8 | 489G>A, 497G>A, 499A>T, 549T>G, 554T>G, 567T>G, 569T>C, 623T>G, 783T>C, 809G>A, 821G>A, 918T>G, 957G>A, 971A>C, 1003G>A, 1029T>C | — | — | 456G>A, 652A>C, 669T>C, 748A>T, 757A>G | 4682G>A, 4698C>T, 4993A>G, 5027C>G | |||||
| 2257 | AIDS-PCNSL | VH4 (V4.38) | 91.1 | 445C>G, 500T>A | — | — | — | — | |||||
. | . | IgV gene analysis* . | . | Mutations (position)† . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case . | Histology . | VH family (gene) . | % homology . | BCL-6 . | PIM-1 . | PAX-5 . | RhoH/TTF . | c-MYC . | |||||
| 110 | AIDS-DLBCL | VH4 (DP-63) | 77.4 | 440T>G, 476C>G, 480C>T, 822A>C | 1916G>A | 1018G>C, 1400G>A | ΔGTTG (604-607) | — | |||||
| 112 | AIDS-DLBCL | VH4 (DP-63) | 90.0 | 480C>G | — | — | — | — | |||||
| 120 | AIDS-DLBCL | NA | NA | 528A>G, ΔTTCT (784-787) | 1363C>A | Δ199 bp (1069-1267) | — | — | |||||
| 568 | AIDS-DLBCL | NA | NA | — | — | — | — | 4524C>A, 4537G>T, 4547C>G, 4595C>G, 4600A>C, 4606A>G, 4619G>T, 4623C>G, 4814G>A, 4832G>C, 4902A>G, 4975A>T, 5009C>A | |||||
| 1001 | AIDS-DLBCL | NA | NA | — | — | — | 819A>T, 820C>T, 898A>C | 2778C>T | |||||
| 1138 | AIDS-DLBCL | VH2 (S12-9) | 97.8 | — | 1764G>A, 1765G>C | — | — | 4996G>T | |||||
| 1139 | AIDS-DLBCL | NA | NA | 419T>A, 426C>A, 504G>A, 602C>A, 679G>A | — | — | — | — | |||||
| 1902 | AIDS-DLBCL | VH1 (2M27) | 100.0 | — | — | — | — | — | |||||
| 1904 | AIDS-DLBCL | NA | NA | — | — | — | — | — | |||||
| 1905 | AIDS-DLBCL | NA | NA | 480C>G, 610A>T | — | — | — | ‡ | |||||
| 2378 | AIDS-DLBCL | VH3 (DP-48) | 97.3 | 886C>T | 1720C>T, 1721C>G | — | — | 4686C>T, 4689C>T | |||||
| 2410 | AIDS-DLBCL | VH4 (DP-63) | 84.3 | 480C>G, 534G>A, 538A>T, 789C>G | — | 904G>A | — | — | |||||
| 2413 | AIDS-DLBCL | VH1 (DP-75) | 95.3 | 734G>T, 745C>T | — | — | — | — | |||||
| 2414 | AIDS-DLBCL | NA | NA | ΔCGCGC (764-768) | — | — | ΔGTA (1026-1028), 1034G>C | — | |||||
| 2768 | AIDS-DLBCL | VH4 (V4-34) | 93.7 | — | — | — | — | — | |||||
| 2771 | AIDS-DLBCL | VH4 (DP64) | 85.6 | 602C>T, 686C>T, 759G>C, 781T>C, ΔT837, 876C>G, 921C>G, 955T>C, 976G>A, 1024T>C | — | 1324A>T | 448C>T, 558T>C, 694G>A, 783T>C, 784C>T, 945C>A, 987T>C | — | |||||
| 2772 | AIDS-DLBCL | VH4 (DP63) | 86.8 | 780C>T | — | — | — | 2594G>A | |||||
| 2790 | AIDS-DLBCL | NA | NA | — | — | — | — | — | |||||
| 642 | AIDS-BL | VH2 (VII-5) | 94.7 | 480C>T, 518C>A | — | — | — | ‡ | |||||
| 645 | AIDS-BL | NA | NA | — | — | Δ25 bp (805-829) | 557T>G, 563A>G, 638C>T, 643A>G | ‡ | |||||
| 1142 | AIDS-BL | VH4 (DP-79) | 85.1 | +G707, ΔT1099 | — | — | — | ‡ | |||||
| 1143 | AIDS-BL | VH3 (DP-47) | 85.4 | 706T>C, 718T>A, 757G>C | — | — | — | ‡ | |||||
| 1220 | AIDS-BL | VH4 (V4-34) | 96.7 | 563T>G, 836G>A | — | 1436C>G | Δ31 bp (418-448), 987T>C | ‡ | |||||
| 1246 | AIDS-BL | NA | NA | — | — | — | — | ‡ | |||||
| 1248 | AIDS-BL | VH4 (DP-66) | 91.4 | — | 1496G>A | — | — | ‡ | |||||
| 1249 | AIDS-BL | NA | NA | — | — | — | — | ‡ | |||||
| 1907 | AIDS-BL | VH3 (DP-33) | 93.0 | — | — | — | — | ‡ | |||||
| 2106 | AIDS-BL | NA | NA | — | — | 1394C>T | — | ‡ | |||||
| 2415 | AIDS-BL | VH4 (DP-63) | 87.8 | 563T>C | — | — | — | ‡ | |||||
| HBL-6 | AIDS-PEL | VH5 (DP-73) | 94.3 | 821C>G, 1085T>G | — | — | 658A>G, 854A>G | — | |||||
| BC-2 | AIDS-PEL | VH3 (3-73) | 94.0 | 605T>C | — | 1436C>T | — | — | |||||
| BCBL-1 | AIDS-PEL | NA | NA | 843C>G, 850T>A | — | — | — | — | |||||
| CRO-AP/3 | AIDS-PEL | VH3 (DP-47) | 93.5 | 447T>C, 480C>T, 522T>C, 539C>G, 564T>C, 794G>A | — | — | 573G>A | — | |||||
| CRO-AP/5 | AIDS-PEL | NA | NA | 543T>C, 549T>A | — | — | — | 2678T>A, 2691G>A | |||||
| CRO-AP/6 | AIDS-PEL | VH4 (DP-79) | 94.3 | — | — | — | — | — | |||||
| 2254 | AIDS-PCNSL | VH3 (DP-47) | 93.9 | 480C>T, 538A>G | — | — | — | — | |||||
| 2255 | AIDS-PCNSL | VH6 (DP-74) | 91.3 | — | — | — | — | — | |||||
| 2256 | AIDS-PCNSL | VH4 (DP-63) | 78.8 | 489G>A, 497G>A, 499A>T, 549T>G, 554T>G, 567T>G, 569T>C, 623T>G, 783T>C, 809G>A, 821G>A, 918T>G, 957G>A, 971A>C, 1003G>A, 1029T>C | — | — | 456G>A, 652A>C, 669T>C, 748A>T, 757A>G | 4682G>A, 4698C>T, 4993A>G, 5027C>G | |||||
| 2257 | AIDS-PCNSL | VH4 (V4.38) | 91.1 | 445C>G, 500T>A | — | — | — | — | |||||
NA indicates not amplified; Δ, deletion; +, insertion; —, wild-type sequence.
Only in-frame rearrangements are listed, and the % homology to the closest germ line V gene is given. Rearrangements of cases 1138, 2768, and 2771 were found to harbor crippling mutations.
Numbering according to GenBank accession nos. AF386792 (PIM-1), AF386791 (PAX-5), AF386789 (Rho-H/TTF), X00364 (c-MYC), and AY189709 (BCL-6).
c-MYC mutations in all BLs and DLBCL case 1905 were excluded from analysis because these cases carry a t(8;14) translocation and the mutations were attributed to the juxtaposed immunoglobulin elements.
Features of PIM-1, PAX-5, RhoH/TTF, c-MYC, and BCL-6 mutations in AIDS-NHL
Locus . | Mutation frequency per 100 bp (range)* . | Single bp substitutions . | Deletions or insertions . | Transitions-transversions (ratio) . | G+C/A+T (ratio) . |
|---|---|---|---|---|---|
| PIM-1 | 0.08 (0.06-0.11) | 7 | 0 | 4/3 (1.33) | 7/0 |
| PAX-5 | 0.07 (0.06-0.12) | 7 | 2 | 4/3 (1.33) | 6/1 |
| RhoH/TTF | 0.18 (0.06-0.41) | 24 | 3 | 17/7 (2.42) | 9/15 |
| c-MYC† | |||||
| Exon 1/intron 1 | 0.05 (0.04-0.08) | 4 | 0 | 3/1 (3.0) | 3/1 |
| Exon 2 | 0.43 (0.09-1.12) | 20 | 0 | 8/12 (0.66) | 15/5 |
| All genes (PIM-1, PAX-5, RhoH/TTF, c-MYC) | NA | 62 | 5 | 36/26 (1.38) | 40/22 (1.82) |
| P < .0001 | |||||
| BCL-6 | 0.20 (0.07-1.08) | 71 | 5 | 36/35 (1.03) | 38/33 |
Locus . | Mutation frequency per 100 bp (range)* . | Single bp substitutions . | Deletions or insertions . | Transitions-transversions (ratio) . | G+C/A+T (ratio) . |
|---|---|---|---|---|---|
| PIM-1 | 0.08 (0.06-0.11) | 7 | 0 | 4/3 (1.33) | 7/0 |
| PAX-5 | 0.07 (0.06-0.12) | 7 | 2 | 4/3 (1.33) | 6/1 |
| RhoH/TTF | 0.18 (0.06-0.41) | 24 | 3 | 17/7 (2.42) | 9/15 |
| c-MYC† | |||||
| Exon 1/intron 1 | 0.05 (0.04-0.08) | 4 | 0 | 3/1 (3.0) | 3/1 |
| Exon 2 | 0.43 (0.09-1.12) | 20 | 0 | 8/12 (0.66) | 15/5 |
| All genes (PIM-1, PAX-5, RhoH/TTF, c-MYC) | NA | 62 | 5 | 36/26 (1.38) | 40/22 (1.82) |
| P < .0001 | |||||
| BCL-6 | 0.20 (0.07-1.08) | 71 | 5 | 36/35 (1.03) | 38/33 |
NA indicates not applicable.
Mutation frequencies were calculated on the entire region analyzed and on mutated cases only, considering 2 alleles/gene/case.
Mutations of c-MYC exon 1 and exon 2 were considered only if they occurred in AIDS-NHL devoid of gross rearrangements of the gene to the immunoglobulin loci.
A total of 68 sequence variants were detected in 19 AIDS-NHL cases. The average frequency of mutations, calculated taking into account only mutated cases, ranged from 0.05 × 10–2/bp in the case of c-MYC exon 1 to 0.43 × 10–2/bp in the case of c-MYC exon 2 (Table 3), with no significant differences among the clinicopathologic categories of AIDS-NHL. The overwhelming majority of the mutations included single base-pair substitutions (n = 62), whereas deletions/insertions of a short DNA stretch were observed in 5 cases (Table 3). Of the 62 single base-pair substitutions observed, 36 were transitions and 26 were transversions, with a transition-to-transversion ratio of 1.38 (expected 0.5; P < .0001; chi-square test) (Table 3). Analysis of the nucleotide exchange pattern, when considering together all the nucleotide substitutions observed in PIM-1, PAX-5, RhoH/TTF, and c-MYC, showed a G+C/A+T ratio of 1.82 (Table 3), similar to that reported in human out-of-frame IgV rearrangements and in 2 in vitro mutating BL cell lines.30,34,35 Differences in the individual transition/transversion and G+C/A+T ratios (eg, c-MYC exon 2 and RhoH/TTF, respectively) may be related to the limited number of mutational events counted. Analogously, although the low number of mutations prevents the assessment of statistical significance, the frequency of mutations targeting G residues within an RGYW motif was in all genes higher than expected (PIM-1, 14% versus 3.4%; RhoH/TTF, 9% versus 4.5%; PAX-5, 14% versus 4.2%; c-MYC exons 1 and 2, 21% versus 3.4%).
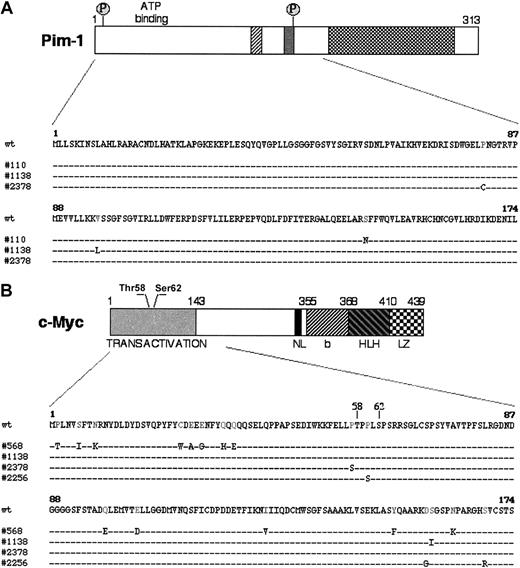
In PIM-1 and c-MYC, a number of the mutations were located in coding exons and led to amino acid substitutions, with potential functional consequences. In particular, 4 PIM-1 mutations in 3 AIDS-NHL cases represented missense mutations affecting the gene exon 4 (Figure 2A). Because this exon also encodes for the serine-threonine kinase domain, the mutations predict a change in the structure and, possibly, the function of the Pim-1 protein (Figure 2A).36,37 In c-MYC, 18 missense mutations were found in 4 cases within the sequences of exon 2, which encode the transactivation domain and represent a mutational hot spot in translocated c-MYC alleles in BL and mouse plasmacytoma (Figure 2B).33,38-40 Notably, 2 of these changes (Pro57Ser in AIDS-DLBCL case 2378 and Pro60Ser in AIDS-PCNSL case 2256) cluster around the evolutionary conserved and functionally critical phosphorylation sites Thr58 and Ser62 (Figure 2B).39,41 Several studies have shown that mutations affecting or flanking these residues abolish phosphorylation of the c-Myc transactivation domain, resulting in defective regulation by p107, protein stabilization, and increased transforming activity.41-43 Thus, alterations introduced by aberrant hypermutation are likely to have a profound effect on the function of this proto-oncogene.
Missense mutations in PIM-1 and c-MYC. (A) Schematic representation of the Pim-1 protein: hatched regions contain catalytic domains highly conserved in most protein kinases. The 2 putative phosphorylation sites (P) are also indicated. The Pim-1 protein sequence, residues 1-174, is expanded in the lower panel to show the distribution of the amino acid changes introduced by mutations in 3 AIDS-NHL cases. (B) Diagram of the c-Myc protein, with its relevant functional domains and the 2 N-terminal residues (Thr58 and Ser62) involved in the response to p107-mediated suppression of activity and in the control of protein stability by phosphorylation-dependent ubiquitination.41-43 The region analyzed is aligned with the sequences of 4 mutated AIDS-NHL cases. NL indicates nuclear localization signal; bHLHLZ, basic Helix Loop Helix Leucine Zipper domain.
Missense mutations in PIM-1 and c-MYC. (A) Schematic representation of the Pim-1 protein: hatched regions contain catalytic domains highly conserved in most protein kinases. The 2 putative phosphorylation sites (P) are also indicated. The Pim-1 protein sequence, residues 1-174, is expanded in the lower panel to show the distribution of the amino acid changes introduced by mutations in 3 AIDS-NHL cases. (B) Diagram of the c-Myc protein, with its relevant functional domains and the 2 N-terminal residues (Thr58 and Ser62) involved in the response to p107-mediated suppression of activity and in the control of protein stability by phosphorylation-dependent ubiquitination.41-43 The region analyzed is aligned with the sequences of 4 mutated AIDS-NHL cases. NL indicates nuclear localization signal; bHLHLZ, basic Helix Loop Helix Leucine Zipper domain.
Relationship between aberrant hypermutation, IgVH gene status, and BCL-6 mutations in AIDS-NHL
A functional immunoglobulin VHDJH rearrangement was obtained from 22 of 39 (56.4%) AIDS-NHL cases, whereas in 3 additional cases a stop-codon was detected within the originally productive VH rearrangement, leading to a crippled immunogloublin sequence (Table 2). In one case, only an out-of-frame IgVHDJH rearrangement was amplified. No PCR product could be obtained in the remaining 13 cases. The rearranged IgVHDJH genes were found to be somatically mutated in all but one case, at frequencies ranging from 2.2% to 22.6% of the nucleotides examined (Table 2).
Mutations of BCL-6 were detected in 24 of 39 (61.5%) AIDS-NHL cases, including 11 of 18 (61.1%) AIDS-DLCL, 5 of 11 (45.4%) AIDS-BL, 3 of 4 (75.0%) AIDS-PCNSL, and 5 of 6 (83.3%) AIDS-PEL. Mutations are detailed in Table 2.
Overall, the average mutation frequency of PIM-1, PAX-5, RhoH/TTF, c-MYC exon 1, and c-MYC exon 2 was 50- to 100-fold lower than that observed in the IgVH genes and 2- to 5-fold lower than that observed in BCL-6 (Table 3), as already observed in DLBCL of the immunocompetent hosts. Comparison of the mutation frequencies within each case revealed that the occurrence of PIM-1, PAX-5, RhoH/TTF, and c-MYC mutations did not correlate with an increased frequency of mutations in the IgVH genes, supporting the hypothesis that aberrant hypermutation may be due to a qualitative rather than to a quantitative defect of the somatic hypermutation mechanism.11
Ongoing aberrant hypermutation of PIM-1, PAX-5, and RhoH/TTF in AIDS-NHL
To investigate whether the aberrant mutational activity is ongoing in AIDS-NHL, we selected a subset of cases (n = 7) shown by direct sequencing to carry mutations in PIM-1, PAX-5, and/or RhoH/TTF. These cases were analyzed for the presence of intraclonal heterogeneity in the 4 proto-oncogenes as well as in the IgVH genes by sequencing cloned PCR products generated using the proofreading Pfu polymerase. As a control for background error rate, the fibroblast IMR91 cell line and naive B cells purified from human tonsils were used.11 Statistical significance was assessed by the Student t test.
The results showed that, in all tumors analyzed, 1 or 2 predominant alleles recapitulate the mutations observed by direct sequencing of the PCR product, confirming their presence in the tumor clone and revealing their frequent biallelic distribution (not shown). In addition, a significant number of intraclonal variants were found in the PIM-1 and RhoH/TTF sequences from 2 of the 7 cases examined. In the PIM-1 gene of AIDS-DLCL 2378, 27 of 65 clones (41.5%) carried one or more additional mutations (corresponding to 34 events in 46 475 bp sequenced) (Table 4 and Figure 3). Accordingly, this same case also displayed ongoing mutations in the rearranged IgVH gene (Table 4), consistent with a derivation of the mutations from a common mechanism. A second case, AIDS-DLBCL 1220, exhibited a total of 5 point mutations, each unique to a single clone, which were distributed in 5 of 50 sequences analyzed and, therefore, did not differ significantly from the expected polymerase error frequency (0.004 to 0.008 × 10–2/bp). This result prevents us from conclusively assessing whether there was low-rate ongoing mutation activity in this case. No evidence of intraclonal diversification was found in the PIM-1, PAX-5, and RhoH/TTF genes of the remaining cases (Table 4). These data indicate that the aberrant hypermutation activity may be ongoing in at least some cases of AIDS-NHL.
Intraclonal heterogeneity of PIM-1, PAX-5, RhoH/TTF, and IgVH genes in AIDS-NHL
Sample . | Histology . | Gene . | No. of mutations (position) in the tumor clone* . | No. of DNA clones analyzed . | Total no. of DNA clones with additional mutations† . | Total no. of additional mutations . | Total no. of bp sequenced . |
|---|---|---|---|---|---|---|---|
| 1138 | AIDS-DLBCL | PIM-1 | 2 (1764G>A, 1765G>C) | 34 | 2 | 2 | 24310 |
| VH2 (S12-9) | 97.8% | 26 | 5 | 8 | 6900 | ||
| 1220 | AIDS-DLBCL | RhoH/TTF | 2 (418Δ31 bp, 987T>C) | 50 | 5 | 5 | 43750 |
| 2378 | AIDS-DLBCL | PIM-1 | 2 (1720C>T, 1721C>G) | 65 | 27 | 34 | 46475 |
| VH3 (DP-48/13) | 96.7% | 27 | 13 | 19 | 4140 | ||
| 2256 | AIDS-PCNSL | RhoH/TTF | 5 (546G>A, 652A>C, 699T>C, 748A>T, 757A>G) | 30 | 0 | 0 | 26250 |
| VH4 (DP-63) | 78.8% | 20 | 11 | 25 | 7130 | ||
| 645 | AIDS-BL | RhoH/TTF | 4 (557T>G, 563A>G, 638C>T, 643A>G) | 25 | 0 | 0 | 21875 |
| 1248 | AIDS-BL | PIM-1 | 1 (1496G>A) | 26 | 1 | 1 | 18590 |
| VH4 (DP-66) | 91.4% | 17 | 5 | 5 | 3910 | ||
| 2106 | AIDS-BL | PAX-5 | 1 (1394C>T) | 25 | 0 | 0 | 23300 |
Sample . | Histology . | Gene . | No. of mutations (position) in the tumor clone* . | No. of DNA clones analyzed . | Total no. of DNA clones with additional mutations† . | Total no. of additional mutations . | Total no. of bp sequenced . |
|---|---|---|---|---|---|---|---|
| 1138 | AIDS-DLBCL | PIM-1 | 2 (1764G>A, 1765G>C) | 34 | 2 | 2 | 24310 |
| VH2 (S12-9) | 97.8% | 26 | 5 | 8 | 6900 | ||
| 1220 | AIDS-DLBCL | RhoH/TTF | 2 (418Δ31 bp, 987T>C) | 50 | 5 | 5 | 43750 |
| 2378 | AIDS-DLBCL | PIM-1 | 2 (1720C>T, 1721C>G) | 65 | 27 | 34 | 46475 |
| VH3 (DP-48/13) | 96.7% | 27 | 13 | 19 | 4140 | ||
| 2256 | AIDS-PCNSL | RhoH/TTF | 5 (546G>A, 652A>C, 699T>C, 748A>T, 757A>G) | 30 | 0 | 0 | 26250 |
| VH4 (DP-63) | 78.8% | 20 | 11 | 25 | 7130 | ||
| 645 | AIDS-BL | RhoH/TTF | 4 (557T>G, 563A>G, 638C>T, 643A>G) | 25 | 0 | 0 | 21875 |
| 1248 | AIDS-BL | PIM-1 | 1 (1496G>A) | 26 | 1 | 1 | 18590 |
| VH4 (DP-66) | 91.4% | 17 | 5 | 5 | 3910 | ||
| 2106 | AIDS-BL | PAX-5 | 1 (1394C>T) | 25 | 0 | 0 | 23300 |
Mutations detected by DNA direct sequencing of the PCR product. For the IgVH genes, the percentage of homology to the closest germ line gene is given.
Number of DNA clones containing mutations that are not visible on direct sequencing and may, therefore, represent subclonal variants of the tumor.
Intraclonal heterogeneity of PIM-1 sequences in AIDS-DLBCL 2378. Presumed genealogic tree showing the relationship between PIM-1 mutations identified in the neoplastic population from AIDS-DLBCL case 2378. Sixty-five clones, obtained from 2 independent PCR amplifications, were sequenced. Each circle represents a distinct sequence carrying the mutations detailed on the side (GenBank accession no. AF386792); the number of independent clones sharing those mutations is indicated within the circle. Alleles A and B were defined according to the presence and distribution of the mutations that had been detected by direct sequencing and, therefore, represent the original, most abundant tumor clone (see also Table 4). The dashed circle indicates presumed intermediates in the genealogic tree.
Intraclonal heterogeneity of PIM-1 sequences in AIDS-DLBCL 2378. Presumed genealogic tree showing the relationship between PIM-1 mutations identified in the neoplastic population from AIDS-DLBCL case 2378. Sixty-five clones, obtained from 2 independent PCR amplifications, were sequenced. Each circle represents a distinct sequence carrying the mutations detailed on the side (GenBank accession no. AF386792); the number of independent clones sharing those mutations is indicated within the circle. Alleles A and B were defined according to the presence and distribution of the mutations that had been detected by direct sequencing and, therefore, represent the original, most abundant tumor clone (see also Table 4). The dashed circle indicates presumed intermediates in the genealogic tree.
Discussion
The aim of this study was to investigate the presence of mutations targeting the PIM-1, PAX-5, RhoH/TTF, and c-MYC genes throughout the clinicopathologic spectrum of AIDS-NHL. We report that mutations of these 4 proto-oncogenes represent a frequent event in AIDS-related lymphomas, display molecular features typical of the IgV-associated somatic hypermutation process, associate with different AIDS-NHL histotypes, and may be ongoing in some cases.
On the basis of several parameters, the molecular profile of aberrant hypermutation in AIDS-NHL is similar to that detected in DLBCL of the immunocompetent host and closely resembles the one reported for IgV and BCL-6 mutations.7-14 First, mutations in PIM-1, PAX-5, RhoH/TTF, and c-MYC are predominantly represented by single nucleotide substitutions, with rare deletions and insertions. Second, a preference for transitions over transversions, a bias for specific targeting motifs (RGYW/WRCY), and an elevated ratio of G+C over A+T substitutions was observed in all of the 4 genes. As already reported in DLBCL from non-AIDS patients, the mutation frequency in PIM-1, PAX-5, RhoH/TTF, and c-MYC was lower than that observed in IgV and BCL-6 sequences in the same cases. Finally, mutations appear to be independent of translocations with the immunoglobulin genes, because, as formally documented for c-MYC, they occurred in cases devoid of gross alterations of the respective loci. Because mutations at the 4 loci analyzed have not been found in normal GC B cells, these data provide further support to the hypothesis that these mutations result from an abnormal activity of the SHM mechanism normally active in GC B cells.11
The identification of an aberrant hypermutation activity throughout the clinicopathologic spectrum of AIDS-NHL expands the types of aggressive lymphoma associated with this molecular abnormality. In fact, aberrant hypermutation was found preferentially associated to DLBCL (> 50% of the cases analyzed) in the immunocompetent host, with only few FL and BL cases harboring isolated mutations in the RhoH/TTF and PAX-5 genes.11 Conversely, in the context of AIDS-NHL aberrant hypermutation appears to affect at similar frequencies AIDS-DLBCL (both systemic and primarily localized to the central nervous system), AIDS-BL, and AIDS-PEL. This difference may be related to the relatively small number of BL cases examined to date in immunocompetent as well as in immunodeficient hosts. Alternatively, it may reflect true differences in the pathogenesis of sporadic versus endemic BL, as also suggested by other molecular differences between the 2 disease variants.1,2 The detection of an aberrant SHM activity in AIDS-NHL reinforces the concept that this molecular mechanism is a frequent and selective feature of aggressive lymphomas, because mutations of PIM-1, PAX-5, RhoH/TTF, and c-MYC are rare or absent in indolent lymphomas.11
The genes targeted by aberrant hypermutation in AIDS-NHL encode proteins with different functions, including both signal transducers (PIM-1 and RhoH/TTF) and transcription factors (PAX-5 and c-MYC).33,36,44-46 As previously noted, these 4 genes represent known proto-oncogenes that have been already implicated in lymphoma-associated chromosomal translocations. On the basis of the type and distribution of the mutations, it can be predicted that they may have relevant functional consequences, with 2 distinct modalities. First, because they cluster around the gene 5′ regulatory regions, they may deregulate gene transcription, as formally documented in the case of c-MYC.32 Second, in the case of c-MYC and PIM-1, in which the process also affects coding sequences introducing amino acid substitutions, the mutations may alter the biochemical and/or structural properties of the respective protein. In particular, in vitro and in vivo studies have previously shown that mutations in the c-MYC transactivation domain can deregulate its function by interfering with its phosphorylation, protein stability, or the repression of its transactivation activity by the Rb-related protein p107.41-43 These findings suggest that at least some of the translocation-independent mutations in AIDS-DLBCL may have been selected for their ability to alter c-MYC function, as is the case for translocated c-MYC alleles in BL.33
Finally, our results shed some light on the timing of aberrant hypermutation in AIDS-lymphomagenesis. Although in most AIDS-NHL cases the IgV somatic hypermutation process seems to be no longer active, as indicated by the absence of ongoing mutations in approximately 70% of the samples (G.G., unpublished observation, 2001), all of the 4 cases analyzed in this study showed intraclonal heterogeneity in the IgV genes; this finding may be related to the small number of cases investigated or may reflect a selection bias based on the presence of aberrant SHM. In addition, isolated AIDS-NHL cases were found which display intraclonal diversity in both IgV and non-IgV genes (Table 4 and Figure 3). This observation indicates that the tumor clone may continue to be exposed to aberrant SHM and to accumulate additional, intraclonal variants. This prolonged mutational activity, and the likelihood that it may affect many more genes than those identified so far, suggests that aberrant SHM may represent a major contributor to genetic damage during AIDS-associated lymphomagenesis.
Prepublished online as Blood First Edition Paper, April 24, 2003; DOI 10.1182/blood-2002-11-3606.
Supported by National Institutes of Health (grant CA-37295); Istituto Superiore di Sanità, Programma Nazionale di Ricerca sull'AIDS–Progetto Patologia, Clinica e Terapia dell'AIDS, Rome, Italy; Cofin 2002–Ministero dell'Istruzione, Università e Ricerca (MIUR), Rome, Italy; Progetto Strategico Oncologia, Consiglio Nazionale delle Ricerche (CNR)–MIUR, Rome, Italy; and “Fondazione CRT,” Torino, Italy. L.P. is a Special Fellow of the Leukemia & Lymphoma Society. D.C., E.B., and C.D. are currently supported by fellowships from the Fondazione “Piera Pietro Giovanni Ferrero,” Alba, Italy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Daniela Vivenza, Michaela Cerri, and Annarita Conconi, from the Hematology Unit, Division of Internal Medicine, Department of Medical Sciences, Amedeo Avogadro University of Eastern Piedmont, for help in various parts of the study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal