Abstract
Patients with Down syndrome (DS) frequently develop 2 kinds of clonal megakaryocytosis: a common, congenital, spontaneously resolving, transient myeloproliferative disorder (TMD) and, less commonly, childhood acute megakaryoblastic leukemia (AMKL). Recently, acquired mutations in exon 2 of GATA1, an X-linked gene encoding a transcription factor that promotes megakaryocytic differentiation, were described in 6 DS patients with AMKL. The mutations prevent the synthesis of the full-length GATA1, but allow the synthesis of a shorter GATA1 protein (GATA1s) that lacks the transactivation domain. To test whether mutated GATA1 is involved in the initiation of clonal megakaryoblastic proliferation or in the progression to AMKL, we screened 35 DS patients with either AMKL or TMD and 7 non-DS children with AMKL for mutations in exon 2 of GATA1. Mutations were identified in 16 of 18 DS patients with AMKL, in 16 of 17 DS patients with TMD, and in 2 identical twins with AMKL and acquired trisomy 21. Analysis revealed various types of mutations in GATA1, including deletion/insertions, splice mutations, and nonsense and missense point mutations, all of which prevent the generation of full-length GATA1, but preserve the translation of GATA1s. We also show that the likely mechanism of generation of GATA1 isoforms is alternative splicing of exon 2 rather than, or in addition to, alternative translation initiation, as was proposed before. These findings suggest that acquired intrauterine inactivating mutations in GATA1 and generation of GATA1s cooperate frequently with trisomy 21 in initiating megakaryoblastic proliferation, but are insufficient for progression to AMKL.
Introduction
Children with Down syndrome (DS) have an unusual propensity to develop megakaryocytic malignancies. At least 10% of children with DS are born with a megakaryocytosis syndrome commonly called transient myeloproliferative disorder (TMD), transient abnormal myelopoiesis, or transient leukemia.1 As suggested by the different names, the disorder is usually transient and resolves spontaneously within several months. The term “transient leukemia” is occasionally used because the blasts in this transient disorder are mostly of clonal megakaryocytic origin.2,3 The biological mechanism of the spontaneous resolution is unclear. Holt et al4 have recently demonstrated that telomerase activity was diminished in this benign form of congenital leukemia and suggested that this deficiency may explain the spontaneous regression. About 3% of DS patients, however, will develop full-blown malignant acute megakaryoblastic leukemia (AMKL) during their first 4 years of life, which will not regress without chemotherapy.5 In fact, the risk of AMKL is about 400 times higher in children with DS.6-8 The factors underlying the transformation from “benign” TMD into “malignant” AMKL are largely unknown.1,9-11
The peculiar association between DS and childhood megakaryoblastic disorders has led to an intensive search for a gene or genes on chromosome 21 that may initiate the leukemia. The strongest candidate has been RUNX1 (also known as AML1 or CBFA2). RUNX1 is a transcription factor that is required for normal hematopoiesis.12 It is commonly mutated and involved in various translocations in both myeloid and lymphoid leukemias.12,13 Inherited mutations in RUNX1 causing haploinsufficiency with low level of expression in hematopoietic stem cells cause a syndrome of familial thrombocytopenia and increased susceptibility to leukemia.14 However, RUNX1 abnormalities have generally not been detected in AMKL and, except for one case report,15 mutations in RUNX1 have not been found in AMKL associated with DS13,16 (also H.K. et al and S.I. et al, unpublished data, 2001-2002).
Recently, Wechsler et al17 reported the surprising finding that a gene on chromosome X, GATA1, was mutated in the megakaryoblasts from each of 6 DS patients with AMKL. GATA1 encodes a zinc-finger transcription factor that has been shown to be critical for normal erythroid and megakaryocytic development.18-20 Mice lacking GATA1 expression in the megakaryocytic lineage have thrombocytopenia and extensive proliferation of immature megakaryoblasts.21 Inherited missense mutations in GATA1 in humans cause a familial dyserythropoietic anemia and thrombocytopenia.22-24 Thus, GATA1 normally suppresses the proliferation of megakaryocytic and erythroid precursors while promoting their differentiation.
The mutations reported by Wechsler et al17 were specifically associated with AMKL of DS, as they were not found in other cases of AMKL or in other French-American-British (FAB) types of acute myelogenous leukemia (AML) or myelodysplastic syndromes. The mutations were restricted to the leukemic clones and were not detected in remission samples. They were therefore acquired and selected probably because they imparted a clonal advantage. All the mutations were deletions or insertions in exon 2 (where the first ATG is located, exon 1 is not coding) of GATA1 and resulted in an introduction of a premature stop codon. These mutations prevented the synthesis of full-length GATA1, but not the synthesis of a shorter variant of GATA1 protein that lacked the N-terminal transactivation domain. The authors suggested that the loss of the full-length GATA1 might provide the differentiation block and cooperate with the increased dosage of gene(s) on chromosome 21 in the pathogenesis of AMKL.
The major goal of the present study was to determine whether mutational inactivation of GATA1 is involved in the much more frequent congenital TMD in patients with DS. Absence of GATA1 mutations in TMD could imply that the inactivation of GATA1 is the key event in transforming the benign megakaryoblastic proliferation initiated by trisomy 21 into malignant AMKL. Conversely, if the cooperation between GATA1 mutations and trisomy 21 is needed for the initiation of the proliferative process, then GATA1 mutations are expected to be found in TMD. In this large multicenter study we report that the blasts in the congenital TMD of DS carry the same type of mutations in exon 2 of GATA1 that are found in AMKL associated with trisomy 21. We also show that the generation of GATA1s occurs through a mechanism of alternative splicing in addition to, or instead of, the previously reported mechanism of alternative translation.25
Materials and methods
Patient samples
All clinical samples were obtained with informed consent by the participating centers and their use in this study was approved by the Institutional Review Board of Sheba Medical Center, Tel-Hashomer, Israel. DNA was extracted from Ficoll-separated (Amersham Biosciences, Freiburg, Germany) blood or bone marrow cells by standard methods. The diagnosis and other relevant patient details are described in Tables 1 and 2. Fifty DNA samples from healthy individuals26 were used as controls.
Analysis of mutations in exon 2 of GATA1 in AML in DS patients
. | . | . | . | . | Exon 2 mutation (according to NM_002049) . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | Sex . | Age at diagnosis, mo . | DNA source (blasts in PB/BM, %) . | Karyotype . | RNA . | Last normal GATA1 amino acid . | Consequence of mutation on protein . | Clinical status at last follow-up (time since diagnosis, mo) . | ||
| 1 | F | 17 | PB (84) | 47,XX,+8,+21c[20] | 202delAG | Gly31 | Stop codon before Met84 | DOD (6) | ||
| 2 | M | 22 | PB (24) | 48,XY,del(6)(q21q24),+21,+21c[20] | 296ins19 | Ala61 | Stop codon before Met84 | DOD (4) | ||
| 3 | F | 21 | PB (68) | 47,XX,inv(9)(p11q13)c,+21c[1]/47,XX,idem,der(4)t(1;4) (q23;p15)[17]/47,XX,idem, iso(7)(q10)[2] | IVS2+2T>C | Pro73 | Splice mutant | CCR (77) | ||
| 4 | M | 23 | PB (51) | 47,XY,+21c[18]/48,XY,+8,+21c[1]/49,XY,+8,+12,+21c[1] | 262delG | Pro50 | Stop codon after Met84 | CCR (47) | ||
| 5† | M | 13 | PB (8) | 47,XY,t(2;4)(q37;q28),+21c[20] | 297insA | Ala61 | Stop codon before Met84 | CCR (67) | ||
| 6 | F | 23 | BM (24) | 47,XX,+21c[7]/47,XX,der(7)t(1;7)(q22;p22), +21c[11]/47,XX,dup(1)(q22q44), +21c[2] | 272ins10 | Ala53 | Stop codon before Met84 | CCR (75) | ||
| 7 | M | 23 | BM (41) | 47,XY,+21c[16]/48,XY,+8,+21c[4] | 263del43 | Pro50 | Stop codon after Met84 | CCR(66) | ||
| 8 | F | 15 | PB (98) | 47,XX,+21c[20] | 113A>G | Met1Val | No translation from Met1 | DOD (0)* | ||
| 9 | F | 35 | BM (17) | Not determined | 225C>T† | Pro38Leu | Polymorphism† | CCR (8) | ||
| 11B‡ | M | 11 | BM (20) | 47, XY +21 | 202delAG | Gly31 | Stop codon before Met84 | CCR (29) | ||
| 14 | F | 31 | BM (>20) | 48,XX,+8,+21c[9/25] | 332G>T | Pro73 | Splice mutant | CCR (95) | ||
| 21 | F | 31 | BM (38.5) | 48,XX,+8,+21c[7]/47,XX,+21c[2] | 292ins11 | Leu60 | Stop codon after Met84 | CCR (25) | ||
| 24 | F | 11 | BM (33) | 47,XX,+21c | 115G>A | Met1Ile | No translation from Met1 | CCR (67) | ||
| 25 | M | 44 | BM (55) | 48,XY,+21c,+mar1,+mar2[7]/49,X, -Y,+8,+11,+21c,+mar1[2] | 161C>T | Pro16 | Stop codon before Met84 | CCR (24) | ||
| 26 | F | 20 | BM (23) | 47,XX,+21c[4]/46,XX[2] | 301del11,ins22 | Tyr63 | Stop codon after Met84 | Died of sepsis in CCR (4) | ||
| 48 | M | 26 | BM (40) | 47,XY+21,-11,-13,+3mar | Normal | Ser413 | CCR (19) | |||
| 49† | M | 50 | BM (70) | Not determined | 116G>T | Met1 | Stop codon before Met84 | DOD (34) | ||
| 62B‡ | M | 22 | BM (40) | 41-45,del(X)(p22),Y,-4,add(5)(p15),-7[3],-8,-9[8],-11,-12[8],-15,del(17)(p11),+21c,+der(21)add(21)(p?)?,-22[7]+3-4mar[9] | 298dup289-298 | Tyr62 | Stop codon before Met84 | DOD§ | ||
. | . | . | . | . | Exon 2 mutation (according to NM_002049) . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | Sex . | Age at diagnosis, mo . | DNA source (blasts in PB/BM, %) . | Karyotype . | RNA . | Last normal GATA1 amino acid . | Consequence of mutation on protein . | Clinical status at last follow-up (time since diagnosis, mo) . | ||
| 1 | F | 17 | PB (84) | 47,XX,+8,+21c[20] | 202delAG | Gly31 | Stop codon before Met84 | DOD (6) | ||
| 2 | M | 22 | PB (24) | 48,XY,del(6)(q21q24),+21,+21c[20] | 296ins19 | Ala61 | Stop codon before Met84 | DOD (4) | ||
| 3 | F | 21 | PB (68) | 47,XX,inv(9)(p11q13)c,+21c[1]/47,XX,idem,der(4)t(1;4) (q23;p15)[17]/47,XX,idem, iso(7)(q10)[2] | IVS2+2T>C | Pro73 | Splice mutant | CCR (77) | ||
| 4 | M | 23 | PB (51) | 47,XY,+21c[18]/48,XY,+8,+21c[1]/49,XY,+8,+12,+21c[1] | 262delG | Pro50 | Stop codon after Met84 | CCR (47) | ||
| 5† | M | 13 | PB (8) | 47,XY,t(2;4)(q37;q28),+21c[20] | 297insA | Ala61 | Stop codon before Met84 | CCR (67) | ||
| 6 | F | 23 | BM (24) | 47,XX,+21c[7]/47,XX,der(7)t(1;7)(q22;p22), +21c[11]/47,XX,dup(1)(q22q44), +21c[2] | 272ins10 | Ala53 | Stop codon before Met84 | CCR (75) | ||
| 7 | M | 23 | BM (41) | 47,XY,+21c[16]/48,XY,+8,+21c[4] | 263del43 | Pro50 | Stop codon after Met84 | CCR(66) | ||
| 8 | F | 15 | PB (98) | 47,XX,+21c[20] | 113A>G | Met1Val | No translation from Met1 | DOD (0)* | ||
| 9 | F | 35 | BM (17) | Not determined | 225C>T† | Pro38Leu | Polymorphism† | CCR (8) | ||
| 11B‡ | M | 11 | BM (20) | 47, XY +21 | 202delAG | Gly31 | Stop codon before Met84 | CCR (29) | ||
| 14 | F | 31 | BM (>20) | 48,XX,+8,+21c[9/25] | 332G>T | Pro73 | Splice mutant | CCR (95) | ||
| 21 | F | 31 | BM (38.5) | 48,XX,+8,+21c[7]/47,XX,+21c[2] | 292ins11 | Leu60 | Stop codon after Met84 | CCR (25) | ||
| 24 | F | 11 | BM (33) | 47,XX,+21c | 115G>A | Met1Ile | No translation from Met1 | CCR (67) | ||
| 25 | M | 44 | BM (55) | 48,XY,+21c,+mar1,+mar2[7]/49,X, -Y,+8,+11,+21c,+mar1[2] | 161C>T | Pro16 | Stop codon before Met84 | CCR (24) | ||
| 26 | F | 20 | BM (23) | 47,XX,+21c[4]/46,XX[2] | 301del11,ins22 | Tyr63 | Stop codon after Met84 | Died of sepsis in CCR (4) | ||
| 48 | M | 26 | BM (40) | 47,XY+21,-11,-13,+3mar | Normal | Ser413 | CCR (19) | |||
| 49† | M | 50 | BM (70) | Not determined | 116G>T | Met1 | Stop codon before Met84 | DOD (34) | ||
| 62B‡ | M | 22 | BM (40) | 41-45,del(X)(p22),Y,-4,add(5)(p15),-7[3],-8,-9[8],-11,-12[8],-15,del(17)(p11),+21c,+der(21)add(21)(p?)?,-22[7]+3-4mar[9] | 298dup289-298 | Tyr62 | Stop codon before Met84 | DOD§ | ||
The French-American-British (FAB) subtype of patient 1's AML was M0; all others were M7. PB indicates peripheral blood; BM, bone marrow; DOD, dead of disease; and CCR, complete clinical remission
DOD at diagnosis
Same mutation identified in one of 50 healthy controls
Previous TMD (samples from the TMD of 2 patients, 11B and 62B, are included in Table 2 as patients 11A and 62A)
Refused treatment
Analysis of mutations in exon 2 of GATA1 in congenital transient myeloproliferative disorder of DS patients
. | . | . | . | . | Exon 2 mutation (according to NM_002049) . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | Sex . | Age at diagnosis, d . | DNA source (blasts in PB/BM, %) . | Karyotype . | RNA . | Last normal GATA-1 amino acid . | Consequence of mutation on protein . | Clinical status at last follow-up (time since diagnosis, mo) . | ||
| 11A | M | 14 | BM(30) | Not determined | 202delAG | Gly31 | Stop codon before Met84 | AMKL at 10 mo* | ||
| 29 | F | 3 | PB(18) | 47,XX,der(13), t(13;21), +21c | 331A>G | Pro73 | Splice mutant | CCR (28) | ||
| 31 | M | 6 | PB(70) | Not determined | 202delAG | Gly31 | Stop codon before Met84 | CCR (3) | ||
| 35 | F | 4 | PB(28) | 48,XX, +18, +21c[18] | 252del35 | Ser46 | Stop codon before Met84 | CCR (99) | ||
| 36 | F | 1 | PB(85) | 47,XX, +21c[24] | 149G>T | Ser12 | Stop codon before Met84 | DOD (1) | ||
| 37 | M | 2 | PB(50) | 47,XY,21c[43]/47,idem,del(11)(q23)[6] | 299dup278-299 | Tyr61 | Stop codon before Met84 | CCR (91) | ||
| 39 | F | 13 | PB(77) | 47,XX,der(11)(q2?3), +21c/47,XX, +21c | 321delGA | Tyr69 | Stop codon after Met84 | CCR (76) | ||
| 40 | M | 2 | BM(16) | Not determined | 217insC | Pro35 | Stop codon before Met84 | CCR (75) | ||
| 44 | M | 6 | BM(14) | 47,XY,1qh+,der(9)(p22), +21c | 292insT | Leu60 | Stop codon before Met84 | AMKL at 1 y CCR (14) | ||
| 51 | F | 6 | BM(18) | 47,XX, +21c[20] | 202delAG | Gly31 | Stop codon before Met84 | CCR (31) | ||
| 56 | M | 1 | BM(8) | Not determined | 305ins5 | Arg64 | Stop codon after Met84 | CCR (3) | ||
| 57 | F | 9 | PB(65) | 47,XX, +21c[20] | 332G>A | Pro73 | Splice mutant | CCR (12) | ||
| 59 | M | 17 | Not determined | 47,XY, +21c[20] | Normal | CCR (7) | ||||
| 60 | M | 1 | PB(13) | Not determined | 304delG,ins3 | Arg64 | Stop codon before Met84 | CCR (1) | ||
| 61 | F | 14 | PB(38) | 47,XX,9qh+, +21c[20] | 332del10 | Pro73 | Splice mutant | Lost to follow-up | ||
| 62A | M | 8 | BM(13) | 47,XY, +21c[27] | 298dup289-298 | Tyr62 | Stop codon before Met84 | AMKL at 22 mo† DOD (4) | ||
| 63 | M | 2 | PB(20) | 47,XY, +21c[20] | 145dup130-145 | Thr11 | Stop codon before Met84 | CCR (49) | ||
. | . | . | . | . | Exon 2 mutation (according to NM_002049) . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | Sex . | Age at diagnosis, d . | DNA source (blasts in PB/BM, %) . | Karyotype . | RNA . | Last normal GATA-1 amino acid . | Consequence of mutation on protein . | Clinical status at last follow-up (time since diagnosis, mo) . | ||
| 11A | M | 14 | BM(30) | Not determined | 202delAG | Gly31 | Stop codon before Met84 | AMKL at 10 mo* | ||
| 29 | F | 3 | PB(18) | 47,XX,der(13), t(13;21), +21c | 331A>G | Pro73 | Splice mutant | CCR (28) | ||
| 31 | M | 6 | PB(70) | Not determined | 202delAG | Gly31 | Stop codon before Met84 | CCR (3) | ||
| 35 | F | 4 | PB(28) | 48,XX, +18, +21c[18] | 252del35 | Ser46 | Stop codon before Met84 | CCR (99) | ||
| 36 | F | 1 | PB(85) | 47,XX, +21c[24] | 149G>T | Ser12 | Stop codon before Met84 | DOD (1) | ||
| 37 | M | 2 | PB(50) | 47,XY,21c[43]/47,idem,del(11)(q23)[6] | 299dup278-299 | Tyr61 | Stop codon before Met84 | CCR (91) | ||
| 39 | F | 13 | PB(77) | 47,XX,der(11)(q2?3), +21c/47,XX, +21c | 321delGA | Tyr69 | Stop codon after Met84 | CCR (76) | ||
| 40 | M | 2 | BM(16) | Not determined | 217insC | Pro35 | Stop codon before Met84 | CCR (75) | ||
| 44 | M | 6 | BM(14) | 47,XY,1qh+,der(9)(p22), +21c | 292insT | Leu60 | Stop codon before Met84 | AMKL at 1 y CCR (14) | ||
| 51 | F | 6 | BM(18) | 47,XX, +21c[20] | 202delAG | Gly31 | Stop codon before Met84 | CCR (31) | ||
| 56 | M | 1 | BM(8) | Not determined | 305ins5 | Arg64 | Stop codon after Met84 | CCR (3) | ||
| 57 | F | 9 | PB(65) | 47,XX, +21c[20] | 332G>A | Pro73 | Splice mutant | CCR (12) | ||
| 59 | M | 17 | Not determined | 47,XY, +21c[20] | Normal | CCR (7) | ||||
| 60 | M | 1 | PB(13) | Not determined | 304delG,ins3 | Arg64 | Stop codon before Met84 | CCR (1) | ||
| 61 | F | 14 | PB(38) | 47,XX,9qh+, +21c[20] | 332del10 | Pro73 | Splice mutant | Lost to follow-up | ||
| 62A | M | 8 | BM(13) | 47,XY, +21c[27] | 298dup289-298 | Tyr62 | Stop codon before Met84 | AMKL at 22 mo† DOD (4) | ||
| 63 | M | 2 | PB(20) | 47,XY, +21c[20] | 145dup130-145 | Thr11 | Stop codon before Met84 | CCR (49) | ||
PCR and mutation analysis
The oligonucleotides used for amplification of exon 2 of GATA1 were gata1-2F (5′-GGAAGGATTTCTGTGTCTGAG-3′) and gata1-2R (5′-GCACTCAGCCAATGCCAAGA-3′). The polymerase chain reaction (PCR) conditions were initial denaturation for 5 minutes at 95°C, followed by 35 cycles of 60 seconds at 95°C, 60 seconds at 62°C, and 90 seconds at 72°C, using an MJ Research PTC-100 thermal cycler (Waltham, MA). After the last cycle, an additional extension step of 10 minutes at 72°C was performed. Appropriate positive and negative controls were included in all experiments.
The amplified PCR products were subjected to denaturating high-performance liquid chromatography (DHPLC) analysis and direct bidirectional sequencing as described.27 Any mutation was verified at least once on an independent PCR product. PCR products with abnormal chromatograms in the DHPLC that did not yield DNA sequence variation by direct sequencing were subcloned and sequenced. When no mutation was found in exon 2 of GATA1, the other GATA1 exons were amplified (primer sequences are available on request) and subjected to DHPLC analysis. In each experiment normal controls were added.
Reverse transcription PCR (RT-PCR) analysis
Total RNA was extracted with TRIzol Reagent (Gibco-BRL, Paisley, United Kingdom). Purified RNA (0.5 μg) was retrotranscribed with MLV reverse transcriptase (Gibco-BRL) by incubating the reactions with hexa-nucleotide random primers for 2 hours at 37°C and 10 minutes at 65°C. Five percent of each reaction was subjected to 35 cycles of PCR at 63°C using a 5′ primer from exon1 (gata1-RT1F2: 5′-AGGACACCCCCTGGGATC-3′) and a 3′ primer from exon 3 (gata1-RT3R: 5′-CTCCATACAGTTGAGCAATGGG-3′). The PCR products were separated by electrophoresis on 2% agarose gel. All PCR products were sequenced.
Results
Mutations in exon 2 of GATA1 occur frequently in both TMD and AMKL in DS patients
DNA from diagnostic samples from 18 DS patients with either AML-M7 (n = 17) or AML-M0 (n = 1) were analyzed. Mutations in exon 2 of GATA1 were detected in all patients except one (Table 1). In one patient (patient 9), a mutation predicted to result in an amino acid change from Pro38 into Leu was detected. An identical mutation was detected in 1 of 50 healthy controls and in a remission sample from the same patient, suggesting that it is a polymorphism. Unfortunately, no additional DNA was available for analysis of other exons. No mutations in any of GATA1 exons were detected in patient 48. Remission samples were available from 5 patients (patients 3-7). GATA1 sequence in these samples was normal, proving that the mutations were acquired in the leukemic blasts. No mutations were detected in 3 DS patients with acute lymphoblastic leukemia (ALL) and in a 31-year-old woman with DS and AML-M2. Thus, with the exception of one DS patient with AML-M0, it seems that the association of GATA1 mutations and constitutional trisomy 21 is specific to AMKL.
To test whether GATA1 was mutated in TMD, we analyzed DNA samples from 17 patients (Figure 1A, Table 2). Similarly to the patients with AMKL, mutations were detected in 16 of the 17 patients. In 4 patients (patients 31, 37, 51, and 57), samples from remission were normal, proving that the mutation was limited to the malignant clone. The clinical course of the congenital TMD was as described in the literature.9,28-30 One patient (patient 36) died of the disease and another was lost to follow-up. In all the rest the TMD resolved with no chemotherapy. Most of the patients (75%) remained in continuous clinical remission with a median follow-up of 30 months. Three patients (17.6%) developed AMKL, at 10, 14, and 22 months from diagnosis, respectively. For 2 patients, samples from both the TMD (patients 11A and 62A, Table 2) and the AMKL (11B and 62B, Table 1) were available. The same mutations in GATA1 were identified in both samples, proving that in these patients the AMKL originated from the TMD clone. Thus, the GATA1 gene is mutated in most of the DS patients born with TMD but is not sufficient for generation of leukemia after resolution of the congenital disorder.
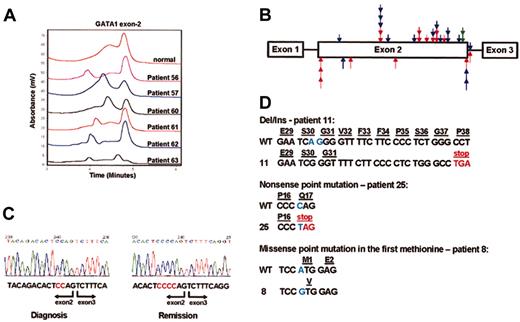
Characterization of mutations in GATA1. (A) DHPLC analysis of PCR products of GATA1 exon 2 from DS patients with TMD. The patients' numbers correspond to the numbers in Table 2. Aberrant chromatogram is seen in all patients. (B) The scatter of mutations in exon 2. Arrows indicate the positions of the different mutations found in the patients screened. Arrows placed above the bar represent insertions/deletions; arrows below the bar represent point mutations. Red arrows indicate DS AMKL patients; blue arrows, DS patients with congenital TMD; green arrows, AMKL with acquired trisomy 21. (C) Sequencing of RT-PCR fragments of GATA1 amplified from bone marrow cells of patient 19 at diagnosis and remission. The mutation 392delCC is clearly seen in the sample taken at diagnosis, but the sample taken at remission is normal. (D) Different types of mutations in exon2 of GATA1.
Characterization of mutations in GATA1. (A) DHPLC analysis of PCR products of GATA1 exon 2 from DS patients with TMD. The patients' numbers correspond to the numbers in Table 2. Aberrant chromatogram is seen in all patients. (B) The scatter of mutations in exon 2. Arrows indicate the positions of the different mutations found in the patients screened. Arrows placed above the bar represent insertions/deletions; arrows below the bar represent point mutations. Red arrows indicate DS AMKL patients; blue arrows, DS patients with congenital TMD; green arrows, AMKL with acquired trisomy 21. (C) Sequencing of RT-PCR fragments of GATA1 amplified from bone marrow cells of patient 19 at diagnosis and remission. The mutation 392delCC is clearly seen in the sample taken at diagnosis, but the sample taken at remission is normal. (D) Different types of mutations in exon2 of GATA1.
GATA1 is mutated in AMKL with acquired trisomy 21
We have screened for GATA1 exon 2 mutations in 7 cases of childhood AMKL not associated with Down syndrome. We found no mutations in 5 additional cases with AMKL and no trisomy 21. Mutations were detected in 2 monozygotic twins (described previously by Stark et al31 ) who developed concordant AMKL associated with acquired trisomy 21 in the blast cells. Both patients (patients 19 and 20) had an identical mutation, Del CC 329-330, resulting in a frame shift introducing a stop codon downstream. The mutation was not detected during remission (Figure 1C). Given an identical acquired mutation in the leukemic cells of monozygotic twins, it is likely that the mutation occurred in one of the twins in utero and that the preleukemic cells migrated to the other twin through embryonic blood connections.
Characterization of GATA1 mutations and the mechanism of GATA1s generation
The detected mutations were spread throughout exon 2 and were generally similar between TMD and AMKL (red and blue arrows, Figure 1B). Deletions/insertions were the most common, detected in 23 (65.7%) of the patients. These mutations are invariably predicted to create a shift in the reading frame of the GATA1, resulting in a premature stop codon. In 2 cases a point mutation introduced a stop codon, and in 2 others a point mutation abolished the first methionine (Figure 1D). All of these mutations prevent the translation of the full-length GATA1 protein and could allow the generation of GATA1s through alternative translation initiation from exon 3.
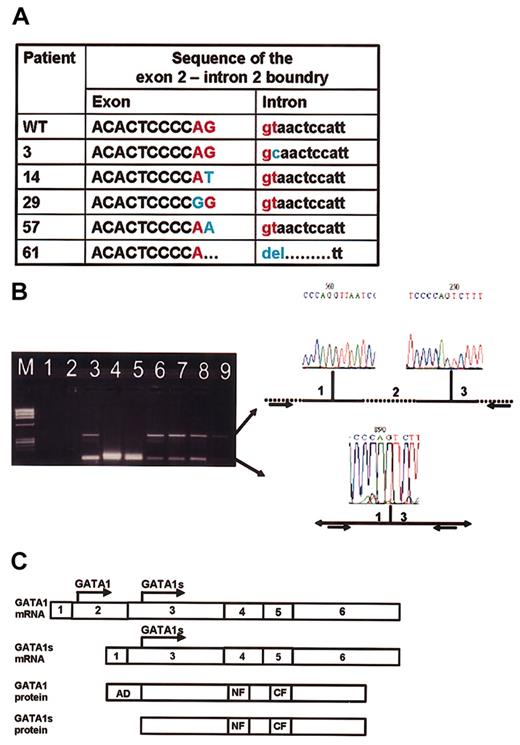
There were 5 additional mutations in the exon2/intron2 boundary (Figure 2A). These were point mutations in 4 patients and a deletion of the exon/intron boundary in 1 patient. To test the effect of these mutations on GATA1 RNA splicing, we obtained RNA from patients 3, 57, and 61, all having splice mutations, and from patient 19, whose mutation is not predicted to affect splicing. We performed RT-PCR using primers from GATA1 exon1 and 3 and sequenced all products. In the 3 patients with the suspected splice mutations an abundant short product consisting of a fusion between exon1 and exon3 was observed (lanes 3-5, Figure 2B). Surprisingly, both splice variants were also detected in leukemic samples without splice mutations or with no mutations in GATA1 (lanes 6, 8, and 9, Figure 2B) as well as in normal bone marrow (lane 7, Figure 2B), suggesting that the wild-type GATA1 mRNA is normally processed by alternative splicing of exon 2. Therefore, there might be 2 mechanisms for generation of GATA1s: alternative translation25 initiation from the full-length GATA1 mRNA and alternative splicing of exon 2 (Figure 2C). The splice mutations observed in DS patients with either TMD or AMKL result in abundant generation of the short exon1/exon3 isoform coding for GATA1s.
Alternative splicing of GATA1. (A) The sequence of the junction between exons 2 and intron 2 in 5 patients compared with wild-type sequence (first row). (B) RT-PCR analysis. M indicates marker. Lane 1: PCR-mix; lane 2: RT-mix (negative controls). Lanes 3-5: patients with splice mutations in GATA1 (patients 3, 57, and 61, respectively). Lane 6: patient 19 at diagnosis (no splice mutation); lane 7: normal bone marrow; lane 8: K562; lane 9: patient 68 (leukemia with no mutation in GATA1). The larger fragment (348 bp) corresponds to the normal splicing of exon 1 to 2 and then exon 2 to3. The smaller fragment (109 bp) corresponds to an alternative splice variant in which exon 1 is spliced to exon 3. (C) Alternative models for generation of GATA1 isoforms. The full GATA1 protein can only be translated from the full GATA1 mRNA, whereas the GATA1s protein can be translated either from the full GATA1 mRNA or from the shorter splice variant in which exon 2 is skipped.
Alternative splicing of GATA1. (A) The sequence of the junction between exons 2 and intron 2 in 5 patients compared with wild-type sequence (first row). (B) RT-PCR analysis. M indicates marker. Lane 1: PCR-mix; lane 2: RT-mix (negative controls). Lanes 3-5: patients with splice mutations in GATA1 (patients 3, 57, and 61, respectively). Lane 6: patient 19 at diagnosis (no splice mutation); lane 7: normal bone marrow; lane 8: K562; lane 9: patient 68 (leukemia with no mutation in GATA1). The larger fragment (348 bp) corresponds to the normal splicing of exon 1 to 2 and then exon 2 to3. The smaller fragment (109 bp) corresponds to an alternative splice variant in which exon 1 is spliced to exon 3. (C) Alternative models for generation of GATA1 isoforms. The full GATA1 protein can only be translated from the full GATA1 mRNA, whereas the GATA1s protein can be translated either from the full GATA1 mRNA or from the shorter splice variant in which exon 2 is skipped.
Discussion
In this multicenter study we confirm and extend the initial findings reported by Wechsler et al17 that exon2 of GATA1 is mutated in almost all patients with DS and AMKL. The detection of GATA1 mutation in AMKL with acquired trisomy 21 further substantiates the association between trisomy 21 and GATA1 mutation. The mutation is specific to megakaryoblastic malignancies, as it does not occur in ALL, another type of leukemia commonly observed in DS patients. The significant new finding is that GATA1 is also mutated in the majority of DS patients with TMD. The presence of GATA1 mutations in a congenital disorder imply that they were acquired in utero. The prenatal timing of GATA1 mutations is further supported by the identification of the same mutation in the leukemic cells of identical twins (patients 19 and 20).
The presence of GATA1 mutations in TMD suggests that cooperation between increased dosage of a gene or genes on chromosome 21 and mutations in exon 2 of GATA1 is associated with the prenatal initiation of the clonal proliferation of megakaryocytic precursors. As also observed in the group of patients described here, in the majority of patients this clonal process resolves spontaneously and never returns. Up to a third of the patients, however, develop full-blown AMKL within the first 4 years of life.5,32 The finding of an identical GATA1 mutation in specimens from TMD and AMKL occurring in the same patients suggests that the leukemia arose from the original TMD clone. The events leading to the progression to leukemia are unknown. Similar to previous reports, we observed additional karyotypic abnormalities in the majority of DS patients with AMKL, with trisomy 8 the most common acquired cytogenetic alteration in the leukemic cells.33-35 Because history of TMD is observed in only a fraction of DS AMKL patients, and since we analyzed only 2 patients for whom both samples from TMD and AMKL were available, it is impossible to rule out de novo emergence of some of the DS AMKLs and their associated GATA1 mutations. The definite answer to this question could come, for example, from analysis of GATA1 mutations in stored neonatal blood spots from DS patients with AMKL. Our data strongly suggest, however, that mutations in GATA1 are probably required for the initiation of clonal proliferation in DS patients but are insufficient for the progression to AMKL.
Since blood counts are not routinely performed on every neonate with DS, TMD may be underdiagnosed. This may also explain the absence of a history of TMD in most of the DS patients with AMKL. The identification of GATA1 mutations in almost all patients with TMD provides a molecular marker of this disease. It is now possible, therefore, to determine the prevalence of this acquired mutation in newborns with DS and to correlate it with the clinical phenotype.
GATA1 has 2 isoforms that are expressed in fetal liver and normal bone marrow.25 The short isoform, GATA1s, lacks the transactivation domain. It has been shown to bind DNA and interact with its essential partner FOG1 similarly to the full-length GATA1, but it has a reduced transactivation potential.17,25 The normal role of GATA1s is unknown. GATA1s was shown previously to be generated by alternative translation initiation starting at codon 84 of the full-length GATA1 mRNA.25 This has been revealed experimentally by in vitro transcription/translation of full-length GATA1 cDNA cloned into an expression vector and by its ectopic expression in transiently transfected cells. Here we have observed that both isoforms of wild-type GATA1 are generated by alternative splicing of exon 2. The mutations in the boundary between exon 2 and intron 2 reported here result in skipping of exon 2 and the generation of mRNA coding exclusively for GATA1s.
All the 33 new mutations observed here and the 7 previously reported ones17 prevent the synthesis of the full-length GATA1 but not the synthesis of the GATA1s. Why would a weaker allele be selected instead of mutations that abolish GATA1 function completely? Such mutations in mice and humans promote survival and proliferation of megakaryocytic precursors and block their differentiation.21-24 We speculate that the unusual selection for mutations that retain GATA1s may imply that the disruption of the normal balance between GATA1 and GATA1s has a direct oncogenic role in the setting of trisomy 21. Alternatively, GATA1s may be required for survival of the leukemic blasts and the oncogenic effect may be achieved by the loss of the full-length GATA1. Another possibility is that this type of mutation may reflect on the specific mechanisms of selection or generation of this mutation in the presence of trisomy 21.
Cooperation between mutations that promote proliferation and survival and mutations that block normal differentiation are necessary for development of leukemia.36-38 For example, activating mutations in the FLT3 kinase in AML are frequently associated with specific recurrent chromosomal translocations or a dominant negative mutation in the granulocyte differentiation factor CEBPA.38,39 The collaboration between a gene or genes on chromosome 21 and mutated GATA1 in megakaryocytic malignancies of DS is unique in its intrauterine occurrence and in its putative initiating role in a common and generally reversible clonal hematopoietic proliferation syndrome.
Prepublished online as Blood First Edition Paper, March 20, 2003; DOI 10.1182/blood-2002-11-3599.
Supported by the Gregorio and Dora Shapiro Chair for Hematology Malignancies, Sackler School of Medicine, Tel-Aviv University (G.R.); Österreischische Kinderkrebshilfe (S.S., O.A.H.); Associazione Italiana Ricerca sul Cancro, Progetto CNR Oncologia (A.B.); and the M. K. Humanitarian Fund (D.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to the clinicians and the laboratory personnel in the participating centers for the collection and processing of the samples and for providing the clinical information. We thank Dr Avi Orr-Urtreger for introducing and allowing us to use the DHPLC facility, and Drs Peter Aplan and Yoram Groner and the members of the Izraeli laboratory for lively discussions and comments on the paper.
The participating centers are members of the International BFM Study Group (I-BFM-SG).
This work was performed in partial fulfillment of the requirements for the PhD degree for Liat Rainis, Sackler Faculty of Medicine, Tel-Aviv University.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal