Abstract
After antigenic challenge, naive T lymphocytes enter a program of proliferation and differentiation during the course of which they acquire effector functions and may ultimately become memory cells. In humans, the pathways of effector and memory T-cell differentiation remain poorly defined. Here we describe the properties of 2 CD8+ T-lymphocyte subsets, RA+CCR7–27+28+ and RA+CCR7–27+28–, in human peripheral blood. These cells display phenotypic and functional features that are intermediate between naive and effector T cells. Like naive T lymphocytes, both subsets show relatively long telomeres. However, unlike the naive population, these T cells exhibit reduced levels of T-cell receptor excision circles (TRECs), indicating they have undergone additional rounds of in vivo cell division. Furthermore, we show that they also share effector-type properties. At equivalent in vivo replicative history, the 2 subsets express high levels of Fas/CD95 and CD11a, as well as increasing levels of effector mediators such as granzyme B, perforin, interferon γ, and tumor necrosis factor α. Both display partial ex vivo cytolytic activity and can be found among cytomegalovirus-specific cytolytic T cells. Taken together, our data point to the presence of T cells with intermediate effector-like functions and suggest that these subsets consist of T lymphocytes that are evolving toward a more differentiated effector or effector-memory stage.
Introduction
As a result of an antigenic challenge, due, for example, to a viral infection, naive T lymphocytes undergo changes in the expression of cell surface molecules and in their migratory properties and proliferate. They acquire effector functions and eventually some of them become memory cells.1 Effector CD8+ T cells kill antigen-bearing target cells by secreting granules containing granzyme and perforin or through the engagement of Fas on the target cell. Effector lymphocytes have the capacity to migrate to extralymphoid sites and to home to the site of infection. Once the antigenic challenge subsides, the number of effector cells decreases drastically, but another class of antigen-specific cells appears, the memory T lymphocytes. Memory T cells produced after a primary immune response are long-lived. They can be distinguished from naive T cells because they respond more efficiently to antigenic recall (due, probably, to less stringent requirements for activation), secrete enhanced levels of cytokines, and express other cell surface molecules.1 Thus, in response to a second encounter with antigen, memory T cells can usually eliminate pathogens before any disease symptoms are detectable. The understanding of the lineage relationships between naive, effector, and memory T cells is a central question in immunology. Although the formation of naive cells is reasonably well understood, the signals and pathways that control the generation of effector and memory T cells are still controversial.
To understand the mechanisms that underlie memory and effector formation, various phenotypic cell surface markers have been used to distinguish primed T lymphocytes from naive cells. In humans, CD45RA and CD27 monoclonal antibodies (mAbs) have been used to define naive (N; RA+27+), effector (E; RA+27–), and memory (M; RA–27+) CD8+ subpopulations.2 More recently, based on simultaneous staining for CD45RA and CCR7, lymphocytes described as memory cells could be further segregated into so-called central-memory (RA–CCR7+) and effector-memory (RA–CCR7–) cells.3,4 Whereas the term “central-memory” (CM) refers to cells that lack immediate effector function and express lymph node homing receptors, the term “effector-memory” (EM) refers to cells that share numerous features with effector (RA+27–) T lymphocytes. Indeed, on in vitro stimulation EM cells rapidly produce effector cytokines such as interferon γ (IFN-γ) and express perforin granules, and they home preferentially to peripheral tissues.3 However, the precise function of each of these subsets as well as their lineage relationship still remains elusive.
Longitudinal studies performed in mice suggest a linear model for effector and memory T-cell formation according to which memory T cells are formed by a small fraction of effector cells that do not undergo apoptosis (N→E→M). However, accumulating data tend to support an alternative model according to which effector and memory cells are formed by distinct pathways (for a review, see Kaech et al5 ). In this model, naive T cells can bypass the effector cell stage and develop directly into memory or CM T cells. Indeed, Sallusto and Lanzavecchia6 have proposed that the duration of antigenic stimulation and the type and the amount of cytokines present during priming determine whether lymphocytes differentiate into effector cells (E or EM or both) or into CM cells that lack effector function and home preferentially to lymph nodes. Yet another model7,8 proposes that CD8 T cells differentiate from precursor (N) through an intermediate (CM/EM) to a fully differentiated effector (E) stage.
Here, by combining phenotypic cell surface marker characterization with assays allowing us to trace the relative numbers of cell divisions in vivo, we identify 2 subsets of circulating CD8+ T lymphocytes (RA+CCR7–27+28+/–) whose proliferative history places them between naive and effector T cells. Both subsets express intermediate levels of effector mediators, such as granzyme B, perforin, and IFN-γ, and display relative potent cytolytic activity. Our data suggest that these subsets descend from recently activated T cells and are committed to become differentiated effector or effector-memory T cells or both.
Materials and methods
mAbs and major histocompatibility complex (MHC)/peptide multimers
The following mAbs were purchased from Becton Dickinson or BD PharMingen (San Diego, CA): anti–CD27-fluorescin isothiocyanate (FITC) and -phycoerythrin (PE), anti–CD28-PE and -allophycocyanin (APC), anti–CD8-FITC and -APC/Cy7, anti–CD57-PE, anti–HLA-DR-PE, anti–CD11a-PE, anti–CD95-FITC, anti–CD56-PE, anti–CD94-PE, and goat antirat IgG-APC. Other sources of mAbs were: Beckman Coulter, Marseille, France (anti–CD158b-PE and anti–CD45RA-ECD) and Caltag Laboratories, Burlingame, CA (goat antirat IgG-PE). Anti-CCR7 rat IgG mAb 3D12 was provided by Drs M. Lipp and R. Forster (Max Delbrück Institute, Berlin, Germany). Antigranzyme B–FITC and antiperforin-FITC mAbs were obtained from Hölzel Diagnostika (Köln, Germany) and Alexis (Lausen, Switzerland), respectively. Synthesis of PE- and APC-labeled HLA-A*0201/CMV pp65495-503 (NLVPMVATV; HCMV-19 ) multimers was performed as previously described.10,11
Cell preparation and flow cytometry
Peripheral blood samples were collected from 20 healthy donors, aged 20 to 70 years, with a normal proportion of CD8+ T lymphocytes (average, 22%; range, 12%-34%). Peripheral blood mononuclear cells (PBMCs) were obtained by density centrifugation using Ficoll-Hypaque (Pharmacia, Uppsala, Sweden). Our experimental procedures involve 2 steps that exclude contamination with natural killer (NK) cells. First, CD8+ T lymphocytes were positively enriched from cryopreserved or fresh PBMCs using anti-CD8–coated magnetic microbeads (Miltenyi Biotech, Bergish Gladbach, Germany), a procedure that eliminates most NK cells because they are not efficiently retained by the magnet. Cells were stained with appropriate mAbs in phosphate-buffered saline (PBS), 0.2% bovine serum albumin (BSA), and 50 μM EDTA (ethylenediaminetetraacetic acid) for 20 minutes at 4°C and either directly analyzed or sorted into defined populations on a FACSVantage SE, using CellQuest software (Becton Dickinson). Immediate reanalysis of the isolated populations revealed more than 95% purity (CD3+CD8+). Second, FACS analysis and sorting was performed on gated CD8bright T cells, allowing the exclusion of any residual contaminating NK cells in the sorted populations (< 2%). In the experiments with cytomegalovirus (CMV)–specific T lymphocytes, cells were first stained with either PE- or APC-labeled CMV multimers for 1 hour at room temperature (RT) in PBS, 0.2% BSA, 50 μM EDTA, and then with appropriate mAbs. Intracellular content of granzyme B and perforin was measured in freshly isolated CD8+ T lymphocytes without previous stimulation. In brief, after staining with appropriate mAbs cells were fixed for 20 minutes at RT in PBS containing 1% formaldehyde, 2% glucose, and 5 mM sodium azide. Fixation was followed by permeabilization with PBS/0.1% saponin (Fluka, Buchs, Switzerland)/0.2% BSA/50 μM EDTA and staining with granzyme B–FITC or perforin-FITC mAbs (both for 20 minutes at RT).
Quantification of TRECs by real-time PCR
The amount of signal joint (sj) T-cell receptor excision circles (TRECs) in 5 to 15 × 104 sorted CD8+ T subsets was determined by real-time quantitative polymerase chain reaction (PCR) using the ABI PRISM 7700 Sequence Detector TaqMan system (Applied Biosystems, Rotkreuz, Switzerland) as previously described.12,13 In brief, after cell lysis in 100 mg/L proteinase K (Roche Diagnostics, Mannheim, Germany) for 2 hours at 56°C followed by 15 minutes at 95°C, PCR was performed in a final volume of 25 μL containing 5 μL cell extract, 12.5 μL TaqMan Universal Master Mix including AmpliTaq Gold (Applied Biosystems), 500 nM of each primer (sj-5′ forward: CACATCCCTTTCAACCATGCT; sj-3′ reverse: GCCAGCTGCAGGGTTTAGG), and 125 nM TaqMan probe (FAM-ACACCTCTGGTTTTTGTAAAGGTGCCCACT-TAMRA). After one cycle of 2 minutes at 50°C followed by an initial 10 minutes of denaturation at 95°C, 40 cycles of 30 seconds at 95°C and 1 minute at 65°C were performed. The number of TRECs in a given sample was estimated by comparing the CT value obtained with a standard curve obtained from PCRs performed with 10-fold serial dilutions of an internal standard provided by Dr Daniel Douek (Vaccine Research Center, National Institute of Allergy and Infectious Diseases, Bethesda, MD). The dilutions contained between 107 and 101 copies of sjTREC, and 4 reactions were run with each dilution. Thus, the lowest limit of quantification was considered to be 10 copies of the target sequence (< 10 copies/sample was quoted as below the quantification limit of the assay). In all PCR assays, the correlation coefficient of the standard curve was more than or equal to 0.997, whereas the slope varied between –3.52 and –3.67. The TREC analysis was performed on young healthy individuals (< 36 years of age) because aging inversely correlates with the TREC levels.12 Because the TREC content of naive cells from younger individuals is significantly higher compared to that of elderly donors, this allows an increased resolution in the quantification of the TREC levels within the different CD8 T-cell subpopulations (data not shown).
cDNA amplification and 5-cell RT-PCR
We first purified CD8+ T cells in 2 rounds of positive sorting by magnetic beads and a MiniMACS device (Miltenyi Biotech). The resulting cells were more than 98% CD3+CD8+. Second, to avoid contamination of small populations by more abundant subsets, 10 × 103 T cells of each subset were sorted by flow cytometry and 5-cell aliquots of the purified subsets were then resorted directly into wells of 96-V bottom plates. The procedures for cDNA preparation, cDNA amplification as well as the reverse transcription-PCR (RT-PCR) were recently described in detail.14 We used the following primers: CD3: 5′-CGTTCAGTTCCCTCCTTTTCTT-3′, rev-5′-GATTAGGGGGTTGGTAGGGAGTG-3′; CCR7: 5′-CCAGGCCTTATCTCCAAGACC-3′, rev-5′-GCATGTCATCCCCACTCTG-3′; granzyme B: 5′-GCAGGAAGATCGAAAGTGCGA-3′, rev-5′-GCATGCCATTGTTTCGTCCAT-3′; perforin: 5′-TTCACTGCCACGGATGCCTAT-3′, rev-5′-GCGGAATTTTAGGTGGCCA-3′; FasL, 5′-GAGCCAGACAAATGGAGGAA-3′, rev-5′-GAAGTGAAGATGCTGCCAGTG-3′; IFN-γ: 5′-GCCAACCTAAGCAAGATCCCA-3′, rev-5′-GGAAGCACCAGGCATGAAATC-3′; tumor necrosis factor α (TNF-α): 5′-CTGCCTTGGCTCAGACATGTT-3′, rev-5′-CAGTTGGTCACCAAATCAGCA-3′; CD94: 5′-GTGGGAGAATGGCTCTGCAC-3′, rev-5′-TGAGCTGTTGCTTACAGATATAACGA-3′. Typically, we used either H20 or Daudi B-cell line extract as negative PCR control; 103 PBMCs from a healthy individual was used as positive control.
Cytolytic activity
Cytolytic activity was tested in a CD3 mAb-mediated 51Cr-release assay. In brief, FcR-bearing P815 target cells were radiolabeled with Na51CrO4 (Perkin Elmer, Boston, MA) for 1 hour at 37°C. Sorted CD8+ T subsets were incubated with P815 target cells (103 cells/well) at varying effector-target ratios in the presence or absence of 300 ng/mL anti-CD3 mAb (OKT3). After 4 hours at 37°C, supernatants were collected and counted on a gamma counter. Percent of lysis was calculated as (experimental release – spontaneous release) × 100/(total release – spontaneous release).
Telomere fluorescence in situ hybridization and flow cytometry
All procedures have been described previously.15,16 Telomere fluorescence was calculated by subtracting the mean fluorescence of the background control (no probe) from the mean fluorescence obtained from cells hybridized with the telomere probe after calibration with FITC-labeled fluorescent beads (Quantum TM-24 Premixed; Bangs Laboratories, Fishers, IN) and conversion into molecules of equivalent soluble fluorochrome (MESF) units. The following equation was used to estimate the telomere length in base pair: bP = MESF × 0.495.15
Telomerase repeat amplification protocol assay
Telomerase activity was measured with the telomerase repeat amplification protocol (TRAP) assay using a telomerase substrate (TS) primer as described.17 Cell extracts were obtained from 5 × 104 to 15 × 104 sorted CD8+ T-cell subsets. As positive control we used extracts from CD8+ T lymphocytes stimulated for 5 days with 1 μg/mL phytohemagglutinin (PHA; Sodiag, Losone, Switzerland) and 150 U/mL recombinant interleukin 2 (rIL-2) in the presence of 1 × 106/mL irradiated feeder cells. Extension of the TS primer by telomerase was performed for 30 minutes at 30°C in the presence of [α-32P] deoxyguanosine triphosphate (dGTP) and the products generated were amplified by 27 cycles of PCR at 94°C for 30 seconds and 60°C for 30 seconds using the ACX anchored return primer. One half of the amplified products were resolved on a 15% polyacrylamide gel and visualized by a phosphoimaging system.
Results
Identification of an RA+CCR7–27+ T-cell subset within the circulating CD8+ compartment of healthy individuals
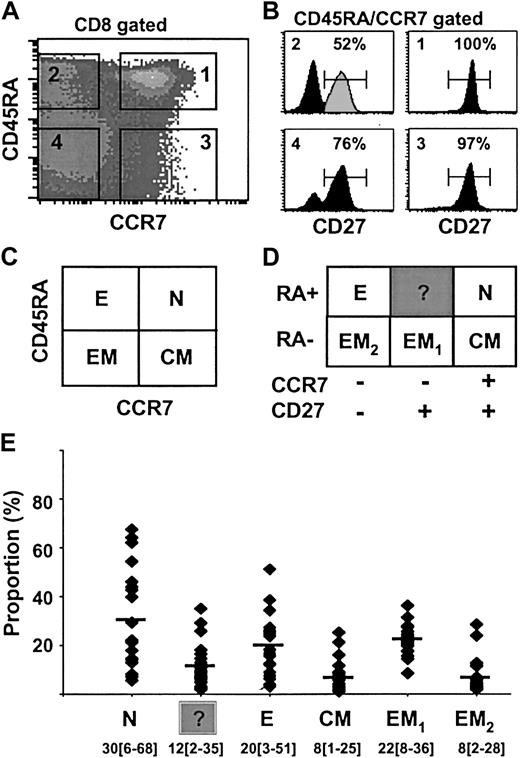
Human antigen–specific T lymphocytes can be separated into functionally different populations using combinations of cell surface markers such as CD45RA and CCR7 or CD27.2,3 However, the relationship between CCR7 and CD27 expression on peripheral blood CD8 lymphocytes has not yet been precisely determined. To do so we analyzed the distribution of the CD27 surface antigen in the 4 different subsets of CD8+ T lymphocytes previously defined on the basis of CD45RA and CCR7 expression3 : naive (N; RA+CCR7+), effector (E; RA+CCR7–), central-memory (CM; RA–CCR7+), and effector-memory (EM; RA–CCR7–; Figure 1A,C). Staining of peripheral blood CD8+ T lymphocytes with antibodies to CD45RA, CCR7, and CD27 revealed the presence of 6 discrete subpopulations in the blood from a representative healthy individual (Figure 1B,D). Naive RA+CCR7+ T cells (N) uniformly expressed CD27. Similarly, 97% of CM (RA–CCR7+)T cells were CD27+. In contrast, E (RA+CCR7–) T cells were split into 2 distinct subsets; 52% were CD27+ and 48% CD27–. The latter subset presumably is identical with the differentiated effector T-cell subpopulation identified by van Lier's group,2,18 whereas the characteristics of RA+CCR7–27+ T cells have not been analyzed thus far. Note that CD27 cell surface expression on these cells is lower than that on naive T cells (Figure 1B). Finally, among EM T cells, 76% were CD27+ (referred to as EM1) and 24% were CD27– (referred to as EM2). Both populations were recently described by others.8,19 The distribution of the 6 CD8+ T-cell subsets defined by different patterns of CD45RA, CCR7, and CD27 expression was analyzed in 20 healthy individuals ranging in age from 20 to 70 years (Figure 1E). The CD8+RA+CCR7–27+ subset was found in all individuals (mean ± SD, 12% ± 9%; range, 2%-35%) and its size was comparable to that of CM (mean ± SD, 8% ± 6%; range, 1%-25%) or EM2 (mean ± SD, 8% ± 7%; range, 2%-28%) CD8+ populations. Thus, our results revealed the widespread presence in peripheral blood of a CD8+ T-cell subset, RA+CCR7–27+, that displays phenotypic features intermediate between those of naive and of effector T cells.
Differential expression of CD45RA, CCR7, and CD27 cell surface molecules on total CD8+ T cells from healthy individuals. CD8+ gated cells were separated into 4 subsets (N, E, CM, EM) based on CD45RA and CCR7 labeling as previously described3 (A,C). Each of these subsets was analyzed for CD27 expression (B), and 6 subpopulations of CD8+ T cells could be distinguished (D). The percentage of CD27+ cells is indicated. Analysis performed on a representative healthy donor is depicted by the black histogram. The RA+CCR7–27+ T-cell subset (gray peak) is referred to as “?” (unknown). The distribution of the 6 defined CD8+ T-cell subsets among 20 healthy individuals ranging in age from 20 to 70 years is shown in panel E as mean percentage (range). N indicates naive; E, effector; CM, central-memory; and EM, effector-memory.
Differential expression of CD45RA, CCR7, and CD27 cell surface molecules on total CD8+ T cells from healthy individuals. CD8+ gated cells were separated into 4 subsets (N, E, CM, EM) based on CD45RA and CCR7 labeling as previously described3 (A,C). Each of these subsets was analyzed for CD27 expression (B), and 6 subpopulations of CD8+ T cells could be distinguished (D). The percentage of CD27+ cells is indicated. Analysis performed on a representative healthy donor is depicted by the black histogram. The RA+CCR7–27+ T-cell subset (gray peak) is referred to as “?” (unknown). The distribution of the 6 defined CD8+ T-cell subsets among 20 healthy individuals ranging in age from 20 to 70 years is shown in panel E as mean percentage (range). N indicates naive; E, effector; CM, central-memory; and EM, effector-memory.
The CD8+RA+CCR7–27+ T-cell subset displays lower levels of TRECs than naive T cells and expresses genes involved in effector functions
To gain more insight into the relationship between the RA+CCR7–27+ T cells and the other CD8+ subsets, we assessed the replicative history of these cells by quantifying their content of TRECs. TRECs are stable DNA episomes formed during T-cell receptor α (TCR-α) gene rearrangement. Because they are not replicated during mitosis, they are diluted out with each cell division.12 Purified CD8+ T cells isolated from 8 young healthy individuals were sorted into RA+CCR7+27+ (N), RA+CCR7–27+ (?), and RA+CCR7–27– (E) subsets (Figure 2A). In all individuals tested, naive cells had the highest level of TRECs, whereas the number in effectors was below the quantification limit of the assay (≥ 0.02 TREC copies/100 cells). Interestingly, the RA+CCR7–27+ T-cell subset contained detectable TRECs in all healthy individuals (mean ± SD, 1.5% ± 1.1%), their level corresponding to 12% of the amount in naive cells. These data indicate that the RA+CCR7–27+ cells have, on average, undergone 3 more cell divisions than the bulk of naive cells. Furthermore, these results demonstrate that CD45RA+CCR7– T cells do not exclusively define the effector CD8 subset. Indeed, RA+CCR7–27+ (?) T cells revealed quantifiable TREC levels, whereas this was not the case for RA+CCR7–27– (E) T cells and thus could account for the measurable content of TRECs previously described within the RA+CCR7– subset.13
The RA+CCR7–27+ T-cell subset contains reduced but detectable TREC levels and expresses genes associated with effector functions. (A) Real-time PCR quantification of TRECs was performed on sorted RA+CCR7+27+ (N), RA+CCR7–27+ (?), and RA+CCR7–27– (E) CD8+ T subsets from 8 healthy young individuals (age range, 20-36 years). Asterisk indicates not detectable (sorted cell number was 5 × 104 to 105, lower quantification limit = 0.01%-0.02%). (B) Gene expression analysis was performed on sorted N, (?), and E CD8+ T cells using a modified RT-PCR protocol (see “Materials and methods”). Data from 10 independent 5-cell aliquots as well as negative (–) and positive (+) controls are depicted. Comparable results were obtained in 5 healthy individuals.
The RA+CCR7–27+ T-cell subset contains reduced but detectable TREC levels and expresses genes associated with effector functions. (A) Real-time PCR quantification of TRECs was performed on sorted RA+CCR7+27+ (N), RA+CCR7–27+ (?), and RA+CCR7–27– (E) CD8+ T subsets from 8 healthy young individuals (age range, 20-36 years). Asterisk indicates not detectable (sorted cell number was 5 × 104 to 105, lower quantification limit = 0.01%-0.02%). (B) Gene expression analysis was performed on sorted N, (?), and E CD8+ T cells using a modified RT-PCR protocol (see “Materials and methods”). Data from 10 independent 5-cell aliquots as well as negative (–) and positive (+) controls are depicted. Comparable results were obtained in 5 healthy individuals.
The RA+CCR7–27+ subset may consist of naive T cells that have expanded in the course of the homeostatic maintenance of T-cell numbers that has recently been described for human T-helper cells.20 Alternatively, this T-lymphocyte class may comprise cells that are transiting from a naive to a more differentiated stage following antigenic challenge. To distinguish between these hypotheses, we then compared the expression level of genes involved in effector or regulatory functions in the RA+CCR7–27+ subset with that in naive and effector populations. For this purpose we used a modified RT-PCR protocol that relies on the detection of specific cDNAs after global amplification of expressed mRNAs.21,22 Because the method yields sufficient cDNA from as few as 5 cells, it allows the analysis of gene expression in small purified subpopulations. In a recent study, we showed that this method could detect CD3ϵ transcripts in 60% to 90% of extracts from single CD3+ cells.14 This is consistent with the finding that all samples of CD8+ cells analyzed in the present study gave a CD3-specific PCR product. All naive T-cell samples but none of the aliquots of effector T cells yielded a detectable CCR7-specific product (Figure 2B). With a single exception this correlates with CCR7 cell surface expression and rules out significant contamination of the naive (RA+CCR7+27+) subpopulation with the RA+CCR7–27+ (?) and RA+CCR7–27– (E) T cells. As expected, naive CD8+ T lymphocytes, which are not cytolytic and do not produce cytokines, did not contain detectable granzyme B, perforin, FasL, IFN-γ, or NK-receptor CD94 mRNA, and only rarely gave a TNF-α signal. In contrast, these mRNA transcripts were found in most (granzyme B, CD94) or a significant fraction of effector T-lymphocyte aliquots. Transcript analysis of RA+CCR7–27+ T cells revealed the presence of all effector function–associated mRNAs in a significant fraction of 5-cell aliquots from this population. We have not determined the efficiency of the PCR amplification of the different mRNAs nor do we have any information on the distribution of the number of transcripts among the cells within a given subpopulation. Thus, the data shown cannot be used to derive reliable estimates of the fraction of cells expressing a particular gene. Among RA+CCR7–27+ cell aliquots there is no obvious correlation between the presence of PCR products corresponding to different effector function–associated genes. For instance, one sample gave PCR products for granzyme B, IFN-γ, and CD94, whereas another was positive for FasL, TNF-α, and CD94 (Figure 2B). At this stage we cannot decide whether this observation reflects a stochastic element in the PCR-based amplification from very small transcript numbers, cell biologic heterogeneity among RA+CCR7–27+ T cells, or heterogeneity of gene expression among individual cells from a homogeneous population.23
We conclude that, unlike naive cells, a proportion of RA+CCR7–27+ T cells express genes associated with cytolytic T-cell effector functions. Moreover, based on the TREC data, RA+CCR7–27+ T cells display a replicative history that places them between naive and effector T lymphocytes. These results suggest that the RA+CCR7–27+ T-cell subset comprises cells that are evolving from a naive toward a more differentiated stage (E or EM stage or both). We refer to this population from here on as pre-effector (pE) T lymphocytes. In previous reports, granzyme B–positive and perforin-positive cells or CMV-specific cells have been observed among RA+27+ cells.2,24,25 Together with our data and those reported by Wills et al,8 this indicates that the RA+27+ phenotype is not sufficient to identify naive CD8+ T cells. Based on the gene expression profiles presented, we conclude that at present naive T lymphocytes are best defined as RA+CCR7+.
The CD8+ pE population includes CD28+ and CD28– cells
To further characterize the pE subset, the expression of the cell surface molecules involved in costimulatory and activation functions (CD57, CD28, HLA-DR), in lymphocyte migration (CD11a), in differentiation processes (CD95/Fas), and in functions of NK cells (CD56, CD94, and CD158b) was analyzed (Figure 3). In line with our previous observations, the RA+CCR7–27+ (pE) T cells shared phenotypic characteristics with antigen-primed cells; they highly expressed CD11a and CD95 in all analyzed individuals (n = 8). A fraction of these cells also expressed HLA-DR or NK receptors, whereas CD57 and early T-cell activation markers such as CD69 and CD25 could hardly be detected (Figure 3A and data not shown). Finally, pE cells showed intermediate side-scatter (SSC)/granularity properties when compared to naive and effector T cells.
Expression of cell surface molecules. (A) The pE T-cell subset shares cell surface molecules with effector T lymphocytes. Purified CD8 T cells were stained with CD8, CCR7, CD45RA, CD27 mAbs and other mAbs as indicated. Histograms show the proportion of positive cells among RA+CCR7+27+ (N), RA+CCR7–27+ (pE), and RA+CCR7–27– (E) gated subsets. These results were consistent in 8 healthy individuals. Note that our experimental procedure (see “Materials and methods” for details) to analyze N, pE, and E CD8 T subpopulations allows the exclusion of any residual contaminating NK cells as shown by anti-CD3 costaining. (B) Analysis of CD28 expression on N, pE, and E CD8+ T-cell subsets. Naive and effector T cells displayed, respectively, RA+CCR7+27+28+ and RA+CCR7–27–28– phenotypes. In contrast, the pE CD8 subset contained CD28+ as well as CD28– cells. Accordingly, pE T cells are split into pE1 (RA+CCR7–27+28+) and pE2 (RA+CCR7–27+28–) subsets.
Expression of cell surface molecules. (A) The pE T-cell subset shares cell surface molecules with effector T lymphocytes. Purified CD8 T cells were stained with CD8, CCR7, CD45RA, CD27 mAbs and other mAbs as indicated. Histograms show the proportion of positive cells among RA+CCR7+27+ (N), RA+CCR7–27+ (pE), and RA+CCR7–27– (E) gated subsets. These results were consistent in 8 healthy individuals. Note that our experimental procedure (see “Materials and methods” for details) to analyze N, pE, and E CD8 T subpopulations allows the exclusion of any residual contaminating NK cells as shown by anti-CD3 costaining. (B) Analysis of CD28 expression on N, pE, and E CD8+ T-cell subsets. Naive and effector T cells displayed, respectively, RA+CCR7+27+28+ and RA+CCR7–27–28– phenotypes. In contrast, the pE CD8 subset contained CD28+ as well as CD28– cells. Accordingly, pE T cells are split into pE1 (RA+CCR7–27+28+) and pE2 (RA+CCR7–27+28–) subsets.
Recent studies have proposed that CD27+CD28+ T cells differentiate through a CD27+CD28– to a CD27–CD28– stage.26 According to this model, CD8+ T cells sequentially down-regulate CCR7, CD28, and CD27 surface expression, while up-regulating expression of molecules that confer cytolytic activity. We found that around half of the pE T cells (mean ± SD, 48% ± 15%; range, 29%-78%; n = 8) expressed CD28, forming a distinct CD28+ subpopulation (Figure 3B). We refer to the RA+CCR7–27+28+ pE cells as pE1, and to the RA+CCR7–27+28– population as pE2. The goal of the next studies was to determine whether pE1 and pE2 cells are 2 functionally distinct CD8+ T-cell subsets or 2 sequential stages of the T-cell differentiation pathway (pE1→pE2→E/EM).
Both pE1 and pE2 subsets exhibit ex vivo killing activity, but pE2 T cells express increased levels of granzyme B and perforin
To investigate whether pE1 and pE2 subsets differed in the expression of genes involved in T-cell effector functions, we compared the presence of the corresponding mRNAs in 5-cell aliquots of these populations (Figure 4A). The proportion of samples containing detectable IFN-γ and TNF-α transcripts was similar in both subsets. However, the number of samples positive for granzyme B and CD94 mRNA was higher in the pE2 population. Thus, the gene expression profile of pE2 cells resembles more closely that of the effector population. These results correlated with the analysis of granzyme B expression by intracellular staining for this enzyme (Figure 4B). Around 40% of total pE (RA+CCR7–27+), but only 16% of pE1 (RA+CCR7–28+) contained detectable granzyme B protein. Note that these percentages clearly exceed the minimum estimates of the frequency of granzyme B mRNA-containing cells, suggesting that the RT-PCR analysis significantly underestimates the proportion of granzyme B–expressing cells in these populations. According to both mRNA analysis (Figure 4A) and intracellular staining (Figure 4B), both pE subsets express perforin but levels are lower than in the E subset. An extended study of 4 healthy donors confirmed that there is a gradual increase of granzyme B and perforin protein from pE1 through pE2 to E T cells, as summarized in Table 1.
Expression of effector mediators in pE1 and pE2 T cells. (A) Gene expression analysis was performed on sorted RA+CCR7+27+28+ (N), RA+CCR7–27+28+ (pE1), RA+CCR7–27+28– (pE2), and RA+CCR7–27–28– (E) CD8+ T cells by RT-PCR. The same set of primers as described in Figure 2B was used. Data from 6 or 8 independent 5-cell aliquots are shown. These results are representative of 2 healthy individuals; (–), negative; (+), positive controls. (B) The proportion of granzyme B–positive or perforin-positive cells among N (27+), pE1 (28+), pE (27+), pE2 + E (28–), and E (27–) T cells was determined by immunofluorescence. The pE population includes pE1 (27+28+) and pE2 (27+28–) T cells. Note that the perforin signal is considerably lower in pE than in E cells. (C) Ex vivo–sorted CD8+,N,pE1,pE2, and E T cells were tested in a redirected cytolytic assay against 51Cr-labeled P815 target cells. The pE and E subsets were unable to lyse P815 cells in the absence of CD3 mAbs (lysis ≥ 2%, data not shown). Data are representative of 2 healthy donors.
Expression of effector mediators in pE1 and pE2 T cells. (A) Gene expression analysis was performed on sorted RA+CCR7+27+28+ (N), RA+CCR7–27+28+ (pE1), RA+CCR7–27+28– (pE2), and RA+CCR7–27–28– (E) CD8+ T cells by RT-PCR. The same set of primers as described in Figure 2B was used. Data from 6 or 8 independent 5-cell aliquots are shown. These results are representative of 2 healthy individuals; (–), negative; (+), positive controls. (B) The proportion of granzyme B–positive or perforin-positive cells among N (27+), pE1 (28+), pE (27+), pE2 + E (28–), and E (27–) T cells was determined by immunofluorescence. The pE population includes pE1 (27+28+) and pE2 (27+28–) T cells. Note that the perforin signal is considerably lower in pE than in E cells. (C) Ex vivo–sorted CD8+,N,pE1,pE2, and E T cells were tested in a redirected cytolytic assay against 51Cr-labeled P815 target cells. The pE and E subsets were unable to lyse P815 cells in the absence of CD3 mAbs (lysis ≥ 2%, data not shown). Data are representative of 2 healthy donors.
Cytolytic properties and replicative history of peripheral blood pE CD8+ T lymphocytes
. | N RA+CCR7+ 27+28+ . | pE1 RA+CCR7- 27+28+ . | pE2 RA+CCR7- 27+28- . | E RA+CCR7- 27-28- . |
|---|---|---|---|---|
| Fas/CD95*, % | 5 | 78 | 66‡ | 72 |
| Granzyme B*, % | 1 | 25 | 49‡ | 93 |
| Perforin*, % | 0 | 23 | 41‡ | 95 |
| Cytolysis 51Cr, % | < 3§ | 12 | 12 | 100 |
| IFN-γ/TNF-α | - | + | + | + |
| NKR/CD94 | - | +/- | ++ | +++ |
| TRECs† | 13 | 2 | 1.5 | ND |
| Telomere length, kb | 7 | 10 | 10 | 5.5 |
. | N RA+CCR7+ 27+28+ . | pE1 RA+CCR7- 27+28+ . | pE2 RA+CCR7- 27+28- . | E RA+CCR7- 27-28- . |
|---|---|---|---|---|
| Fas/CD95*, % | 5 | 78 | 66‡ | 72 |
| Granzyme B*, % | 1 | 25 | 49‡ | 93 |
| Perforin*, % | 0 | 23 | 41‡ | 95 |
| Cytolysis 51Cr, % | < 3§ | 12 | 12 | 100 |
| IFN-γ/TNF-α | - | + | + | + |
| NKR/CD94 | - | +/- | ++ | +++ |
| TRECs† | 13 | 2 | 1.5 | ND |
| Telomere length, kb | 7 | 10 | 10 | 5.5 |
NKR indicates NK cell receptor; ND, not detectable; and kb, kilobase.
Percentage of positive cells by FACS analysis (mean percent of analysis performed in 5 healthy individuals).
Mean numbers of TRECs per 100 cells from 4 healthy individuals (Figure 6A).
Expression of Fas, granzyme B, and perforin in pE2 T cells was calculated based on the proportion of pE2 T cells in the total pE population.
Comparison of the lytic activity found in N, pE1, and pE2 subsets with that found in the E subset (Figure 4C).
Our results are in agreement with a model according to which there is a differentiation pathway, N→pE1→pE2→E, from naive to effector cells. The observation that CD95/Fas expression was high in both pE1 and pE2 subsets and similar to the level in effector T cells (Table 1) suggests that CD95/Fas is an early event in the differentiation process that occurs before granzyme B up-regulation. When we compared the cytolytic activity of these cell populations, using a CD3 mAb-mediated redirected 51Cr-release assay (Figure 4C), we found, as expected, that effector T cells displayed high lytic activity, whereas naive T cells were inactive. Both pE1 and pE2 subsets had comparable cytolytic activity, which was about 10 times lower than that of the effector population.
pE2 and effector T-cell subsets contain CMV-specific T lymphocytes
The common herpes virus, CMV, persists at low frequencies in healthy individuals and provides a chronic stimulus to the immune system. CMV-specific CD8+ T cells can eliminate virus-infected cells and may play a major role in the control of these persistent infections. Previous studies reported that CMV-specific CD8+ T cells belonged to 2 distinct subsets, effector (E; RA+CCR7–CD27–) and effector-memory (EM1;RA–CCR7–CD27+) cells.7,24,27 To test whether CMV-specific T cells would be present as well in the 2 pE subsets, we analyzed the phenotype of T cells staining with HLA-A2/CMVpeptide tetramers from a healthy individual with a relatively high frequency (∼ 6% of total CD8 T cells) of such cells. We observed that most of the antigen-specific cells displayed the phenotype of pE2 (33%; RA+CCR7–27+28–) or E (62%; RA+CCR7–27–28–) CD8+ T cells (Figure 5A). Only a small proportion (< 5%) had pE1 (RA+CCR7–27+28+) characteristics. RT-PCR analysis of sorted CMV-specific cells with pE and E phenotypes showed that both subsets had a gene expression profile similar to that of bulk pE2 and E CD8+ T-cell populations; they contained granzyme B, perforin, IFN-γ, TNF-α, and CD94 mRNA. These data demonstrate that a considerable fraction of CMV-specific T cells from a healthy individual displays the salient properties of the newly described pE2 subset. These results were confirmed by analysis of other healthy individuals with lower percentages of HLA-A2/CMV peptide tetramers–specific CD8+ T cells (data not shown). Our findings are consistent with those of Wills et al,8 who observed the presence of CMV-specific T cells in the CD45RA+27+28– subpopulation.
Most of CMV-specific CD8+ T cells are found among pE2 and effector subsets. (A) Phenotypic analysis of purified CD8+ T cells from a healthy CMV carrier following staining with CMV multimers and the indicated mAbs. (B) Gene expression analysis on sorted RA+CCR7–27+ (pE) and RA+CCR7–27– (E) CMV+ T-cell subsets by RT-PCR. The same set of primers as described in Figure 2B was used. Data from 7 independent 5-cell aliquots are shown. The phenotype as well as the pattern of gene expression of these CMV-specific T cells was stable over an interval of 8 months. The (–) indicates negative and (+) positive controls.
Most of CMV-specific CD8+ T cells are found among pE2 and effector subsets. (A) Phenotypic analysis of purified CD8+ T cells from a healthy CMV carrier following staining with CMV multimers and the indicated mAbs. (B) Gene expression analysis on sorted RA+CCR7–27+ (pE) and RA+CCR7–27– (E) CMV+ T-cell subsets by RT-PCR. The same set of primers as described in Figure 2B was used. Data from 7 independent 5-cell aliquots are shown. The phenotype as well as the pattern of gene expression of these CMV-specific T cells was stable over an interval of 8 months. The (–) indicates negative and (+) positive controls.
Both pE1 and pE2 subsets show reduced level of TRECs but extended telomere lengths compared with naive T lymphocytes
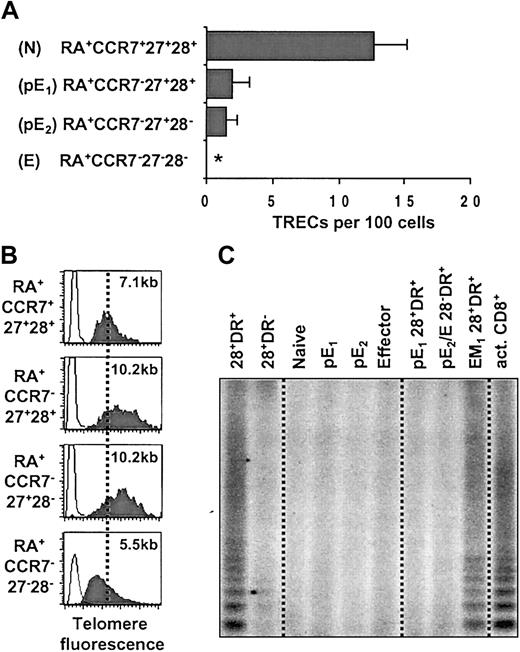
To analyze the replicative history of the pE subsets we compared TREC levels in RA+CCR7+27+28+ (N), RA+CCR7–27+28+ (pE1), RA+CCR7–27+28– (pE2), and RA+CCR7–27–28– (E) populations sorted from 4 young healthy individuals (Figure 6A). As expected, naive T cells displayed high levels of TRECs, whereas the TRECs in effector T cells were undetectable (≥ 0.01 TREC copies/100 cells). Both pE1 and pE2 subsets showed a similar reduction of TREC levels, to about 14% of those in naive cells in all individuals. These results indicate that both pE1 and pE2 subsets have undergone 3 to 4 more divisions than naive cells, implying that pE2 cells are not derived from pE1 cells by further cell divisions.
Both pE CD8+ T subsets display the same distinctive replicative history. (A) Real-time PCR quantification of TRECs was performed on sorted RA+CCR7+27+28+ (N), RA+CCR7–27+28+ (pE1), RA+CCR7–27+28– (pE2), and RA+CCR7–27–28– (E) CD8 subsets from 4 healthy individuals (< 35 years old). Asterisk indicates not detectable (sorted cell number was 105, lower quantification limit = 0.01%). (B) Telomere fluorescence analysis in sorted N, pE1, pE2, and E CD8+ T cells. The telomere fluorescence was converted to kilobase as described in “Materials and methods.” (C) Telomerase activity in cell extracts of sorted 28+DR+, 28+DR–, N, pE1, pE2, E, pE1 (28+DR+), pE2 + E (28–DR+), and EM1 (28+DR+) CD8+ T cells. As positive control, we used cell extracts of in vitro PHA-activated CD8+ T cell (act. CD8+). Data are representative of 4 healthy individuals.
Both pE CD8+ T subsets display the same distinctive replicative history. (A) Real-time PCR quantification of TRECs was performed on sorted RA+CCR7+27+28+ (N), RA+CCR7–27+28+ (pE1), RA+CCR7–27+28– (pE2), and RA+CCR7–27–28– (E) CD8 subsets from 4 healthy individuals (< 35 years old). Asterisk indicates not detectable (sorted cell number was 105, lower quantification limit = 0.01%). (B) Telomere fluorescence analysis in sorted N, pE1, pE2, and E CD8+ T cells. The telomere fluorescence was converted to kilobase as described in “Materials and methods.” (C) Telomerase activity in cell extracts of sorted 28+DR+, 28+DR–, N, pE1, pE2, E, pE1 (28+DR+), pE2 + E (28–DR+), and EM1 (28+DR+) CD8+ T cells. As positive control, we used cell extracts of in vitro PHA-activated CD8+ T cell (act. CD8+). Data are representative of 4 healthy individuals.
Telomeres progressively shorten as a function of cell division.28 Previous reports have shown that telomere length measurements are particularly useful to assess the replicative in vivo history of lymphocytes.16,29 When we measured the average telomere length in naive, pE1, pE2, and effector CD8 subsets by flow cytometry–fluorescence in situ hybridization15 (flow FISH), we observed a reduction in mean telomere fluorescence in effector compared with naive T cells that corresponded to a telomere shortening of about 1.6 kb (Figure 6B). This agrees with previously published results.15,30 Surprisingly, both pE1 and pE2 subsets displayed brighter telomere fluorescence compared with naive cells corresponding to an increase in average telomere length by about 3 kb. Telomere length measurements performed on another healthy donor also showed telomere length elongation by about 1 kb in the pE subset. In a third individual, the average telomere fluorescence of pE T cells was between that of naive and effector cells (data not shown). These results are consistent with the conclusion from the TREC data (Figure 6A) that pE1 and pE2 subsets have a similar in vivo replicative history. The finding that in the pE T cells of some healthy donors telomeres are longer than in their naive T cells strongly suggests that telomeres can be elongated during differentiation of naive into pE cells (Figure 6B) as a result of telomerase expression.
TRAP assays with extracts of FACS-sorted cells revealed no detectable telomerase activity in naive, pE1, pE2, and E cells (Figure 6C). Since Speiser et al31 have recently reported that, among CD8+ T lymphocytes, telomerase activity is detectable in cells that express CD28 and the activation marker HLA-DR, we carried out TRAP assays with the HLA-DR+ fraction of pE T cells (Figure 3A) but failed to detect telomerase activity (Figure 6C). On the other hand, the EM1 (RA–CCR7–28+DR+) T-cell subset was clearly telomerase positive, indicating that this population accounts for the previously described telomerase activity in CD8+28+DR+ T cells.31
Discussion
For many years antigen-experienced T lymphocytes have been classified into 2 distinct subpopulations—effector and memory cells.32 Effectors are generated early during the onset of an immune response and are capable of migrating to the site of infection. Such cells are short-lived, produce cytolytic effector mediators, and are capable ex vivo of killing target cells; they are also more susceptible to activation-induced cell death (AICD). In contrast, memory cells appear later in the immune response, lack ex vivo cytotoxicity, and are long-lived. However, many questions concerning the role of each subset as well as their lineage relationship to each other remain unanswered. Recently, Sallusto and coworkers have used a mAb that recognizes the chemokine receptor CCR7 to distinguish 2 subpopulations, designated central-memory and effector-memory T cells, respectively, among CD45RO+ T cells.3 Although the role of the so-called effector-memory cells is, at present unclear, these observations, together with the characterization of the properties of antigen-experienced T cells present after primary or during chronic infection with influenza virus, Epstein-Barr virus (EBV), CMV, or HIV7,8,24,26,27,33,34 challenge the simple classification of primed T cells into effector or memory subsets.
In the present study we describe 2 populations of CD45RA+CCR7–27+28+ and CD45RA+CCR7–27+28– cells within the circulating pool of CD8+ T cells of healthy individuals. They share phenotypic and functional characteristics, intermediate between those of naive and of effector cells. Their properties suggest the existence of additional heterogeneity of primed T lymphocytes. Both populations have a similar replicative history; they have undergone more cell divisions than naive cells but fewer than effector cells. In some individuals, their telomeres are, on average, longer than those of naive cells, although they do not contain detectable telomerase activity. Thus, unless they are derived from precursors with telomeres longer than those of the majority of naive T cells, they must have passed through a stage in which telomerase elongated their telomeres. They express mediators characteristic of effector cells and have detectable cytolytic activity, albeit less than that of effectors. The properties of these cells suggest that they represent intermediate stages in the differentiation of naive to effector cells; hence we propose the designation “pre-effectors,” or pE cells. Both CD28+ and CD28– pE subsets, referred to as pE1 and pE2, respectively, have a similar in vivo replicative history but the gene expression profile of the pE2 cells resembles more closely that of effector T cells. These results suggest that pE2 cells have acquired further effector characteristics without undergoing additional cell divisions.
Our data are also in agreement with a model according to which there is a differentiation pathway, N→pE1→pE2→E, from naive to effector T lymphocytes. One way to test the validity of such a model is to measure the TREC levels within the memory subpopulation. In this regard, we found an average of 0.4 ± 0.2 TRECs/100 cells (n = 4, N.R., A.Z. et al, unpublished data, December 2002) in sorted CM (CD8+RA–CCR7+27+) T cells from young healthy individuals. These levels are within the same range as those found for the pE cells (n = 8, 1.5 ± 1.1 TRECs/100 cells, Figure 2A). Unless pE T cells differentiate from CM T cells without cell division, these observations support the notion that pE cells may descend from naive T cells rather than from memory T cells. Their increased average telomere length, compared with naive cells, indicates that activation was accompanied by induction of telomerase expression.35 In line with these results, Plunkett et al36 have recently reported that during acute viral infection with EBV, virus-specific CD8+ T lymphocytes did not have shorter telomeres after in vivo clonal expansion. The telomeres of germinal center B cells that have undergone extensive clonal expansion and selection are also longer than those of their naive B-lymphocyte precursors.37 Transient expression of telomerase was observed in both studies.36,37 Because we could not detect telomerase activity in pE T lymphocytes from peripheral blood, it would be of interest to find out whether such activity is present in T lymphocytes isolated from lymph nodes. We observed a marked telomere shortening in CD8+ effector T cells (Figure 6C), and Plunkett et al36 found short telomeres in EBV-specific T cells several years after acute infection. This implies that effector T cells have undergone further extensive cell proliferation, without expressing telomerase (Figure 6C, see lines effector and pE2/E 28–DR+).
The comparison of the diversity of the T-cell repertoires in different CD8+ T-cell subsets using spectratyping,38 which measures the size heterogeneity of the β chain of the TCR hypervariable CDR3 region (BV), is a tool to dissect the process of CD8+ T-cell differentiation. Spectratypes from CD8+RA+27– effector T cells reveal a limited TCR repertoire with large expansions of particular BV families (Hamann et al30 and data not shown). In contrast, pE T cells express a more complex but still skewed T-cell repertoire (data not shown). Observations made by others8 have revealed CMV-specific T cells with the same TCR clonotype sequence among CD27+CD28– (that includes the pE2 subset) and CD27–CD28– (that includes the E subset) CD8+ T cells. Although preliminary, these results further support the model according to which pE and E cells represent different stages of the same lineage.
As proposed by Appay and colleagues,26 CD8+ T-cell differentiation may correlate with progressive loss of CCR7, CD28, and CD27 surface expression and up-regulation of cytolytic capacity. Our data are in agreement with this view, in that we have identified CD8+ T lymphocytes with intermediate cell surface phenotypes and partial effector functions in the peripheral blood from healthy individuals. The same authors described that during primary infection with HIV-1, EBV, or hepatitis C virus, the virus-specificT cells in the peripheral blood display a 27+28+/– perforinlow phenotype,26 which resembles that of the pE T cells described in this report. Thus, peripheral blood pE T cells may be in transit from the draining lymph nodes to the site of inflammation where they complete their differentiation into differentiated effector or effector-memory cells or both.
Longitudinal studies in which the turnover of the pE T cells can be determined are necessary to account for the relative high proportion of these cells in the total CD8+ compartment (∼ 10%; Figure 1C) or in the CMV-specific compartment (∼ 33%, Figure 5A) of healthy individuals. Our observations are consistent with the hypothesis that pE T cells are antigen-specific cells that have not encountered all the signals required for differentiation into effectors. They may, for example, accumulate in the circulating pool over time in response to persistent but low levels of antigen. A recent study of CD4+ T-cell responses to antigenic stimulation in the mouse,33 showing that those effector T cells that have divided and differentiated the most are recruited to the site of inflammation, supports this hypothesis. Other partially activated subsets observed during the acute infection phase persist after viral clearance.33 A conclusive answer to the questions concerning the function of CD8+ pE cells and their relationship with other subpopulations requires the tracking of antigen-specific T cells in individuals during and after specific immune responses against CMV, EBV, or influenza viruses.
In conclusion, by combining simultaneous analysis of 5-cell surface markers (by flow cytometry) with analysis of gene expression (by RT-PCR), of replicative history (by measurement of TRECs), and of telomere length (by flow FISH) in sorted cells we have identified 2 subpopulations among primed CD8+ T cells, with characteristics distinct from those of effector as well as of memory T cells. Understanding the position of these subsets in the process of CD8+ T-cell differentiation and their role in immune responses will help us to improve vaccination and adoptive immunotherapy strategies.
Prepublished online as Blood First Edition Paper, May 15, 2003; DOI 10.1182/blood-2003-02-0420.
Supported by the National Center of Competence in Research (NCCR) Molecular Oncology program of the Swiss National Science Foundation (N.R.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are thankful to Drs H. Robson MacDonald, Daniel Speiser, and Joachim Lingner for critically reading the manuscript; Dr Immanuel Lüscher and Philippe Guillaume for synthesis of multimers; Dr Daniel Douek for providing signal joint internal standard; and Dr Martin Lipp and Dr Reinhold Forster for the anti-CCR7 mAb. We thank Séverine Reynard, Solange Visher, and Martine van Overloop for excellent technical support and Dr Anne-Lyse Ducrest for helpful discussions.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal