Abstract
Activating mutations of receptor tyrosine kinases (RTKs) and their downstream affectors are common in acute myeloid leukemia (AML). We performed mutational analysis of FLT3, c-kit, c-fms, vascular endothelial growth factor (VEGF) receptors (Flt-1, KDR [kinase domain receptor]), and ras genes in a group of 91 pediatric patients with AML treated on Children's Cancer Group clinical trial CCG-2891. Forty-six percent of patients had activating mutations of FLT3 (24.5%), c-kit (3%), or ras (21%) genes. Mutation-positive patients had a higher median diagnostic white blood cell (WBC) count (71.5 vs 19.6 × 109/L; P = .005) and lower complete remission rate (55% versus 76%; P = .046) than mutation-negative patients. The Kaplan-Meier estimate of overall survival (OS) for patients with and without an activating mutation was 34% versus 57%, respectively (P = .035). However, within this group, patients with FLT3/ALM (activation loop mutation) had good outcomes (OS, 86%). Exclusion of the FLT3/ALM from analysis decreased the OS for the remaining mutation-positive patients to 26% (P = .003). Ten of the 23 mutation-positive and 11 of the 34 mutation-negative patients received an allogeneic bone marrow transplant (BMT) in first complete remission (CR). In the mutation-positive group, the disease-free survival (DFS) for the allogeneic BMT recipients was 72% versus 23% for the 13 patients who received chemotherapy or autologous BMT (P = .01). DFS for the mutation-free patients with and without allogeneic BM transplantation was 55% and 40%, respectively (P = .38). Activating mutations in the RTK/ras signaling pathway are common in pediatric AML, and their presence may identify a population at higher risk of poor outcome who may benefit from allogeneic BM transplantation.
Introduction
Receptor tyrosine kinases (RTKs) and their downstream affectors (ras, JAK/STAT [Janus kinase/signal transducer and activator of transcription]) have emerged as significant components in the pathogenesis of malignancies.1-3 Whether it is the constitutive activation of the receptor by an intrinsic receptor mutation (FLT3, kit, and fms mutations),4-6 the autocrine/paracrine stimulation of the receptor by a ligand secreting tumor (VEGF receptor),7,8 or the activation of the downstream affectors (eg, ras),9-11 such activating events directly contribute to disease pathogenesis and progression. The role of individual receptors in the pathogenesis of acute myeloid leukemia (AML) has been receiving increasing attention. Mutations in the FLT3 receptor gene have been demonstrated to be the most common genetic alteration in AML, and these mutations are associated with rapid disease progression and resistance to conventional therapy.12-15 The FLT3 internal tandem duplication (FLT3/ITD) as well as the FLT3 activation loop mutation (FLT3/ALM) lead to constitutive activation of the RTK.12,16,17 Other members of this class of receptors (c-kit, c-fms, and vascular endothelial growth factor [VEGF] receptors) have also been implicated in the pathogenesis of AML.18,19 Activating mutations in c-kit and c-fms receptor genes involve point mutations in the juxtamembrane or the kinase domains which lead to constitutive activation of their respective receptors and have been identified in 2% to 10% of myeloid malignancies,6,19-21 including mastocytosis, myelodysplastic syndrome (MDS), and AML. Although the presence of such activating mutations has been demonstrated in adult AML, the prevalence and clinical significance of such mutations is unknown in pediatric AML.
Findings implicate vascular endothelial growth factor (VEGF) in pathogenesis of AML,8,17,22-25 and there is evidence of increased marrow vascularization in patients with AML17 as well as high levels of the VEGF ligand associated with poor outcome in AML.8 Although activating mutations of the VEGF receptors have been constructed in vitro,7,26 such mutations have not been identified in malignancies.
In addition to the activating mutations at the receptor level, activating mutations of the secondary mediator of the RTKs (eg, ras gene) have also been reported in AML.6,27 Although mutations in the ras gene have been clearly documented in AML, the clinical significance of these mutations remains in doubt.13,27
There are very few studies that have characterized the prevalence and significance of the RTK/ras pathway in a single population. Therefore, we studied the prevalence of activating mutations of FLT3, c-kit, c-fms, Flt-1, and kinase domain receptor (KDR), as well as N-ras, and K-ras in a group of 91 pediatric patients with AML on Children's Cancer Group clinical trial CCG-2891 and examined the clinical characteristics and outcome of the patients with these mutations compared with the patients without the activating mutations.
Patients, materials, and methods
Patients and treatments
Newly diagnosed patients with de novo AML registered on Children's Cancer Group (CCG) pediatric AML protocol CCG-2891 were included in this study. One hundred available cryopreserved marrow specimens were obtained from CCG-AML reference laboratory. Nine patients with myelodysplastic syndrome (MDS), secondary AML, or Down syndrome were excluded, and the remaining 91 de novo patients' marrow specimens were used in the study. The study was approved by the Fred Hutchinson Cancer Research Center (FHCRC) Institutional Review Board and the CCG Biology Committee. The diagnosis of AML was made according to French-American-British (FAB) classification and was confirmed by the CCG Central Review Committee. The FLT3/ITD status of these patients was reported previously.15
Children's Cancer Group clinical trial CCG-2891 has been described in detail elsewhere.28-30 In brief, CCG-2891 is a prospective randomized trial that accrued 1114 patients from 1989 to 1995. This study randomly assigned pediatric patients with AML at diagnosis to receive 1 of 2 induction regimens involving 4-day cycles of 5 chemotherapeutic agents (daunomycin, cytarabine, etoposide, 6-thioguanine and dexamethasone). The second cycle was administered either after 10 days despite low counts (intensive timing) or after 14 days depending on the marrow status (standard timing). Induction consisted of a total of 4 cycles in both groups. At the end of induction, patients who achieved remission and had a compatible matched related donor went on to receive an allogeneic bone marrow transplant (BMT). Patients without related donors were randomly assigned to receive either nonmyeloablative chemotherapy or an autologous BMT.
Of the 91 patients tested, 59 were randomly assigned to the intensive timing and 30 to the standard timing induction arms. Two patients received both standard and intensive timing.
Mutational analysis
Mutational analysis of the FLT3 (FLT3/ITD and FLT3/ALM) and ras genes are detailed elsewhere.15,16,31
c-kit. The identification of exon 17 mutations in the c-Kit receptor were determined as previously described.18,32,33 Briefly, 180-bp polymerase chain reaction (PCR) products of c-Kit exon 17 were generated and subjected to direct sequencing to determine the presence of sequence changes. For samples that demonstrated both wild-type and a possible mutation, individual clones of the PCR products were sequenced to rule out false positives. We have observed no false-negative results using positive control samples at different concentrations.
Activating mutations of the c-fms gene have been previously described in codons 301 and 969. For c-fms mutational analysis total RNA was extracted from diagnostic marrow specimens using TRIzol protocol (Invitrogen, Carlsbad, CA). Juxtamembrane (JM) and the tyrosine kinase (TK) domain of the c-fms genes were amplified by one-step, duplex reverse transcriptase (RT)–PCR. RT-PCR amplification was performed using primers 301XLF, 5′-CCTCAACCTCGATCAAGTAGATTT-3′, and 301XLR, 5′-GTCCC AGGTAGGTCCA GTTAAAA-3′, for the JM region and 969FMSF, 5′-CTGACCTGCTGCGAGCAA-3′, and 969FMSR, 5′-ATCCATGGAGGAGTTGAAGTTTG-3′, for the TK region. Beta2 microglobulin control gene was also amplified with primers 5′B2, 5′-ATGTCTCGCTCCGTGGCCTTAGCT-3′, and 3′B2, 5′-CCTCCATGATG CTGCTTACATGTC-3′. The RT-PCR mixture contained 400 ng RNA, 12.5 pMol of each primer, 0.25 pMol of each 5′B2 and 3′B2 primers, and 3 mM MgCl2. Denaturing, annealing, and extension steps were performed at 94°C for 30 seconds, 55°C for 1 minute, and 72°C for 1 minute, respectively, for 35 cycles on a GeneAmp PCR system 2400 (Perkin Elmer, Emeryville, CA). There was an initial RT step at 42°C for 30 minutes, followed by an initial denaturation step at 94°C for 5 minutes, and a final extension step at 72°C for 7 minutes. Products were resolved on 5% polyacrylamide gel, and molecular weight bands corresponding with the c-fms gene were cut out for direct nucleotide sequencing. The beta2 bands were used as a reference for RNA integrity.
For VEGF receptor (Flt-1 and KDR) mutation analysis, juxtamembrane (JM) and the tyrosine kinase (TK) domain of the VEGF receptor genes were amplified by one-step RT-PCR. RT-PCR amplification was performed using conditions described as follows.
KDR kinase domain. Primers used were FKDR kinase, 5′-TCCACTTACCTGAGGAGCAAGA, and RKDR kinase, 5′-CCTTAGCCACTTGGAAGCTGT. RT-PCR was performed under the following conditions: 42°C for 30 minutes, 94°C for 5 minutes, 94°C for 30 seconds, 60°C for 1 minute for 35 cycles, 72°C for 2 minutes, and 72°C for 7 minutes.
KDR JM domain. Primers used were FKDRJM, 5′-CCATGTTCTTCTGGCTACTTCTT, and RKDRJM, 5′-CCAAGAGGCTTACCTAGCTTCA. RT-PCR was performed under the same conditions as for the kinase domain described in “KDR kinase domain.”
Flt-1 kinase domain. Primers used were FfltK, 5′-GGCCAGAGGCATGGAGTT, and RfltK, 5′-CGCTCTTGGTGCTGTAGATTTT. RT-PCR was performed under the following conditions: 42°C for 30 minutes, 94°C for 5 minutes, 94°C for 30 seconds, 56°C for 1 minute for 35 cycles, 72°C for 2 minutes, and 72°C for 7 minutes.
Flt-1 JM domain. Primers used were FfltJM, 5′-TCTGGCTCCTATTAACCCTCTTT, and RfltJM, 5′-AAGCCCCTCTTCCAAGTGAT. RT-PCR was performed under the same conditions as for the Flt-1 kinase domain.
All products were resolved on 5% polyacrylamide gel, and molecular weight bands corresponding with the Flt-1 or KDR genes were cut out for direct nucleotide sequencing.
Direct nucleotide sequencing. Bands cut out from polyacrylamide gel were incubated overnight at room temperature in diethylpyrocarbonate (DEPC)–H2O to extract dsDNA from the gel. The extracted dsDNA was directly sequenced using the Thermo Sequence Dye Terminator sequencing reaction.34
Statistical methods
Data obtained in CCG-2891 through July 31, 2001, were used to compare patients with activating mutations with patients without such mutations. The significance of observed differences in proportions was tested using the chi-squared test and Fisher exact test when data were sparse. For continuous data, the Mann-Whitney test was used to compare the medians of distributions.35
Patients lost to follow-up were censored at their last known point of study, with a cutoff of January 31, 2001. Actuarial estimates of overall survival (OS) from on study and disease-free survival (DFS), the time from achieving remission to marrow relapse or death, were calculated using the Kaplan-Meier method.36 Standard errors were calculated using Greenwood's formula.37 Differences in OS and DFS were tested for significance using the log-rank statistic.38
Results
Patient characteristics of study population versus CCG-2891
The clinical characteristics and outcome of the study population have previously been compared with that of the CCG-2891 study population.15 In brief, the study population had a higher median white blood cell (WBC) count at diagnosis than the rest of the CCG-2891 (37.5 versus 20.4, P = .007). Comparison of diagnostic WBC count by categories (0-20 × 109/L, 20-50 × 109/L, > 50 × 109/L) showed no difference between the study population and the rest of the CCG-2891 study (P = .25). The median age at presentation was higher for the patients in the study group compared with the rest of the CCG-2891 population (10.5 years versus 7.6 years, P = .05). Overall and disease-free survival were not different between the study population and the rest of the CCG-2891 population (OS of 46% versus 43%, P = .82; DFS of 45% versus 45%, P = .6).
Mutational analysis of the RTK genes
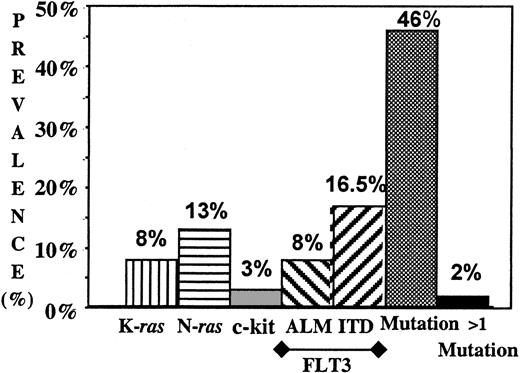
DNA samples of 91 pediatric patients with AML were tested for the presence of mutations in the JM or kinase domain of the FLT3, c-fms, Flt-1, and KDR receptor genes. The kinase domain (exon 11) for the c-kit gene was also tested for mutations. Of the 91 patients tested, FLT3/ITD was previously demonstrated in 15 patients (16.5%). Additionally, we identified FLT3/ALM and c-kit mutations of the kinase domains in 7 (8%) of 91 and 3 (3%) of 91 patients, respectively. All FLT3/ALMs were identified in codon 835. Five of the 7 mutations were a G>T (Asp→Tyr, Asp835Tyr), and the other 2 were G>C (Asp→His, Asp835His) transversions. Each of the 3 AML samples that had c-Kit mutations showed the previously described Asp816Val change.18,20,32,39 Twenty-five patients had a mutation of the FLT3 or c-kit gene. No mutations were identified in the tested regions of the c-fms, Flt-1, or KDR receptor genes. Thus, collectively, RTK mutations were identified in 25 (27%) of 91 patients tested (Figure 1).
Prevalence of the mutations of ras (N-ras and K-ras), c-kit, and FLT3 (ITD and ALM) in pediatric AML. Prevalence of all mutations as well as prevalence of patients with more than one mutation are presented.
Prevalence of the mutations of ras (N-ras and K-ras), c-kit, and FLT3 (ITD and ALM) in pediatric AML. Prevalence of all mutations as well as prevalence of patients with more than one mutation are presented.
Mutational analysis of the ras gene (N-ras and K-ras)
Exon 1 and 2 of the N-ras, and K-ras were evaluated for point mutations by genomic PCR, single-strand conformation polymorphism (SSCP), and direct sequencing. A total of 19 (21%) patients had a mutation in codon 12 or 13 of either N-ras 1 (n = 12) or K-ras 1 (n = 7) with no mutations identified in N-ras 2, or K-ras 2 genes. No patient had more than one ras mutation. One patient with N-ras mutation had a concomitant FLT3/ITD, and one more patient with K-ras mutation had a c-kit mutation. Otherwise, 40 (95%) of 42 patients with mutations had only a single activating mutation, and an additional 2 (5%) patients had 2 activating mutations (Figure 1). Thus, in summary, 42 of 91 (46%) patients tested had at least one activating mutation.
Clinical characteristics of the study population
The parameters of age, diagnostic WBC count, bone marrow blast percentage, cytogenetics, and FAB classification are shown in Table 1. There was no significant difference in the median age of the patients with and without activating mutations. Patients with FLT3/ITD, N-ras, and K-ras had an elevated diagnostic WBC count, and, as a collective group, patients with activating mutations had a median diagnostic white count of 71.5 × 109/L compared with 19.6 × 109/L for the patients without mutations (P = .005). Patients with activating mutations also had a higher marrow blast percentage (median, 87%) compared with the patients without activating mutations (median, 70%; P = .003).
Laboratory characteristics of patients with and without activating mutations
. | Flt3/ITD N = 15 . | Flt3/PM N = 7 . | c-kit N = 3 . | N-ras N = 12 . | K-ras N = 7 . | All mutations N = 42 . | No mutation N = 49 . | P (mutation vs. no mutation) . |
|---|---|---|---|---|---|---|---|---|
| Median age, y (range) | 8.3 (3.1-18.7) | 7.3 (2.3-13.3) | 15.2 (13.0-17.6) | 10.6 (1.1-18.7) | 5.5 (0.5-17.9) | 8.2 (0.5-18.7) | 11.2 (0.2-17.2) | .8 |
| Median WBC, × 109/L (range) | 73.8 (4.9-453.1) | 37.5 (2.2-480) | 41 (9-106.7) | 56.8 (4.2-351.2) | 80.8 (1.6-345.2) | 71.4 (1.6-480) | 19.6 (0.5-329) | .005 |
| Marrow blast, % (range) | 89 (48-99) | 84 (25-95) | 62 (57-87) | 78 (40-99) | 87 (57-95) | 87 (25-99) | 70 (5-99) | .003 |
| FAB class | .462 | |||||||
| M0, n (%) | 1 (7) | 0 | 0 | 0 | 0 | 1 (2) | 0 | |
| M1, n (%) | 2 (13) | 2 (29) | 1 (33) | 3 (25) | 2 (29) | 9 (21) | 6 (12) | |
| M2, n (%) | 6 (40) | 0 | 0 | 4 (33) | 1 (14) | 10 (24) | 16 (33) | |
| M3, n (%) | 2 (13) | 2 (29) | 0 | 0 | 0 | 4 (10) | 5 (10) | |
| M4, n (%) | 4 (27) | 1 (14) | 1 (33) | 4 (33) | 2 (29) | 12 (29) | 15 (31) | |
| M5, n (%) | 0 | 1 (14) | 1 (33) | 1 (8) | 2 (29) | 5 (12) | 4 (8) | |
| M6, n (%) | 0 | 0 | 0 | 0 | 0 | 0 | 2 (4) | |
| M7, n (%) | 0 | 1 (14) | 0 | 0 | 0 | 1 (2) | 1 (2) | |
| Karyotype | N = 12 | N = 2 | N = 1 | N = 9 | N = 5 | N = 27 | N = 36 | .688 |
| Normal, n (%) | 5 (47) | 0 | 1 (100) | 4 (44) | 1 (20) | 10 (37) | 10 (28) | |
| t(15;17), n (%) | 2 (17) | 2 (100) | 0 | 0 | 0 | 4 (15) | 4 (11) | |
| t(8;21), n (%) | 1 (8) | 0 | 0 | 3 (33) | 0 | 3 (11) | 5 (14) | |
| Inv 16, n (%) | 1 (8) | 0 | 0 | 0 | 0 | 1 (4) | 5 (14) | |
| Abn 11, n (%) | 0 | 0 | 0 | 1 (11) | 2 (40) | 3 (11) | 5 (14) | |
| -7, n (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| -5, n (%) | 0 | 0 | 0 | 0 | 0 | 0 | 2 (6) | |
| Other, n (%) | 3 (25) | 0 | 0 | 1 (11) | 2 (40) | 6 (22) | 5 (14) |
. | Flt3/ITD N = 15 . | Flt3/PM N = 7 . | c-kit N = 3 . | N-ras N = 12 . | K-ras N = 7 . | All mutations N = 42 . | No mutation N = 49 . | P (mutation vs. no mutation) . |
|---|---|---|---|---|---|---|---|---|
| Median age, y (range) | 8.3 (3.1-18.7) | 7.3 (2.3-13.3) | 15.2 (13.0-17.6) | 10.6 (1.1-18.7) | 5.5 (0.5-17.9) | 8.2 (0.5-18.7) | 11.2 (0.2-17.2) | .8 |
| Median WBC, × 109/L (range) | 73.8 (4.9-453.1) | 37.5 (2.2-480) | 41 (9-106.7) | 56.8 (4.2-351.2) | 80.8 (1.6-345.2) | 71.4 (1.6-480) | 19.6 (0.5-329) | .005 |
| Marrow blast, % (range) | 89 (48-99) | 84 (25-95) | 62 (57-87) | 78 (40-99) | 87 (57-95) | 87 (25-99) | 70 (5-99) | .003 |
| FAB class | .462 | |||||||
| M0, n (%) | 1 (7) | 0 | 0 | 0 | 0 | 1 (2) | 0 | |
| M1, n (%) | 2 (13) | 2 (29) | 1 (33) | 3 (25) | 2 (29) | 9 (21) | 6 (12) | |
| M2, n (%) | 6 (40) | 0 | 0 | 4 (33) | 1 (14) | 10 (24) | 16 (33) | |
| M3, n (%) | 2 (13) | 2 (29) | 0 | 0 | 0 | 4 (10) | 5 (10) | |
| M4, n (%) | 4 (27) | 1 (14) | 1 (33) | 4 (33) | 2 (29) | 12 (29) | 15 (31) | |
| M5, n (%) | 0 | 1 (14) | 1 (33) | 1 (8) | 2 (29) | 5 (12) | 4 (8) | |
| M6, n (%) | 0 | 0 | 0 | 0 | 0 | 0 | 2 (4) | |
| M7, n (%) | 0 | 1 (14) | 0 | 0 | 0 | 1 (2) | 1 (2) | |
| Karyotype | N = 12 | N = 2 | N = 1 | N = 9 | N = 5 | N = 27 | N = 36 | .688 |
| Normal, n (%) | 5 (47) | 0 | 1 (100) | 4 (44) | 1 (20) | 10 (37) | 10 (28) | |
| t(15;17), n (%) | 2 (17) | 2 (100) | 0 | 0 | 0 | 4 (15) | 4 (11) | |
| t(8;21), n (%) | 1 (8) | 0 | 0 | 3 (33) | 0 | 3 (11) | 5 (14) | |
| Inv 16, n (%) | 1 (8) | 0 | 0 | 0 | 0 | 1 (4) | 5 (14) | |
| Abn 11, n (%) | 0 | 0 | 0 | 1 (11) | 2 (40) | 3 (11) | 5 (14) | |
| -7, n (%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| -5, n (%) | 0 | 0 | 0 | 0 | 0 | 0 | 2 (6) | |
| Other, n (%) | 3 (25) | 0 | 0 | 1 (11) | 2 (40) | 6 (22) | 5 (14) |
Cytogenetic information on the study population is presented in Table 1. Twenty-seven (64%) of the 42 patients with activating mutation had informative cytogenetics. Ten (37%) of the 27 patients had normal cytogenetics, 8 (30%) of 27 had favorable cytogenetics (4 with t(15;17), 3 with t(8;21), and 1 with inv16), and 3 (11%) patients had abnormal chromosome 11. No patient with activating mutation had monosomy 7 or deletion 5. In the mutation-free group, 36 (73%) of 49 patients had informative cytogenetics. Twenty-four (67%) of the 36 patients had either normal (28%) or favorable (39%) cytogenetics. Five patients (14%) had abnormal chromosome 11, and 2 patients had monosomy 5 (6%).
Clinical outcome
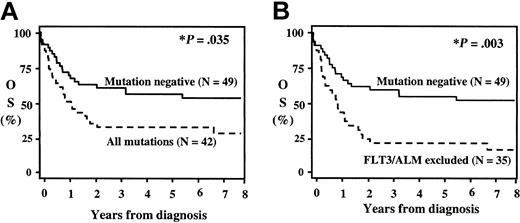
Complete remission (CR) rate and overall survival (OS) for patients with and without activating mutations is shown in Figure 2 and Table 2. As presented previously,15 patients with FLT3/ITD had a significantly worse CR and OS. With the exception of patients with FLT3/ALM, the remainder of patients with activating mutations had a lower CR rate and overall survival than the mutation-free patients. FLT3/ALM-positive patients had a remission induction rate of 86%. Patients with activating mutations as a group had a CR rate of 55% compared with 76% for the mutation-free patients (P = .046). Exclusion of FLT3/ALM from analysis reduced the remission induction for the remainder of mutation-positive population to 49% (P = .035). Twenty-five (60%) of the 42 patients with a mutation had received an intensively timed induction regimen compared with 34 (69%) of 49 of the mutation-free population. Remission rate with intensively timed regimen for patients with mutation was 55% compared with 76% in the mutation-free group (P = .046). Sixteen mutation-positive (39%) and 14 mutation-negative (29%) patients received standard timed induction regimen. Remission rate with standard timed regimen for the mutation-positive group was 56% versus 45% for the mutation-negative group (P = .58).
Actuarial overall survival (OS) from diagnosis for patients with and without RTK/ras-activating mutations. (A) Actuarial OS from diagnosis for all patients with and without activating mutations; (B) actuarial OS from diagnosis for patients without and with activating mutations after exclusion of FLT3/ALM patients. *P reflects log-rank comparison.
Actuarial overall survival (OS) from diagnosis for patients with and without RTK/ras-activating mutations. (A) Actuarial OS from diagnosis for all patients with and without activating mutations; (B) actuarial OS from diagnosis for patients without and with activating mutations after exclusion of FLT3/ALM patients. *P reflects log-rank comparison.
Clinical characteristics of patients with and without activating mutations
. | Flt3/ITD N = 15 . | Flt3/ALM N = 7 . | c-kit N = 3 . | N-ras N = 12 . | K-ras N = 7 . | All mutations N = 42 . | No mutation N = 49 . | P (mutation vs. no mutation) . |
|---|---|---|---|---|---|---|---|---|
| Median WBC, × 109/L (range) | 73.8 (4.9-453.1) | 37.5 (2.2-480) | 41 (9-106.7) | 56.8 (4.2-351.2) | 80.8 (1.6-345.2) | 71.4 (1.6-480) | 19.6 (0.5-329) | .005 |
| CR (%) | 6 (40) | 6 (86) | 2 (67) | 7 (58) | 3 (43) | 23 (55) | 34 (76)* | .046 |
| 8-y OS (%) | 2 (13) | 6 (86) | 1 (33) | 4 (33) | 2 (29) | 14 (34) | 28 (57) | .035 |
| HR for death† (P) | 2.5 (0.01) | 0.3 (0.23) | 2.1 (0.33) | 1.7 (0.2) | 3.4 (0.02) | 1.8 (0.04) | — | — |
| Allo-BMT in 1st CR | 1 allo BMT, 0 alive | 4 allo BMT, 4 alive | 1 allo BMT, alive | 4 allo BMT, 3 alive | 1 allo BMT, alive | 10 allo BMT, 8 alive | 11 allo BMT, 8 alive | .68 |
. | Flt3/ITD N = 15 . | Flt3/ALM N = 7 . | c-kit N = 3 . | N-ras N = 12 . | K-ras N = 7 . | All mutations N = 42 . | No mutation N = 49 . | P (mutation vs. no mutation) . |
|---|---|---|---|---|---|---|---|---|
| Median WBC, × 109/L (range) | 73.8 (4.9-453.1) | 37.5 (2.2-480) | 41 (9-106.7) | 56.8 (4.2-351.2) | 80.8 (1.6-345.2) | 71.4 (1.6-480) | 19.6 (0.5-329) | .005 |
| CR (%) | 6 (40) | 6 (86) | 2 (67) | 7 (58) | 3 (43) | 23 (55) | 34 (76)* | .046 |
| 8-y OS (%) | 2 (13) | 6 (86) | 1 (33) | 4 (33) | 2 (29) | 14 (34) | 28 (57) | .035 |
| HR for death† (P) | 2.5 (0.01) | 0.3 (0.23) | 2.1 (0.33) | 1.7 (0.2) | 3.4 (0.02) | 1.8 (0.04) | — | — |
| Allo-BMT in 1st CR | 1 allo BMT, 0 alive | 4 allo BMT, 4 alive | 1 allo BMT, alive | 4 allo BMT, 3 alive | 1 allo BMT, alive | 10 allo BMT, 8 alive | 11 allo BMT, 8 alive | .68 |
—indicates not applicable.
Excludes 4 patients who withdrew without knowledge of induction outcome.
Hazard ratio for death (mutation positive vs. mutation negative).
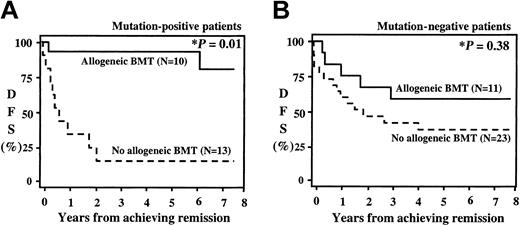
The actuarial OS at 8 years for the entire study population was 46%. Patients with activating mutations had an OS at 8 years of 34% versus 57% for the patients without mutations (P = .035, Figure 2A). Patients with FLT3/ALM had an excellent outcome (OS, 86%). Exclusion of the FLT3/ALM from survival analysis decreased the OS for the remainder of the mutation-positive patients to 26% (P = .003). All 6 of the FLT3/ALM-positive patients who are long-term survivors had received allogeneic BMT. Four of the 6 patients had received allogeneic BMT in first CR, and an additional 2 patients had received an unrelated donor BMT after relapse. As patients with FLT3/ALM had such an excellent outcome with BM transplantation, we evaluated the effect of BM transplantation for the entire study population (Figure 3). The actuarial DFS from end of induction for the entire study population was 45%, and DFS was comparable between the patients with and without activating mutations (46% versus 45%, P = .91). However, postinduction therapy varied, depending on the availability of an appropriate donor for transplantation. Patients in CR received an allogeneic BMT if a family donor was available. In the absence of an appropriate donor, patients were randomly assigned to conventional chemotherapy versus autologous BM transplantation. Of the 42 patients with activating mutations who achieved CR (23 patients), 10 (43%) received a matched related donor transplant, and the remaining 13 patients received conventional chemotherapy or an autologous stem cell transplant. Actuarial DFS from remission at 8 years for the mutation-positive group who received an allogeneic BMT was 72% compared with 23% for patients treated with chemotherapy or autologous transplantation (Figure 3A; P = .01). DFS for the mutation-free population based on the postinduction therapy is presented in Figure 3B. Actuarial DFS from remission for the 23 mutation-free patients who received conventional chemotherapy or autologous BMT was 40% compared with 55% for the 11 patients who received allogeneic BMT (P = .38).
Actuarial disease-free survival from remission for patients with (A) or without (B) activating mutations on the basis of postinduction therapy. Patients with matched family donor received an allogeneic bone marrow transplant (allogeneic BMT). Those without a family donor received either autologous BMT or chemotherapy (no allogeneic BMT). *P reflects log-rank comparison.
Actuarial disease-free survival from remission for patients with (A) or without (B) activating mutations on the basis of postinduction therapy. Patients with matched family donor received an allogeneic bone marrow transplant (allogeneic BMT). Those without a family donor received either autologous BMT or chemotherapy (no allogeneic BMT). *P reflects log-rank comparison.
Discussion
An extensive mutational analysis of the RTK/ras signal transduction pathway in a group of pediatric patients with AML treated on CCG-2891 revealed that nearly half of the patients tested harbored an activating mutation of either the RTK (FLT3, c-kit) or the ras pathway and that patients with such activating mutations as a collective group have a high diagnostic WBC count, lower CR, and lower survival. The data also show heterogeneity within the mutation-positive population and suggest that mutation-positive patients may have improved outcome with allogeneic BM transplantation.
The signal transduction pathway involving the mitogen-activated protein (MAP) kinase or the STAT pathway has been shown to be constitutively activated in a significant proportion of the patients with AML11,40,41 and may correlate with more aggressive disease and worse outcome.41 Activation of the signal transduction pathway may be due to the constitutive activation at the receptor level (FLT3/ITD, c-kit mutation, etc) or due to an intrinsic activating mutation of a downstream mediator (ras mutation). It has been demonstrated that the signal generated by the activated FLT3 receptor is transduced via both the ras and STAT pathways, thus constitutively activating both systems.42-44 Similar findings have been demonstrated for the c-kit and c-fms activating mutations.18,21,32,33,45 As the role of additional RTKs is being evaluated for involvement in myeloid leukemogenesis (VEGF, platelet-derived growth factor receptor [PDGFR]), and targeted therapies are being designed for counteracting their affects, the need for determination of the prevalence and significance of such mutations grows. As demonstrated, although activating mutations of the RTK/ras pathway are detected in nearly half of the AML cases, rare patients harbor more than one mutation, arguing that, if an activating mutation is involved in leukemic evolution, there may not be a biologic advantage to the redundancy of a second activating mutation. Patients with activating mutations had higher diagnostic WBC counts and marrow blast percentages than the patients without the activating mutations, and, as a collective group, this difference is statistically significant. These clinical correlations may be relevant to the in vitro observation that such mutations lead to activation of the signal transduction pathway and to autonomous proliferation in cell culture. Additionally, patients with activating mutations predominantly have normal or favorable cytogenetics and do not segregate with any particular FAB classification.
The clinical significance of FLT3/ITD has been clearly demonstrated in previous studies13,15,46-48 ; however, the clinical signifi-cance of other activating mutations (FLT3/ALM, c-kit, and ras mutations) has not been individually established. In this study, we evaluated the clinical significance of the FLT3/ITD, FLT3/ALM, c-kit, N-ras, and k-ras mutations as a collective group as a model for evaluating the role of activating mutations in AML. The caveat of this approach is that, until prognostic significance of each mutation is individually established, this approach would not be appropriate for implementation in the clinical setting. This argument is strengthened by the fact, in this group of 91 patients, that patients with FLT3/ALM had a significantly different outcome than the remaining group of patients with mutations. Patients with FLT3/ITD, c-kit, N-ras, and k-ras mutations had a worse overall survival than patients without mutations and patients in each mutation group had a higher likelihood of death (hazard ratio [HR], 1.7-3.4). In contrast, patients with FLT3/ALM had an excellent outcome compared with patients with other activating mutations or patients without mutations (HR, 0.3). However, given the fact that 6 of 7 patients with FLT3/ALM had received allogeneic stem cell transplants, the prognostic importance of FLT3/ALM in patients who are treated with chemotherapy cannot be addressed in this study. Given the variation in the outcome within the mutation groups, the clinical significance of each mutation must be individually established before combining these mutations into a single group for clinical decision making.
In addition, we demonstrate that patients with activating mutations who are at high risk of poor outcome may benefit from allogeneic stem cell transplantation. Validation of such an approach in a large cohort of patients is under way. Determination of markers of high-risk disease and identification of treatment options for patients who are at high relapse risk may enable risk-based treatment when patients at high risk of relapse would be identified at diagnosis and their treatment altered (allogeneic stem cell transplantation or targeted therapy with small molecule inhibitors) to improve the outcome.
Prepublished online as Blood First Edition Paper, April 17, 2003; DOI 10.1182/blood-2003-01-0137.
Supported by grants from Fundación Internacional José Carreras (FIJC)–99/INT (S.M.), Child Health Research Center (CHRC) NIA 62-4532 (S.M.), CA18029 (J.P.R.), K23CA92405-01 (D.L.S.), CA 30969; Doris Duke Charitable Foundation Clinical Scientist Development Award (D.A.S.), and American Society of Hematology Junior Faculty Scholar Award (D.A.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal