The t(4;14) translocation in multiple myeloma (MM) is identified in 15% of patient samples and dysregulates both FGFR3 and multiple myeloma SET domain (MMSET).1 Activating FGFR3 mutations are observed in cell lines and late-stage MM, but the incidence of mutation in newly diagnosed patients, although not well characterized,2 is likely pivotal in MM progression.3 We therefore selected CD138+ cells from the bone marrow of 150 newly diagnosed MM patients. cDNA from selected cells was hybridized to HuGeneFL GeneChip microarrays (Affymetrix, Santa Clara, CA).4 FGFR3 overexpression was noted in 24 patients (16%). Reverse transcriptase–polymerase chain reaction (RT-PCR) and fluorescence in situ hybridization (FISH) subsequently confirmed that all of the FGFR3-positive patients carried the t(4;14) translocation (Figure 1A). The entire open reading frame of FGFR3 was then amplified and bidirectionally sequenced in triplicate, which identified mutation in 2 patients. One patient harbored a 6-bp insertion 5′-AACAGT-3′ at nucleotide position 1306, which was unique to the FGFR3 IIIb isoform, implying a splice variant (Figure 1Bi). In a second patient, a point mutation was observed at nucleotide number 761A>G resulting in a Tyr241Cys (tyrosine to cysteine change) located in the linker region between immunoglobulin-like domain I and immunoglobulin-like domain II (Figure 1Bii). This genetic change is in close proximity to other previously identified activating mutations. Together these data indicate that mutation of FGFR3 is observed in less than 5% of t(4;14)-positive and in only 1 of 150 newly diagnosed MM patients overall. Incorporating findings from the literature5-9 with our larger study, 52 (15%) of 348 myeloma patients have a t(4;14) and 3 (6%) of the 52 translocation-positive patients have a potential activating mutation of FGFR3, or 3 (< 1%) of 348 overall.
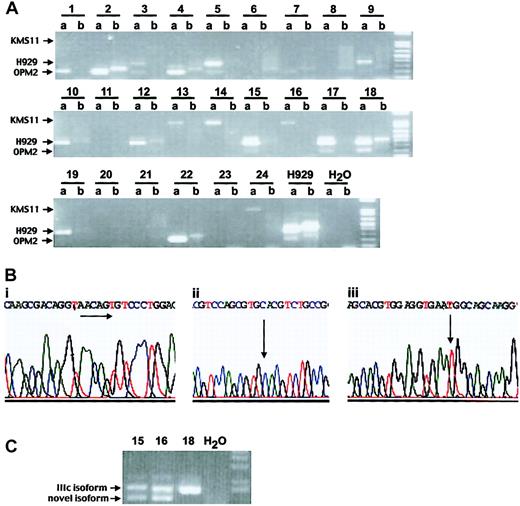
FGFR3 gene translocation and mutation screening in MM patients. (A) Detection of immunoglobulin heavy chain (IgH)/MMSET fusion product using translocation-specific RT-PCR5 (a lanes represent MMSET + JH PCR, whereas b lanes represent MMSET + 1 m PCR). Of 24 patient samples, 22 are positive: 5 patients have a translocation breakpoint as in the myeloma cell line KMS11, 9 patients have a breakpoint like H929, and 8 patients have an OPM2 breakpoint. (B) Genetic changes of FGFR3 gene in patient samples. (i) Sequence analysis of cloned cDNA of patient no. 1 revealed a 6-bp insertion of 5′-AACAGT-3′ at nucleotide position 1306, resulting in an insertion of amino acids threonine and cysteine. (ii) Point mutation at nucleotide 761A>G resulting in tyrosine to cysteine change in FGFR3 codon 241 (patient no. 23, sequencing from reverse direction of the gene). (iii) Of 22 patient samples, 17 carry C to T transition in FGFR3 codon 294. (C) Different isoforms of FGFR3 are detected in patient nos. 15 and 16. Patient nos. 15 and 16 expressed the FGFR3 IIIc isoform (upper band) and novel isoforms devoid of Ig-like domain I and of both domain I and the acid-box, respectively, (lower band) while patient no. 18 expressed only FGFR3 IIIc isoform.
FGFR3 gene translocation and mutation screening in MM patients. (A) Detection of immunoglobulin heavy chain (IgH)/MMSET fusion product using translocation-specific RT-PCR5 (a lanes represent MMSET + JH PCR, whereas b lanes represent MMSET + 1 m PCR). Of 24 patient samples, 22 are positive: 5 patients have a translocation breakpoint as in the myeloma cell line KMS11, 9 patients have a breakpoint like H929, and 8 patients have an OPM2 breakpoint. (B) Genetic changes of FGFR3 gene in patient samples. (i) Sequence analysis of cloned cDNA of patient no. 1 revealed a 6-bp insertion of 5′-AACAGT-3′ at nucleotide position 1306, resulting in an insertion of amino acids threonine and cysteine. (ii) Point mutation at nucleotide 761A>G resulting in tyrosine to cysteine change in FGFR3 codon 241 (patient no. 23, sequencing from reverse direction of the gene). (iii) Of 22 patient samples, 17 carry C to T transition in FGFR3 codon 294. (C) Different isoforms of FGFR3 are detected in patient nos. 15 and 16. Patient nos. 15 and 16 expressed the FGFR3 IIIc isoform (upper band) and novel isoforms devoid of Ig-like domain I and of both domain I and the acid-box, respectively, (lower band) while patient no. 18 expressed only FGFR3 IIIc isoform.
To determine the single nucleotide polymorphism (SNP) profile of FGFR3 we next examined the Celera database and identified 21 SNPs pertaining to the coding region of FGFR3. One of these SNPs, 921C>T, was identified in 17 of 22 FGFR3 sequences (Figure 1Biii). Using the SNP human database (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi) this 294C>T polymorphism has an allele frequency of 0.877. The 77% SNP incidence in the patient samples is therefore not statistically different from that of the general human population.
Our patient samples expressed different FGFR3 isoforms by sequencing, with the FGFR3 IIIc isoform being most prevalent. We also identified 2 novel FGFR3 isoforms in MM plasma cells: an isoform devoid of immunoglobulin-like domain I and an isoform that is devoid of both domain I and the acid-box (Figure 1C). Interestingly, although both isoforms have ligand-binding properties similar to those of the full-length receptor, they have previously been reported to have a higher affinity for the FGFR ligand.10
In summary, our comprehensive analysis of 150 newly diagnosed MM patients has demonstrated that overexpression of FGFR3 is seen in 16% of newly diagnosed patients, overexpression is observed only in the presence of the t(4;14), and activating mutations of FGFR3 are rare in newly diagnosed MM patients. The 294C>T polymorphism is common in MM patients and a number of different isoforms of the gene are expressed (the significance of which is uncertain).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal