Abstract
Pseutarin C is a group C prothrombin activator from the venom of the eastern brown snake Pseudonaja textilis. It is a multi-subunit protein complex consisting of catalytic and nonenzymatic subunits similar to coagulation factor Xa and factor Va, respectively. Here we describe the complete sequence of the nonenzymatic subunit. Based on the partial amino acid sequence of the nonenzymatic subunit, degenerate primers were designed. Using a “walking” strategy based on sequentially designed primers, we determined the complete cDNA sequence of the nonenzymatic subunit. The cDNA encodes a protein of 1461 amino acid residues, which includes a 30-residue signal peptide, a mature protein of 1430 amino acid residues, and a stop codon. cDNA blot analysis showed a single transcript of approximately 4.6 kb. The deduced amino acid sequence shows approximately 50% identity to mammalian factor V and by homology has a similar domain structure consisting of domains A1-A2-B-A3-C1-C2. Interestingly, the B domain of pseutarin C is shorter than that of mammalian factor V (FV). Although most of the proteolytic activation sites are conserved, 2 of 3 proteolytic sites cleaved by activated protein C are mutated, and thus activated protein C is not able to inactivate this procoagulant toxin. The predicted posttranslational modifications, including disulfide bonds, N-glycosylation, phosphorylation, and sulfation, in pseutarin C are significantly different compared with bovine factor V. Thus, our data demonstrate that the nonenzymatic subunit of group C prothrombin activators is structurally similar to mammalian FV.
Introduction
Blood coagulation involves the sequential activation of the plasma zymogens finally culminating in the formation of a fibrin clot. Prothrombin activation, a crucial reaction in the coagulation cascade, occurs when the serine protease factor Xa (FXa) cleaves prothrombin at 2 peptide bonds, Arg271-Thr272 and Arg320-Ile321, leading to the formation of thrombin.1 Prothrombin activation by FXa alone occurs at a very slow rate. For activation at a physiologically relevant rate, FXa must form the “prothrombinase complex” with the nonenzymatic cofactor factor Va (FVa), calcium ions, and phospholipid membranes.1 Phospholipids stimulate prothrombin activation by lowering the Km for prothrombin2-4 and provide a surface on which FXa, FVa, and prothrombin bind in the presence of Ca2+, leading to a productive enzyme-substrate complex. FVa acts as a receptor for FXa by the virtue of its ability to bind to phospholipid membranes. It enhances the ability of FXa to activate prothrombin by increasing the Vmax by approximately 700-fold for thrombin formation.2,5,6 Hence, the formation of the prothrombinase complex results in 105-fold acceleration in the rate of prothrombin activation compared with that by FXa alone.2-4
In addition to its physiologic activator FXa, prothrombin is activated by several exogenous factors including those from snake venom.7,8 These snake venom prothrombin activators are classified into 4 groups based on their cofactor requirements.9 Among these, group C and D prothrombin activators are serine proteinases. Group C prothrombin activators are large (∼250 kDa) protein complexes with multiple subunits. These activators cleave prothrombin to mature thrombin, and their prothrombin converting activity is enhanced by Ca2+ ions and phospholipids but not by FVa. They have been isolated from Oxyuranus scutellatus (oscutarin C) and Pseudonaja textilis (pseutarin C) venoms.10-13 Group D activators are 2-chain serine proteinases.14-16 They convert prothrombin to mature thrombin and their activity is greatly enhanced by FVa, phospholipids, and Ca2+. Functionally, they resemble blood coagulation FXa. Hence, we initiated the study of these group C and D activators as it will significantly contribute to understanding the formation of the prothrombinase complex and FXa-FVa–mediated prothrombin activation.
We recently determined the complete amino acid sequence of group D prothrombin activators trocarin D (Tropidechis carinatus) and hopsarin D (Hoplocephalus stephensi).15,16 Both are glycoproteins and are homologous (62%-70% similarity) to FXa with identical domain architecture. The light chain consists of an N-terminal Gla-domain containing 11 gamma-carboxyglutamic acid (Gla) residues followed by 2 epidermal growth factor–like domains; the heavy chain is a serine proteinase. Thus, group D prothrombin activators are true structural and functional homologues of coagulation FXa.
Recently, we have shown that pseutarin C, a group C activator from the venom of the eastern brown snake Pseudonaja textilis, is a multimeric protein complex consisting of catalytic and nonenzymatic subunits similar to mammalian FXa-FVa complex.12,13 In this paper, we present the complete cDNA sequence and the translated amino acid sequence of the nonenzymatic subunit of pseutarin C and its structural similarity to mammalian FV. These studies indicate that the nonenzymatic subunit of pseutarin C has a domain architecture identical to mammalian FV (A1-A2-B-A3-C1-C2). However, the nonenzymatic subunit differs significantly in its potential posttranslational modifications and the size of B domain. This is the first sequence of an FV-like protein from a nonhepatic and nonmammalian origin.
Materials and methods
Materials
Pseudonaja textilis venom and venom gland were obtained from Venom Supplies (Tanunda, Australia). The RNA isolation kit, one-step reverse transcription–polymerase chain reaction (RT-PCR), Qiaquick gel extraction, and plasmid miniprep kits were purchased from Qiagen (Valencia, CA). pDrive cloning kit was obtained from Qiagen. The 5′ and 3′ rapid amplification of cDNA ends (RACE) kits were obtained from Invitrogen (Carlsbad, CA). Digoxigenin (DIG) nucleic acid labeling and detection kits were from Roche Diagnostics (Indianapolis, IN). The ABI PRISM BigDye terminator-cycle sequencing ready-reaction kit was purchased from Perkin Elmer (Foster City, CA). Oligonucleotides were custom synthesized from GENSET (Singapore Biotech, Singapore). Bovine prothrombin, factor Xa, factor Va, and activated protein C were purchased from Hematologic Technologies (Essex Junction, VT). S-2238 was purchased from Chromogenix (Mölndal, Sweden). All other chemicals and reagents were of the purest grade available.
Methods
RNA isolation, RT-PCR. Total RNA was isolated from the venom gland using the Qiagen RNA isolation kit according to the manufacturer's instructions. One-step RT-PCR was carried out in a final volume of 25 μL using 100 ng total RNA, and degenerate primers were designed based on the peptide sequences12 (sense primers KIVYRE [IP1: 36-41]; WEYFIA [IP2: 323-328]; RPYSIYV [IP3: 414-420]; AAKTTF [N-ter: 821-826] and antisense primers RPYSLHA [A2R: 877-883]; FPAING [A3R: 1015-1020]). The RT-PCR conditions were as follows: reverse transcription at 50°C for 30 minutes; followed by inactivation of reverse transcriptase and activation of Taq polymerase at 95°C for 15 minutes; and PCR for 35 cycles consisting of 94°C for 1 minute, 50°C for 1 minute, 72°C for 1 minute, followed by final extension at 72°C for 10 minutes. PCR products obtained were subjected to 1% agarose gel electrophoresis and visualized by ethidium bromide staining. PCR product was purified according to the manufacturer's instructions using the Qiagen PCR purification kit.
Cloning of pseutarin FV cDNA. In order to determine the complete sequence of the pseutarin C nonenzymatic subunit, primer corresponding to A1 domain from 604 to 623 (A1fwd) (5′-AATGCAAATGGTTCACAAAA-3′) and the 3′-adapter primer (5′-GGCCACGCGTCGACTAGTAC(T) 17-3′) were used in PCR. The product obtained (∼4 kb) was purified, ligated with pDrive vector (Qiagen), and transformed into competent Escherichia coli cells (JM109) by the heat-shock method. The transformants were selected on Luria-Bertani (LB) amp plates supplemented with isopropylthio-β-d-galactoside (IPTG) and X-gal and the inserts were sequenced as described in “DNA sequencing.”
cDNA blot analysis. cDNA blotting was carried out according to the protocol of Jaakola et al.17 Briefly, total RNA (∼2 μg) isolated from the venom gland was reverse-transcribed at 42°C for 50 minutes in a 20-μL reaction volume containing Moloney murine leukemia virus–reverse transcriptase (MMLV-RT; Invitrogen) and adapter primer (5′-GGCCACGCGTCGACTAGTAC(T) 17-3′). The cDNA was run on a 1% agarose gel at 22 V for 10 hours. The gel was then stained with ethidium bromide and blotted onto the nylon membrane (Hybond-N; Amersham Biosciences, Chalfont St Giles, United Kingdom) overnight in 20 × SSC (standard saline citrate). After cross-linking, the membrane was hybridized with a 924-bp PCR fragment (region 1854-2778 of pseutarin C) that was labeled with DIG dUTP (2′-deoxyuridine-5′-triphosphate). Hybridization was performed overnight at 50°C. The blot was treated with anti-DIG antibody conjugated to alkaline phosphatase and detected using the DIG nucleic acid detection kit according to the manufacturer's instructions.
In situ hybridization. The slides containing the venom gland sections were treated with proteinase K (1 μg/mL in phosphate-buffered saline [PBS]) for 10 minutes at 37°C, and the slides were then washed with PBS at room temperature. Prehybridization was carried out using DIG prehybridization solution for 2 hours at 42°C. The DIG-labeled probe, which covered bases 1854 to 2778 of pseutarin C, was denatured by heating at 85°C for 15 minutes and quickly chilled on ice before adding it to the hybridization solution. Hybridization was carried out at 42°C for 16 hours. Detection of the DIG-labeled probe was carried out according to manufacturer's instructions.
3′ rapid amplification of cDNA end. cDNA was synthesized from approximately 1.5 μg total RNA using Superscript II reverse transcriptase (Invitrogen) and 3′ RACE adapter primer (5′-GGCCACGCGTCGACTAGTAC(T)17-3′) as described earlier. Following RT, the reaction was treated with RNase H (2 U) for 30 minutes at 37°C. The RT product (2 μL) was used in first round PCR (94°C, 1 minute; 50°C, 1 minute; 68°C, 1 minute, 35 cycles; followed by final extension at 68°C for 10 minutes) with a pseutarin C nonenzymatic subunit forward primer corresponding to region 3273 to 3291 (5′-GAGGGAAGGGAAGATAAT-3′) and an antisense 3′ RACE abridged universal amplification primer (5′-GGCCACGCGTCGACTAGTAC-3′). The PCR product obtained was visualized using ethidium bromide staining following agarose gel electrophoresis (1%). Sequencing of the PCR product was performed as described below.
5′ RACE. The 5′ RACE was carried out essentially according to the manufacturer's instructions (Invitrogen). For the synthesis of the cDNA, the following gene-specific primer (GSP; A2 domain 1646-1667) (5′-CAGTCTTCGCAGTGTTTGATGA-3′) was used. cDNA (5 μL) was used as a template for PCR along with 5′-abridged anchor primer (5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′) and an antisense GSP2 (A2 domain 1318-1337) (5′-TAAGGTCGACTGGCCAGATT-3′), and amplification was carried out as described in the previous section. The PCR product was visualized on an ethidium bromide–stained agarose gel (1%). PCR product was then purified and sequenced.
DNA sequencing. DNA sequencing was carried out using the ABI PRISM 377 automated DNA sequencer. All sequencing reactions were carried out using the ABI PRISM BigDye terminator-cycle sequencing ready-reaction kit according to the manufacturer's instructions. All the PCR products and the pseutarin C nonenzymatic subunit clone were sequenced at least 4 times in both directions.
Preparation of peptide digest and mass spectrometry. Pseutarin C from Pseudonaja textilis venom was purified to homogeneity by gel filtration on Superdex 75 column (Pharmacia Biotech, Uppsala, Sweden) followed by hydroxyapatite column chromatography as described earlier.12 Purified pseutarin C was subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; 4%-20% gradient gel) under reducing conditions according to the method of Laemmli.18 Following electrophoresis, the gel was stained with Coomassie blue. Individual bands were cut out and, in gel, digested with trypsin. The sequences of the peptides were obtained by electrospray ionization tandem mass spectrometry (MS/MS) using a Q-TOF (Micromass, Manchester, United Kingdom).
Sequence analysis. Sequence analysis and domain search were carried out using the BLASTX program and Conserved Domain Database, respectively, at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). Sequence alignments were carried out using the GeneDoc program (http://www.psc.edu/biomed/genedoc). Signal peptide and N-glycosylation site prediction was carried out using the PSORT and NetNGlyc prediction site, respectively, at the Expasy website (http://www.expasy.ch).
Activated protein C (APC) resistance assay. Pseutarin C (8 nM) was incubated in 50 mM Tris-HCl, pH 7.5, containing 100 mM NaCl, 5 mM CaCl2, and 0.5 mg/mL bovine serum albumin (BSA). APC (28, 84, 240, and 480 nM) was added, and the reaction mixture was incubated for 30 minutes at room temperature (∼25°C). Prothrombin activation was initiated by addition of prothrombin to a final concentration of 2.8 μM. At 20 minutes, 2.5 μL of the reaction mixture was withdrawn and diluted with 187.5 μL 50 mM Tris-HCl, pH 7.5, containing a 100 mM NaCl, 25 mM EDTA (ethylenediaminetetraacetic acid) solution (stop buffer) in a 96-well microtiter plate. The amount of thrombin formed was determined by the hydrolysis of thrombin-specific chromogenic substrate S-2238 (10 μL; final concentration 100 μM) measured at 405 nm. Measurements were made in triplicates using 96-well mictrotiter plates on a Ceres UV 900C ELISA (enzyme-linked immunosorbent assay) plate reader (Bio-Tek Instruments, Winooski, VT).
For the control assay, bovine FXa (42 nM) was dissolved in 50 mM Tris-HCl buffer, pH 7.5, containing 100 mM NaCl, 5 mM CaCl2, 0.5 mg/mL BSA. Bovine FVa (2 nM) was added and the reaction mixture was incubated for 15 minutes at room temperature. Then, different concentrations of APC (28, 84, 240, and 480 nM) were added and incubated further for 30 minutes. Prothrombin was subsequently added to a final concentration of 2.8 μM and the thrombin formation was measured as described in the previous paragraph.
Results and discussion
cDNA sequence of pseutarin C nonenzymatic subunit
In an earlier study, we had determined the amino terminal sequences of several internal peptides of the nonenzymatic subunit of pseutarin C.12 Degenerate primers were designed based on these peptide sequences. The complete cDNA sequence of pseutarin C nonenzymatic subunit was determined by RT-PCR followed by 5′ and 3′ RACE. The first set of RT-PCR was carried out using degenerate primers (see “Materials and methods” for details). Using the primer combinations of IP2/A2R we got PCR products of size 2 kb and 700 bp; with IP3/A2R a 2-kb fragment was obtained and for Nter/A2R we got a 600-bp product (data not shown). The PCR products were either gel-eluted or purified directly and their sequences were determined. The deduced amino acid sequences showed similarity (∼60%) to the A2 and A3 domains of mammalian coagulation FV.19-21 Based on this sequence information, a new sense primer corresponding to A1 domain 604 to 623 (5′-AATGCAAATGGTTCACAAAA-3′) was designed. This primer was used along with the 3′ RACE adapter primer in order to amplify an almost full-length pseutarin C nonenzymatic subunit cDNA. An approximately 4-kb PCR product (data not shown) was ligated to pDrive vector to obtain the clone PFV-A9. This clone was further analyzed by restriction enzyme digestion (data not shown). The clone, PFV-A9, was then used to determine the sequence of the nonenzymatic subunit of pseutarin C. The sequence of the 5′ end of pseutarin C nonenzymatic subunit was determined by 5′ RACE. On the basis of the sequence data generated from the clone PFV-A9 and 5′ RACE, the complete cDNA sequence of the pseutarin C nonenzymatic subunit was assembled and the amino acid sequence deduced (GenBank number AY168281). The open reading frame of pseutarin C nonenzymatic subunit is 4383 bp; it encodes a protein of 1461 amino acids, which includes a 30–amino acid signal peptide and a stop codon (Figure 1).
Complete cDNA sequence of pseutarin C nonenzymatic subunit. Sequence was determined using the clone PFV-A9. The sequence of the 5′ and the 3′-untranslated regions was determined by RACE. The deduced amino acid sequence is shown below the nucleotide sequence. The predicted signal peptide is shown in italics. Sequences that were obtained by Edman degradation are underlined; sequences of peptides obtained by Q-TOF are double underlined. The beginning and the end of each domain is marked by an arrow.
Complete cDNA sequence of pseutarin C nonenzymatic subunit. Sequence was determined using the clone PFV-A9. The sequence of the 5′ and the 3′-untranslated regions was determined by RACE. The deduced amino acid sequence is shown below the nucleotide sequence. The predicted signal peptide is shown in italics. Sequences that were obtained by Edman degradation are underlined; sequences of peptides obtained by Q-TOF are double underlined. The beginning and the end of each domain is marked by an arrow.
The amino acid sequences of some of the peptides (19 of 28 total peptides sequenced) determined by Edman degradation12 matched with the deduced amino acid sequence. However, we observed a few differences that are summarized in Table 1. Such differences may indicate the presence of more than one isoform of this protein. Edman degradation of some of the peptides showed heterogeneity. For example, peptide Hc101-104 (LIQY; Hc indicates heavy chain, and Lc indicates light chain) showed both “Q” and “I” at the third position, whereas peptide Hc352-357 showed “H” and “F” at position 4. The existence of multiple isoforms is well known in snake venom proteins. In addition, the crude venom used for the purification of pseutarin C was pooled from different snakes. It is also well established that the composition of the venom varies with age, diet, geographic location, and the season of the year.22 Both of these possibilities could explain the observed minor differences between the peptide sequences and the deduced amino acid sequence.
N-terminal sequences of pseutarin C peptides
. | Sequence . | . | |
|---|---|---|---|
| Location . | Edman degradation . | Deduced . | |
| Hc79-82 | EKPQ | EEPR | |
| Hc101-104 | LIQY | LIIY | |
| Hc117-124 | PQDAVKIG | PQSAVYNK | |
| Hc144-147 | AVAP | AVPP | |
| Hc155-158 | WEIT | WNIT | |
| Hc349-363 | AQLWEYHIAAQKED | IKNWEYFIAAEEIT | |
| Hc440-444 | DLAVQ | NLASR | |
| Hc521-525 | GILGP | GLIGP | |
| Lc818-827 | QNTGNKLY | INRGNKRR | |
| Lc838-840 | DYI | DYS | |
| Lc876-878 | GKEY | GEY | |
| Lc900-902 | QFR | QFK | |
| Lc943-945 | PFG | PNG | |
| Lc972-975 | YXGX | YSGV | |
| Lc981-995 | VEPGLIGPLYSIAEEAV | IHSGLIGPILICQKG | |
| Lc1040-1044 | QNLGT | QSLHT | |
| Lc1057-1059 | GKT | LQGLT | |
| Lc1081-1085 | VVNXH | VVNFH | |
| Lc1164-1174 | SAAGHVQYWEP | SASGHVGYWEP | |
| Lc1273-1275 | IVA | IIA | |
| Lc1366-1368 | TKK | TKI | |
| Lc1398-1404 | STYKPYG | STWKPYL | |
| Lc1430-1433 | PISL | PILS | |
. | Sequence . | . | |
|---|---|---|---|
| Location . | Edman degradation . | Deduced . | |
| Hc79-82 | EKPQ | EEPR | |
| Hc101-104 | LIQY | LIIY | |
| Hc117-124 | PQDAVKIG | PQSAVYNK | |
| Hc144-147 | AVAP | AVPP | |
| Hc155-158 | WEIT | WNIT | |
| Hc349-363 | AQLWEYHIAAQKED | IKNWEYFIAAEEIT | |
| Hc440-444 | DLAVQ | NLASR | |
| Hc521-525 | GILGP | GLIGP | |
| Lc818-827 | QNTGNKLY | INRGNKRR | |
| Lc838-840 | DYI | DYS | |
| Lc876-878 | GKEY | GEY | |
| Lc900-902 | QFR | QFK | |
| Lc943-945 | PFG | PNG | |
| Lc972-975 | YXGX | YSGV | |
| Lc981-995 | VEPGLIGPLYSIAEEAV | IHSGLIGPILICQKG | |
| Lc1040-1044 | QNLGT | QSLHT | |
| Lc1057-1059 | GKT | LQGLT | |
| Lc1081-1085 | VVNXH | VVNFH | |
| Lc1164-1174 | SAAGHVQYWEP | SASGHVGYWEP | |
| Lc1273-1275 | IVA | IIA | |
| Lc1366-1368 | TKK | TKI | |
| Lc1398-1404 | STYKPYG | STWKPYL | |
| Lc1430-1433 | PISL | PILS | |
Boldface, italics, and underlining indicate those amino acids that are different between the deduced amino sequence from cDNA and those determined by Edman degradation.
In order to further confirm the deduced amino acid sequence, we also determined the sequence of a few of the peptides of both heavy chain and light chain by MS/MS using Q-TOF. The sequences of the peptides obtained by electrospray ionization MS/MS sequencing are shown in Figure 1. While most of the peptide sequences obtained by MS/MS were identical to the deduced amino acid sequence (17 of 20 peptides sequenced), we observed the presence of isoforms as in the case of Edman degradation. For example, the sequence of the peptide Hc487-493 obtained by MS/MS was WTVLDTPFPTAR; however, the deduced amino acid sequence is WTVLDTDEPTVK. Thus, data from mass spectra and Edman degradation strongly support the deduced amino acid sequence of the nonenzymatic subunit.
cDNA blot analysis
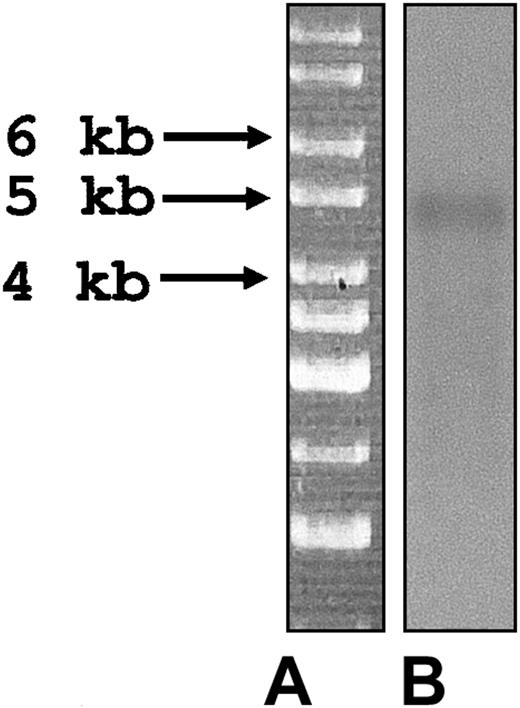
cDNA blot analysis was used to estimate the size and abundance of the pseutarin C nonenzymatic mRNA. A 924-bp gene-specific probe (region 1854-2778 of pseutarin C) was used to probe the cDNA blot. A single band of approximately 4.6 kb was observed (Figure 2) that closely corresponded to the open reading frame (Figure 1) and confirmed that the sequence of the nonenzymatic subunit determined is the complete sequence.
cDNA blot analysis of P textilis cDNA library. (A) Marker. (B) cDNA blot probed with a pseutarin C nonenzymatic subunit gene-specific probe (see “cDNA blot analysis” in “Materials and methods” for details).
cDNA blot analysis of P textilis cDNA library. (A) Marker. (B) cDNA blot probed with a pseutarin C nonenzymatic subunit gene-specific probe (see “cDNA blot analysis” in “Materials and methods” for details).
In situ hybridization
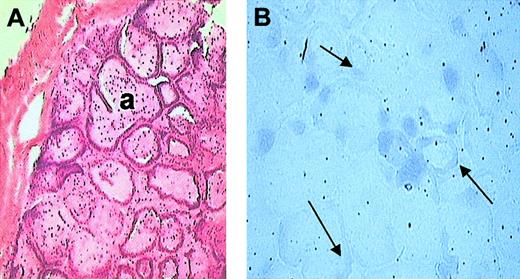
The snake venom gland consists of mainly 2 kinds of cells, tall columnar epithelial cells and horizontal spindle-shaped cells, the principal cells being the columnar epithelial cells. In situ hybridization was carried out to identify the localization of pseutarin C nonenzymatic subunit mRNA in the P textilis venom gland. Hybridization of the sections with the DIG-labeled gene-specific probe (region 1854-2778 of pseutarin C) indicated localization of the pseutarin C nonenzymatic subunit mRNA to columnar epithelial cells (Figure 3). Thus, these studies indicate that pseutarin C mRNA is synthesized in the venom gland, confirming the biosynthesis of pseutarin C nonenzymatic subunit in the venom gland.
In situ hybridization. (A) Hematoxylin and eosin staining. P textilis venom gland showing acini (a) lined with single layer of epithelium. (B) NBT/BCIP (nitro-blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate p-toluidine salt) staining. Localization of nonenzymatic subunit of pseutarin C mRNA in P textilis venom gland by in situ hybridization using DIG-labeled purified PCR product as probe. Arrows indicate gene-specific staining. The mRNA of pseutarin C is localized to the columnar epithelial cells. Original magnification, × 10.
In situ hybridization. (A) Hematoxylin and eosin staining. P textilis venom gland showing acini (a) lined with single layer of epithelium. (B) NBT/BCIP (nitro-blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate p-toluidine salt) staining. Localization of nonenzymatic subunit of pseutarin C mRNA in P textilis venom gland by in situ hybridization using DIG-labeled purified PCR product as probe. Arrows indicate gene-specific staining. The mRNA of pseutarin C is localized to the columnar epithelial cells. Original magnification, × 10.
Sequence homology of pseutarin C with mammalian factor V proteins
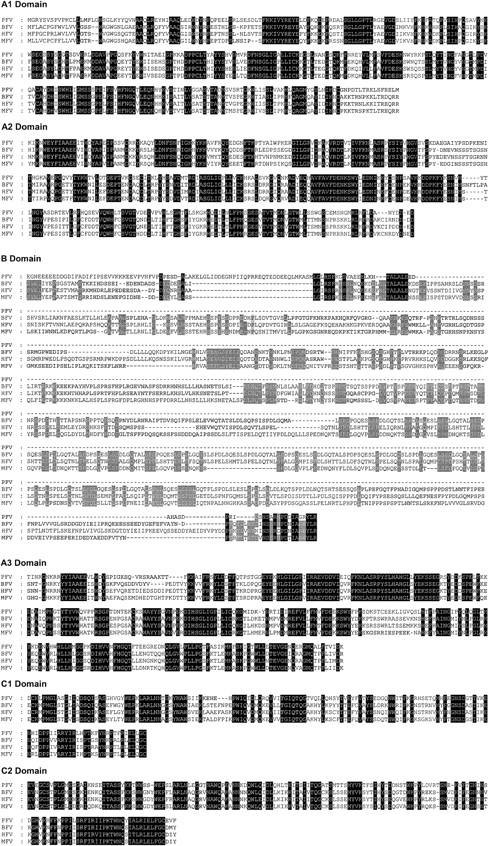
The nonenzymatic subunit of pseutarin C shows significant sequence similarity to mammalian FV. It has similar domain architecture to mammalian FV19-21 consisting of domains A1-A2-B-A3-C1-C2. Alignment of the translated amino acid sequence of pseutarin C nonenzymatic subunit with other mammalian FV sequences shows that it shares overall sequence identity of approximately 50% with the functionally important A and C domains (Figure 4). However, the B domain is significantly shorter.
Sequence alignment of pseutarin C nonenzymatic subunit with mammalian FV. The deduced amino acid sequence of pseutarin C nonenzymatic subunit (AY168281) is compared with bovine FV (Q28107), human FV (P12259), and mouse FV (T42764) sequences. Sequence alignment was done using the GeneDoc program. Gaps (–) have been inserted for optimal alignment. Identical amino acid residues are shaded in black. In B domain, amino acid residues identical in all 3 mammalian sequences are shaded in gray.
Sequence alignment of pseutarin C nonenzymatic subunit with mammalian FV. The deduced amino acid sequence of pseutarin C nonenzymatic subunit (AY168281) is compared with bovine FV (Q28107), human FV (P12259), and mouse FV (T42764) sequences. Sequence alignment was done using the GeneDoc program. Gaps (–) have been inserted for optimal alignment. Identical amino acid residues are shaded in black. In B domain, amino acid residues identical in all 3 mammalian sequences are shaded in gray.
Like the mammalian FV proteins, the A1 and the A2 domains of the nonenzymatic subunit of pseutarin C show similarity to the A domains of ceruloplasmin, a major transport protein of Cu2+ in the blood.23 While both A1 and A2 domains have the Cu-oxidase domain (residues 226-332 and 551-683, respectively), interestingly the A3 domain does not contain the Cu-oxidase domain; this is unlike mammalian FV proteins wherein all three A domains have the Cu-oxidase domain.19-21 Although Mann et al24 have shown the presence of a Cu2+ ion in mammalian FV, the significance of this is not clear.
The B domain is a large, poorly conserved segment in mammalian FV. It is released upon activation of FV by thrombin as 2 activation fragments, 150 kDa and 71 kDa.25 The B domain of pseutarin C is small, consisting of only 127 amino acids as opposed to 827 residues in bovine FV (Figure 4). The size of the transcript obtained by cDNA blot analysis confirmed the small size of the B domain. In addition, it was also confirmed by RT-PCR wherein a 924-bp band was obtained when the region spanning 1854 to 2778 of pseutarin C nonenzymatic subunit was amplified (data not shown). The connecting region of mammalian FV (human) is unusual in that it contains 2 tandem repeats of 17 amino acids and 31 tandem repeats of 9 amino acids.19,20 The B domain of pseutarin C nonenzymatic subunit lacks these repeats. The function of the B domain in mammalian FV is not clear. The poor conservation of this domain across the species (∼35% identity) indicates that it may be a spacer segment. Further, FV-lacking B domain shows markedly increased procoagulant activity.26 The C-terminal region of the B domain has been implicated to play an important role in APC-cofactor activity.27,28
The C1 and C2 domains of pseutarin C show approximately 50% identity to those of the mammalian FV proteins (Figure 4). The C domains are related to the discoidin family of proteins. The slime mold protein discoidin I is a galactose-binding lectin,29 which is essential for cell adhesion in Dictyostelium discoideum. The main function of the C domains (particularly the C2 domain) is to bind to the phosholipid membranes.30
Proteolytic regulation of cofactor activity
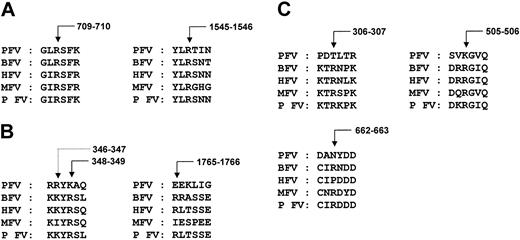
Comparison of sequences at the thrombin and FXa cleavage sites of factor V. Coagulation FV is a single-chain glycoprotein, which has little or no intrinsic procoagulant activity.19-21 It is converted to active FVa by proteolytic cleavage mediated by either thrombin or FXa.31-37 They cleave at the peptidyl bonds of Arg709, Arg1018, and Arg1545 leading to the release of the B domain and generation of FVa. Cleavage at Arg709 and Arg1545 is important for full FVa cofactor activity and sufficient for the complete release of B domain; cleavage at Arg1018 enhances the rate of cleavage at Arg1545.37 Of the 3 thrombin cleavage sites, the nonenzymatic subunit of pseutarin C has only 2 conserved sites (Figure 5A); the third cleavage site Arg1018, which is expected to be within the B domain, is missing. Since cleavage at Arg709 and Arg1545 is sufficient to release the B domain, absence of the third cleavage site may not be of physiologic relevance. Thus, the size of the less important B domain is efficiently reduced in the snake while retaining the critical residues required for the activation of the nonenzymatic subunit.
Proteolytic cleavage sites. (A) Thrombin cleavage sites. Of the 3 thrombin cleavage sites, only 2 are present (Arg709 and Arg1545) in pseutarin C nonenzymatic subunit, but they are sufficient to release the activation peptide (B domain). (B) FXa cleavage sites. Although a similar site is found in pseutarin C nonenzymatic subunit, a proteolytic cleavage was observed at Arg346 (dotted arrow) in native pseutarin C. The additional FXa cleavage site on the light chain (Arg1765) is absent in pseutarin C. Numbering of the amino acids is based on human FVa sequence. (C) APC cleavage sites. Two of the 3 APC cleavage sites are absent in pseutarin C. Arrows indicate the site of cleavage in bovine FV (see “Proteolytic regulator of cofactor activity”).
Proteolytic cleavage sites. (A) Thrombin cleavage sites. Of the 3 thrombin cleavage sites, only 2 are present (Arg709 and Arg1545) in pseutarin C nonenzymatic subunit, but they are sufficient to release the activation peptide (B domain). (B) FXa cleavage sites. Although a similar site is found in pseutarin C nonenzymatic subunit, a proteolytic cleavage was observed at Arg346 (dotted arrow) in native pseutarin C. The additional FXa cleavage site on the light chain (Arg1765) is absent in pseutarin C. Numbering of the amino acids is based on human FVa sequence. (C) APC cleavage sites. Two of the 3 APC cleavage sites are absent in pseutarin C. Arrows indicate the site of cleavage in bovine FV (see “Proteolytic regulator of cofactor activity”).
Earlier studies10 have shown that the nonenzymatic subunit of oscutarin C (from Oxyuranus scutellatus) enhances the prothrombin activation mediated by the FXa-like catalytic subunit, indicating that the nonenzymatic subunit exists in the active form. Further, based on our previous studies, we know that the amino terminal sequencing of the protein bands of pseutarin C obtained on a reducing SDS-PAGE correspond to A1 (heavy chain, 100 kDa), A3 (light chain, 65 kDa), A2 (52 kDa), and A1 (40 kDa) domains of nonenzymatic subunit and the heavy chain of enzymatic subunit (30 kDa).12 The amino terminal of the light chain indicates the cleavage at Arg1545, whereas the sizes of the heavy chain and the A2 domain indicate the (possible) cleavage at Arg709. In addition, we have not found a single peptide from the B domain during sequencing of the internal peptides. Thus, the B domain is apparently absent in the pseutarin C complex.
In addition to thrombin cleavage sites, FXa also cleaves mammalian FV at position Arg348. This additional cleavage site is found in pseutarin C nonenzymatic subunit.12 During N-terminal sequencing of pseutarin C subunits and their fragments,12 we observed that there is proteolytic cleavage at position Arg346 located within the A2 domain (Figure 5B). Since the active form of the nonenzymatic subunit exists in the venom as a complex with the catalytic subunit, which is similar to mammalian FXa, it is possible that this and other proteolytic cleavages may be due to the catalytic subunit of pseutarin C. At high FXa concentrations, it cleaves FVa at position Arg1765 on the light chain.37 This site, however, is missing in pseutarin C (Figure 5B). In addition to the thrombin and FXa cleavage sites, pseutarin C nonenzymatic subunit has 2 additional cleavage sites at position Leu57-Ser58 in the A1 domain and Arg925-Ser926 in the A3 domain.12 However, the proteinase(s) that is responsible for these cleavages is yet to be identified.
Comparison of sequences at the APC cleavage sites of factor V. Since FVa plays a critical role in blood coagulation, it is not surprising that down-regulation of FVa by APC is an effective way to maintain hemostatic balance. APC, which is a vitamin K–dependent serine proteinase, cleaves FVa at positions Arg306, Arg505, and Arg66238-40 in the heavy chain leading to its inactivation. Initial cleavage of FVa occurs at Arg505, followed by cleavage at Arg306 and Arg662. Cleavage of the FVa molecule at Arg505 and Arg662 results only in the partial loss of activity and it can still bind to FXa.38-40 However, the cleavage at Arg306 results in the release of the A2 domain and results in the complete loss of FVa cofactor activity.39 Interestingly, 2 of these 3 sites, including the crucial Arg306, are mutated in the nonenzymatic subunit of pseutarin C (Figure 5C). Further, it is known that binding of FXa to FVa protects it from APC inactivation,41 and because the nonenzymatic subunit of pseutarin C exists as a complex with the FXa-like subunit in the venom, this existence prevents the subunit from being exposed to APC.
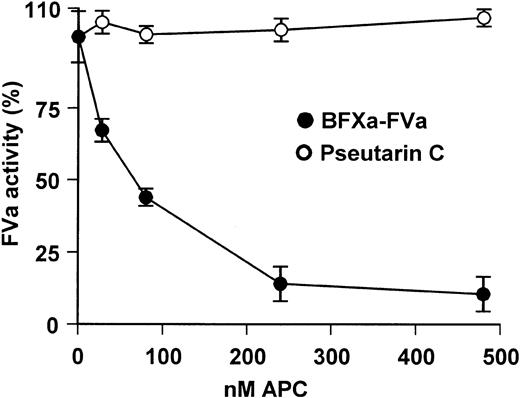
To determine the importance of these mutations we examined the proteolytic inactivation of pseutarin C by APC. Treatment of pseutarin C with APC for 30 minutes did not result in any loss of the cofactor activity of the nonenzymatic subunit of pseutarin C (Figure 6). In contrast, more than 90% of the cofactor activity of bovine FVa was lost. Hence, pseutarin C activity is not susceptible to inactivation by APC. Thus, APC resistance gives an added advantage to pseutarin C in its role as a toxin.
APC resistance assay. Varying concentrations of APC were added either to pseutarin C (○; 8 nM) or bovine FXa-FVa (•; FXa 42 nM, FVa 2 nM) complex diluted in 50 mM Tris-HCl buffer (pH 7.5) containing 100 mM NaCl, 5 mM CaCl2, and 0.5 mg/mL BSA. The reaction mixture was incubated for 30 minutes at room temperature. Prothrombin was added to a final concentration of 2.8 μM and thrombin formed was assayed using thrombin-specific chromogenic substrate S-2238 as described in “Materials and methods.” Each point represents an average of 2 independent experiments each carried out in triplicates. Error bars indicate SEM.
APC resistance assay. Varying concentrations of APC were added either to pseutarin C (○; 8 nM) or bovine FXa-FVa (•; FXa 42 nM, FVa 2 nM) complex diluted in 50 mM Tris-HCl buffer (pH 7.5) containing 100 mM NaCl, 5 mM CaCl2, and 0.5 mg/mL BSA. The reaction mixture was incubated for 30 minutes at room temperature. Prothrombin was added to a final concentration of 2.8 μM and thrombin formed was assayed using thrombin-specific chromogenic substrate S-2238 as described in “Materials and methods.” Each point represents an average of 2 independent experiments each carried out in triplicates. Error bars indicate SEM.
Posttranslational modifications
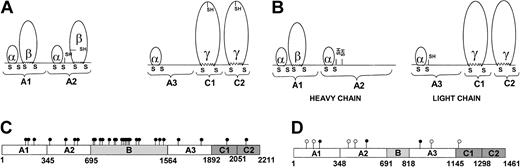
Disulphide linkage. Disulphide bonds play an important role in determining the overall 3-dimensional structure and stability of the protein. Comparison of the disulphide bond pattern between bovine FVa42,43 and pseutarin C nonenzymatic subunit (proposed based on structural similarity) is depicted in Figure 7A-B. In mammalian FVa, the A1 and A2 domains are identical in their disulphide-bonding pattern; both have an α loop (26 amino acids) followed by a β loop of 82 amino acids (Figure 7A). The A2 domain in addition has 2 free cysteine residues. However, the A3 domain has only a α loop. Similar disulphide-bonding pattern exists in A1, A2, and A3 domains of factor VIII (FVIII), and ceruloplasmin. Further, the sequence identity within the α and β loops of the A1, A2, and A3 domains between mammalian FV and FVIII is high (∼75%). Interestingly, in pseutarin C nonenzymatic subunit the pattern of disulphide linkage appears to be significantly different. While the A1 domain has the α and β loops, the A2 and A3 domains have only the α loop (Figure 7B). The amino acids within the α loops of the A1, A2, and A3 domains share 78%, 63%, and 82% identity to the α loops in mammalian FV, respectively.42 In addition, there are 3 free cysteine residues (2 in A2 and 1 in A3 domains); Cys1032 in A3 domain and Cys570 in A2 domain of pseutarin C and Cys566 in bovine A2 domain appear to be in a conserved position.
Posttranslational modifications in bovine V and pseutarin C nonenzymatic subunit. The disulphide pattern in bovine FVa (A) and pseutarin C nonenzymatic subunit (B) are shown. The α, β, and γ loops are labeled according to Xue et al.43 Free cysteines are shown as “- SH.” Both bovine FV and pseutarin C nonenzymatic subunit are glycosylated. The potential N-glycosylation sites in bovine FV (C) and pseutarin C nonenzymatic subunit (D) are shown. ○ represents potential novel N-glycosylation sites in pseutarin C nonenzymatic subunit compared with bovine FV; •, potential N-glycosylation sites (C) the conserved potential N-glycosylation sites (D).
Posttranslational modifications in bovine V and pseutarin C nonenzymatic subunit. The disulphide pattern in bovine FVa (A) and pseutarin C nonenzymatic subunit (B) are shown. The α, β, and γ loops are labeled according to Xue et al.43 Free cysteines are shown as “- SH.” Both bovine FV and pseutarin C nonenzymatic subunit are glycosylated. The potential N-glycosylation sites in bovine FV (C) and pseutarin C nonenzymatic subunit (D) are shown. ○ represents potential novel N-glycosylation sites in pseutarin C nonenzymatic subunit compared with bovine FV; •, potential N-glycosylation sites (C) the conserved potential N-glycosylation sites (D).
C1 and C2 domains in bovine FV have one conserved disulfide bond forming a γ loop (Figure 7A). Both these γ loops contain a single free cysteine residue. Pseutarin C also contains cysteine residues that could form γ loops in its C domains. However, it does not have the free cysteine residues.
Glycosylation. Glycosylation is known to play an important role in the stability as well as the folding of proteins. Mammalian FV and FVa both contain potential glycosylation sites. Most of the glycosylation sites (18 of 29) in bovine FV are present in the B domain (Figure 7C); the heavy chain has 7 potential N-glycosylation sites (3 in the A1 domain and 4 in the A2 domain) and the light chain has 4 sites (2 in the A3 and 1 each in the C1 and C2 domains).21 Since the B domain of pseutarin C nonenzymatic subunit is small, most of the glycosylation sites are deleted. It has only 11 potential N-glycosylation sites (Figure 7D). During sequencing of the internal peptides,12 we determined the complete sequence of 2 peptides (381-386 YLDNFS; 1395-1399 DNSTW) by Edman degradation indicating that these potential sites are not glycosylated. Of the remaining 9 potential N-glycosylation sites, only 3 are conserved and the rest are novel potential N-glycosylation sites (Figure 7D). The heavy chain has 6 potential N-glycosylation sites of which 3 are in the A1 domain and 3 in the A2 domain. The light chain of pseutarin C nonenzymatic subunit has 3 potential N-glycosylation sites, 2 in the A3 and one in the C1 domain.
Altering either the carbohydrate moiety or the glycosylation site can either enhance or cause loss of function of the protein. For example, the α-neurotoxins from snake venoms are targeted against nicotinic acetylcholine receptors (nAChR).44 However, these neurotoxins fail to bind to snake's own nAChR receptor although the critical residues in the binding site are conserved in these receptors. Studies by Takacs et al45 have shown that the presence of the unique N-glycosylation signal in the ligand-binding domain of Naja haje prevents the binding of the α-neurotoxins. Nevertheless, the presence of this carbohydrate moiety does not alter the binding and response of acetylcholine.45 By this strategy, the snake has adapted well to resist the action of its own venom. Since the snake might have a similar, if not identical, coagulation FV present in its plasma where it performs the role of a hemostatic factor, it would be interesting to examine the role of this receptor's glycosylation in pseutarin C nonenzymatic subunit.
Phosphorylation and sulfation sites. Mammalian FV is both sulfated and phosphorylated.46-49 Phosphoamino acid analyses combined with peptide mapping techniques indicate the presence of a phosphoserine (Ser692) at the C-terminal end of the FVa heavy chain.46 It is speculated that phosphorylation may be important for regulating the inactivation of FVa by APC. This phosphorylation site (Ser692) is absent in pseutarin C nonenzymatic subunit. As mentioned above, the APC cleavage sites are not conserved in pseutarin C. The absence of Ser692 may make pseutarin C a less preferred substrate of APC. Although several other potential phosphorylation sites are present in pseutarin C nonenzymatic subunit, it is not clear if any of these sites are indeed phosphorylated.
Sulfation is known to occur in both the mammalian FV heavy chain and in the activation peptide with the greatest amount of sulfate added to the activation peptide.48 There are 6 potential sulfation sites in human FV: residues 696, 698, 1494, 1510, 1515, and 1565 (data not shown). None of these sites are present in the pseutarin C nonenzymatic subunit. It is speculated that sulfation may be important for regulating the activation of FV by thrombin.49 However, in the case of pseutarin C, the nonenzymatic subunit may not be activated by thrombin (or equivalent proteinase), as such proteinases have not been found in P textilis venom.
Pseutarin as a venom toxin
Blood coagulation factors, including FV, are constitutively expressed in the liver and they play a critical role in the hemostatic function. Pseutarin C (and the other snake venom prothrombin activators) are expressed in an unusual tissue, the venom gland. These prothrombin activators are present in activated form in large quantities in the venom. Both group C (pseutarin C)11 and group D (trocarin D)15 activators are lethal when injected into mice indicating that these prothrombin activators play the role of toxin in the venom. Since a snake would also need hemostatic functions for its survival, it is not far fetched to hypothesize that a similar, if not identical, factor might be present in its plasma where it functions like a hemostatic factor. Thus, 2 closely related proteins play distinctly different roles—as a toxin in the venom and as a hemostatic factor when present in the plasma.
In conclusion, this is the first sequence of an FV-like protein from a nonhepatic and nonmammalian source. This nonenzymatic subunit of pseutarin C shows identical domain architecture to mammalian FV consisting of domains A1-A1-B-A3-C1-C2. Unique posttranslational modifications and a resistance to APC make it highly suitable for its role as toxin. The complete sequence of this protein will help us understand structure-function relationships of FVa in prothrombin activation.
Prepublished online as Blood First Edition Paper, May 1, 2003; DOI 10.1182/blood-2002-12-3839.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to acknowledge P. Srinivasa Rao, Department of Biological Sciences, National University of Singapore, for his helpful discussions during cloning.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal