Abstract
Results from experimental models, in vitro studies, and clinical data indicate that granulocyte colony-stimulating factor (G-CSF) stimulation alters T-cell function and induces Th2 immune responses. The immune modulatory effect of G-CSF on T cells results in an unexpected low incidence of acute graft-versus-host disease in peripheral stem cell transplantation. However, the underlying mechanism for the reduced reactivity and/or alloreactivity of T cells upon G-CSF treatment is still unknown. In contrast to the general belief that G-CSF acts exclusively on T cells via monocytes and dendritic cells, our results clearly show the expression of the G-CSF receptor in class I– and II– restricted T cells at the single-cell level both in vivo and in vitro. Kinetic studies demonstrate the induction and functional activity of the G-CSF receptor in T cells upon G-CSF exposure. Expression profiling of T cells from G-CSF–treated stem cell donors allowed identification of several immune modulatory genes, which are regulated upon G-CSF administration in vivo (eg, LFA1-α, ISGF3-γ) and that are likely responsible for the reduced reactivity and/or alloreactivity. Most importantly, the induction of GATA-3, the master transcription factor for a Th2 immune response, could be demonstrated in T cells upon G-CSF treatment in vivo accompanied by an increase of spontaneous interleukin-4 secretion. Hence, G-CSF is a strong immune regulator of T cells and a promising therapeutic tool in acute graft-versus-host disease as well as in conditions associated with Th1/Th2 imbalance, such as bone marrow failure syndromes and autoimmune diseases.
Introduction
Peripheral blood stem cells (PBSCs) obtained from granulocyte colony-stimulating factor (G-CSF)–mobilized donors are increasingly used for allogeneic transplantation. Some of the advantages of PBSC transplantation (PBSCT) compared with conventional bone marrow transplantation (BMT) include easier harvesting of stem cells, improved yield of progenitor cells, and a more rapid and robust engraftment.1,2 Despite the presence of a more than 10-fold higher number of mature T cells in the graft, the incidence and severity of acute graft-versus-host disease (GVHD) is surprisingly low and comparable with BMT.3,4 However, recent studies suggest an increased risk of chronic GVHD after G-CSF–mobilized PBSCT.5,6
Results from experimental model systems, in vitro studies, and clinical data indicate that G-CSF induces particular immunologic changes in healthy donors and patients undergoing PBSCT. In human and animal studies, G-CSF stimulation alters the T-cell function and modulates the balance between Th1 and Th2 immune responses by affecting cytokine production.7-10 After G-CSF stimulation in vivo, human and murine T cells show a reduced cytotoxic activity and proliferative response upon in vitro stimulation.11,12 Monocytes from G-CSF–mobilized human donors have also been shown to suppress T-cell alloreactivity.11 This suppresion may occur through an interleukin-10 (IL-10)–dependent mechanism13 and/or through the inhibition of IL-12 and tumor necrosis factor α (TNF-α) release.9 However, others recently suggested a direct effect of G-CSF on human CD4+ T cells skewing T-cell differentiation toward a Th2 type with an increase of IL-4 and decrease of interferon α (IFN-α) production upon alloantigeneic or mitogeneic stimulation in vitro.8 Additionally, an anti-inflammatory response with a reduction of proinflammatory cytokines such as TNF-α, IL-12, IL-1β, and IL-2 has been reported.9,10 In murine models for GVHD, G-CSF administration of the donor led to a reduced severity and mortality rate of acute GVHD, presumably due to the functional polarization of the donor T cells,3,14 which might be related to the selective mobilization of type 2 dendritic cells upon G-CSF treatment.14,15
Materials and methods
G-CSF treatment of stem cell donors and in vitro stimulation with G-CSF
Blood samples were obtained from 32 stem cell donors after written informed consent before and after treatment with recombinant G-CSF at a dosage of 10 μg/kg/d for 4 consecutive days (filgrastim; Amgen, Thousand Oaks, CA). Approval was obtained from the institutional review board of Hannover Medical School. T-cell subpopulations were freshly isolated and used for further analysis. For in vitro stimulation PBMCs or freshly isolated T cells of healthy donors were incubated in triplicates with G-CSF (100 ng/mL) for 2 to 72 hours at 37°C and 5% CO2 in RPMI 1640 supplemented with antibiotics (Life Technologies, New York, NY) and 10% fetal calf serum. In the case of stimulating PBMCs, CD3+ T cells were isolated after G-CSF stimulation and used for further analysis.
Isolation of CD3+, CD4+, and CD8+ T-cell populations
CD3+ T cells were negatively selected using the pan T-cell isolation kit (Miltenyi-Biotec, Gladbach, Germany) according to the manufacturer's instruction. Isolation of CD4+ and CD8+ T cells was performed with an additional step of positive selection using anti-CD4+ or anti-CD8+ microbeads, respectively. The purity of the isolated T-cell populations was controlled by 3-color fluorescence-activated cell sorter (Becton Dickinson, Mountain View, CA) analysis and was higher than 98%. The residual contaminating cell populations varied in the isolations from the different donors and represented natural killer cells (1%-2%), dendritic cells (< 1%), B cells (< 1%), and monocytes (< 1%). In order to have sufficient starting material (cell number and total RNA amount) the molecular analysis (cDNA arrays and real-time reverse transcription–polymerase chain reaction [RT-PCR]) of CD4+ T-cell subpopulations required pooling of donor T cells, whereas CD3+ T cells could be analyzed in individual donors.
Single-cell RT-PCR for G-CSF receptor (G-CSFR) expression in T cells
After T-cell isolation, single cells were handpicked under microscopic control using a microcapillary system. cDNA was synthesized after thermic lysis of single cells as previously described.16 Single cells were subsequently analyzed for mRNA expression of G-CSFR and T-cell receptor α (TCR-α) chain. Coamplification of the TCR-α chain in the analyzed single cells allowed identification as T cells. Seminested RT-PCRs for G-CSFR and TCR-α chain were done in separate tubes as follows: the first PCR was performed with a total volume of 85 μL reaction mix containing 15 μL cDNA for amplification of G-CSFR and 5 μL cDNA for amplification of the TCR-α chain; 0.2 mM deoxynucleoside triphosphate; 2 mM MgCl2; 2.5 U Ampli Taq DNA polymerase (Gibco, Eggstein, Germany); and 0.5 μM of each primer. First and second RT-PCRs were amplified for G-CSFR and TCR-α, respectively, under the same conditions: at 94°C for 45 seconds for both G-CSFR and TCR-α; at 62°C for 45 seconds for G-CSFR; at 64°C for 30 seconds for TCR-α; at 72°C for 1 minute 20 seconds for G-CSFR; and at 1 minute for TCR-α—for a total of 38 cycles in the first and 35 cycles in the second round. Primer sequences can be obtained from the corresponding author. Specificity of RT-PCRs was confirmed by DNA sequencing of single-cell RT-PCR products. Efficiencies of the RT-PCRs for amplification of G-CSFR and the TCR-α chain were higher than 90% and 95%, respectively, as determined by RT-PCR of more than 50 single cells of the leukemic cell line U937 (for G-CSFR) and CD3+ T cells (for TCR-α).
cDNA gene expression array
Differential expression of 406 genes was investigated by cDNA array hybridization (Atlas Human Hematology Array; Clontech, Palo Alto, CA) in CD3+ T cells of 3 PBSC donors and in CD4+ T cells of 3 pools of 3 additional donors each before and after G-CSF application. Total RNA was extracted using Tri-Reagent LS (Molecular Research Center, Cincinnati, OH). After DNAse I digestion, a second RNA extraction was performed. Synthesis of 32P-radiolabeled cDNA probes, hybridization, and array analysis were performed as previously described.17
Quantitative real-time RT-PCR
To quantify mRNA levels of GATA-3, IL-4, and IFN-γ in CD3+ and CD4+ T cells, total RNA was isolated using Trizol (Gibco) according to the manufacturer's protocol. Then, DNAse treatment (1 U/10 μg RNA) was performed, followed by re-extraction of RNA. After reverse transcription, real-time PCRs were performed using the GeneAmp5700 Sequence Detection System (Perkin Elmer Biosystems, Warrington, United Kingdom): 2 minutes at 50°C, 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 58°C using SYBR green (Perkin Elmer). Standard curves were generated by serial dilution of cDNA probes. The RPS9 housekeeping gene was used for normalization. The sequences for the primers can be obtained from the corresponding author.
Western blot analysis
Total protein was extracted from T cells and assessed by Western blot analysis using a monoclonal mouse antibody for immunoblotting of GATA-3 (Santa Cruz Biotechnology, Santa Cruz, CA) followed by enhanced chemiluminescence. Ratios of GATA-3 protein expression before and after G-CSF treatment were calculated by applying Scion Image Beta 4.02 software (Frederick, MD) for densitometric quantification of the immunoreactive bands.
ELISPOT assay
The enzyme-linked immunospot assay was performed as previously described.18 In brief, negatively selected CD3+ T cells were transferred by 3 1:10 dilution steps (starting at 1 × 106/well) in triplicates to 96-well microtiter plates (Millipore, Bedford, MA) that had been coated with the capturing monoclonal antibody (mAb) to IL-4 (Autoimmun Diagnostika, Strassberg, Germany). After incubation for 48 hours to allow IL-4 secretion, cells were removed and spot-forming cells were visualized with a biotinylated mAb to IL-4 in conjunction with avidin peroxidase and 3-amino-9-ethylcarbazole (Sigma Chemical, St Louis, MO). The frequency of IL-4–secreting cells was determined using ELISPOT Reader Software Version 2.5 (Autoimmun Diagnostika).
Statistical analysis
Statistical analysis of gene expression in single cells was performed by using the chi-square test. Differences were considered significant for a P value less than .05.
Results
G-CSFR gene expression in T cells identified by single-cell RT-PCR
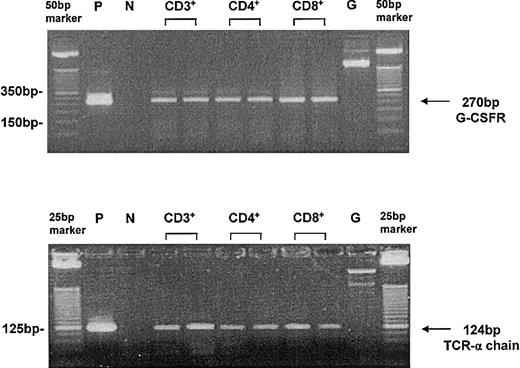
Single T cells of PBSC donors were analyzed for mRNA expression of the G-CSFR before and after G-CSF stimulation both in vivo and in vitro. CD4+ and CD8+ T-cell subpopulations were enriched by microbead technology before single-cell picking and could be identified as T cells because of the coexpression of the TCR-α chain. The efficiency of the single-cell RT-PCR for the G-CSFR and TCR-α chain was higher than 90%; specificity was confirmed by DNA sequencing of the single-cell RT-PCR product. Analysis of more than 150 freshly isolated peripheral single T cells expressing the TCR-α chain demonstrated that G-CSFR gene expression was undetectable before G-CSF stimulation (Table 1). After in vivo treatment of the donors with G-CSF for 4 days, expression of the receptor could be detected in 7 donors tested at the mRNA level in 21% to 50% of the analyzed single T cells in each subpopulation analyzed (Table 1; Figure 1). In total, the G-CSFR was induced after G-CSF treatment in 91 of 268 single donor T cells (34%) without a significant difference among the individual donors or CD4+ and CD8+ T-cell subpopulations (P > .05). The kinetic study of G-CSFR receptor gene expression was performed in more than 200 single T cells after direct stimulation of T cells (purity > 98%) or stimulation of PBMCs with G-CSF in vitro and did not show a significant difference (P = .28; chi-square test) in the frequency of T cells expressing the G-CSFR (Table 2). However, the kinetic analysis demonstrates that G-CSFR mRNA expression in T cells is induced after G-CSF stimulation in a time-dependent manner. G-CSFR–positive T cells could be detected as early as 2 to 4 hours after in vitro incubation with G-CSF. The highest frequency was reached after 12 to 24 hours. After 72 hours of in vitro stimulation with G-CSF, the G-CSFR could no longer be detected by single-cell RT-PCR (Table 2), whereas the TCR-α chain could still be amplified. However, repetitive stimulation with G-CSF maintained G-CSFR expression in T cells in vitro (Table 2).
G-CSFR expression in T cells before and after G-CSF treatment in vivo analyzed by single-cell RT-PCR
Timepoint . | CD3+ T cells (%) . | CD4+ T cells (%) . | CD8+ T cells (%) . |
|---|---|---|---|
| Before G-CSF | 0/89 (0) | 0/36 (0) | 0/32 (0) |
| Donor 1 | 0/28 (0) | — | — |
| Donor 2 | 0/21 (0) | — | — |
| Donor 3 | — | 0/16 (0) | 0/16 (0) |
| Donor 4 | — | 0/20 (0) | 0/16 (0) |
| Donor 5 | 0/13 (0) | — | — |
| Donor 6 | 0/15 (0) | — | — |
| Donor 7 | 0/12 (0) | — | — |
| After G-CSF | 36/102 (35) | 22/80 (27) | 33/86 (38) |
| Donor 1 | 14/36 (38.9) | — | — |
| Donor 2 | 6/19 (31.6) | — | — |
| Donor 3 | — | 6/21 (28.6) | 6/16 (37.5) |
| Donor 4 | — | 4/16 (25) | 8/19 (42.1) |
| Donor 5 | 8/16 (50) | 5/15 (33) | 5/20 (25) |
| Donor 6 | 4/16 (25) | 3/14 (21) | 7/16 (43) |
| Donor 7 | 4/15 (27) | 4/14 (28) | 7/15 (46) |
Timepoint . | CD3+ T cells (%) . | CD4+ T cells (%) . | CD8+ T cells (%) . |
|---|---|---|---|
| Before G-CSF | 0/89 (0) | 0/36 (0) | 0/32 (0) |
| Donor 1 | 0/28 (0) | — | — |
| Donor 2 | 0/21 (0) | — | — |
| Donor 3 | — | 0/16 (0) | 0/16 (0) |
| Donor 4 | — | 0/20 (0) | 0/16 (0) |
| Donor 5 | 0/13 (0) | — | — |
| Donor 6 | 0/15 (0) | — | — |
| Donor 7 | 0/12 (0) | — | — |
| After G-CSF | 36/102 (35) | 22/80 (27) | 33/86 (38) |
| Donor 1 | 14/36 (38.9) | — | — |
| Donor 2 | 6/19 (31.6) | — | — |
| Donor 3 | — | 6/21 (28.6) | 6/16 (37.5) |
| Donor 4 | — | 4/16 (25) | 8/19 (42.1) |
| Donor 5 | 8/16 (50) | 5/15 (33) | 5/20 (25) |
| Donor 6 | 4/16 (25) | 3/14 (21) | 7/16 (43) |
| Donor 7 | 4/15 (27) | 4/14 (28) | 7/15 (46) |
T-cell populations were isolated by microbead technology (purity > 98%) and subsequently handpicked. The analyzed single cells could be identified as T cells because of coamplification of the TCR-α chain mRNA. G-CSFR expression was significantly induced upon G-CSF treatment in vivo (P < .01), but did not differ significantly among the analyzed 7 donors (P > .05).—indicates not determined
Single-cell RT-PCR for G-CSFR expression in T cells after G-CSF treatment in vivo. T cells were freshly isolated by microbead technology (purity > 98%) and single cells were handpicked under microscopic control using a microcapillary system. The leukemia cell line U937 served as a positive control (P). The application of exon–intron spanning primers allowed the discrimination from genomic amplification (G) of the G-CSFR (top blot). Because of the coamplification of the T-cell receptor α (TCR-α) chain, the analyzed cells could be identified as T cells (bottom blot). Peripheral T cells served as a positive control. Efficiency of single-cell RT-PCR for G-CSFR and for TCR-α chain was higher than 90%. P indicates positive control; N, negative control; and G, genomic amplification.
Single-cell RT-PCR for G-CSFR expression in T cells after G-CSF treatment in vivo. T cells were freshly isolated by microbead technology (purity > 98%) and single cells were handpicked under microscopic control using a microcapillary system. The leukemia cell line U937 served as a positive control (P). The application of exon–intron spanning primers allowed the discrimination from genomic amplification (G) of the G-CSFR (top blot). Because of the coamplification of the T-cell receptor α (TCR-α) chain, the analyzed cells could be identified as T cells (bottom blot). Peripheral T cells served as a positive control. Efficiency of single-cell RT-PCR for G-CSFR and for TCR-α chain was higher than 90%. P indicates positive control; N, negative control; and G, genomic amplification.
Kinetic analysis of G-CSFR mRNA expression in CD3+ T cells at single-cell level following G-CSF stimulation in vitro
Time, h . | After single stimulation of T cells (%) . | After single stimulation of PBMCs (%) . | After repetitive stimulation of T cells (%) . |
|---|---|---|---|
| 2 | 1/14 (7) | 2/12 (17) | — |
| 4 | 2/16 (12) | 3/18 (17) | — |
| 8 | 2/20 (10) | 4/17 (23) | — |
| 12 | 5/25 (20) | 5/17 (29) | — |
| 24 | 4/21 (19) | 4/21 (19) | 6/32 (19) |
| 36 | 0/21 (0) | — | 7/22 (32) |
| 48 | 0/22 (0) | — | 4/23 (17) |
| 60 | 0/23 (0) | — | 6/25 (24) |
| 72 | 0/18 (0) | 0/18 (0) | 4/24 (17) |
Time, h . | After single stimulation of T cells (%) . | After single stimulation of PBMCs (%) . | After repetitive stimulation of T cells (%) . |
|---|---|---|---|
| 2 | 1/14 (7) | 2/12 (17) | — |
| 4 | 2/16 (12) | 3/18 (17) | — |
| 8 | 2/20 (10) | 4/17 (23) | — |
| 12 | 5/25 (20) | 5/17 (29) | — |
| 24 | 4/21 (19) | 4/21 (19) | 6/32 (19) |
| 36 | 0/21 (0) | — | 7/22 (32) |
| 48 | 0/22 (0) | — | 4/23 (17) |
| 60 | 0/23 (0) | — | 6/25 (24) |
| 72 | 0/18 (0) | 0/18 (0) | 4/24 (17) |
Kinetic analysis of G-CSFR mRNA expression in CD3+ T cells at single-cell level following G-CSF stimulation in vitro. Direct stimulation of T cells (purity > 98%) compared with stimulation of PBMCs with G-CSF in vitro did not show a significant difference in the frequency of T cells expressing G-CSFR mRNA (P = .28, chi-square test). Repetitive stimulation of T cells every 24 hours led to sustained G-CSFR expression. All of the analyzed single cells expressed TCR-α chain mRNA. — indicates not determined
Gene expression profiling of T cells before and after G-CSF treatment in vivo
Here, we studied data sets derived from 3 independent experiments analyzing CD3+ and CD4+ T cells before and after G-CSF stimulation, respectively. About one third of the 406 genes assessed by the cDNA arrays were expressed to an appreciable extent in all of the T-cell populations tested. The patterns of gene expression changed considerably at different levels of activation, proliferation, and functional differentiation upon G-CSF treatment in vivo. Genes, which were regulated in each independent experiment in CD3+ and CD4+ T cells, are presented in Table 3 as mean values of expression ratios (after versus before G-CSF application). The T cells showed signs of activation with an up-regulation of surface activation markers (CD69 and CD53). In contrast, the mRNA expression of costimulatory molecules (CD5 and CD44) was down-regulated. The gene expression of the adhesion molecule LFA-1α was reduced by 50% in CD3+ and 70% in CD4+ T cells compared with the unstimulated controls. Most importantly, GATA-3, the master regulatory factor for a Th2 immune response, was induced upon G-CSF stimulation. GATA-3 gene expression was increased by more than 7-fold in CD4+ T cells and by 1.5-fold in CD3+ T cells. Gene expression of another transcription factor, Stat5, was up-regulated upon G-CSF stimulation by nearly 2-fold in CD3+ T cells and more than 10-fold in CD4+ T cells. Interestingly, Stat5 is critical for cell proliferation and survival and is known to be regulated by several cytokines including G-CSF. In addition, mRNA expression of the γ-subunit of the transcriptional regulator complex ISGF3, essential for the induction of IFN-γ responsive gene expression, was down-regulated in CD3+ (0.6) and CD4+ T cells (0.7), further underlining the impact of G-CSF stimulation on T-cell function in vivo.
Regulation of gene expression in CD3+ and CD4+ T cells upon G-CSF treatment in vivo
. | Name of gene . | GenBank accession no. . | CD3+ T cells, fold change . | CD4+ T cells, fold change . |
|---|---|---|---|---|
| Activation marker | ||||
| A | CD69 | NM001781 | 2.1 | 5.8 |
| B | CD53 | NM000560 | 1.6 | 1.4 |
| Cell-surface molecules/adhesion marker | ||||
| C | CD44H | AX060538 | 0.5 | 0.7 |
| D | CD5 | X04391 | 0.7 | 0.7 |
| E | LFA1-α | AF114104 | 0.4 | 0.3 |
| Transcription factors | ||||
| F | GATA-3 | NM002051 | 1.5 | 7.6 |
| G | Stat5 | NM003152 | 1.8 | 16 |
| H | ISGF3-γ | NM006984 | 0.6 | 0.7 |
. | Name of gene . | GenBank accession no. . | CD3+ T cells, fold change . | CD4+ T cells, fold change . |
|---|---|---|---|---|
| Activation marker | ||||
| A | CD69 | NM001781 | 2.1 | 5.8 |
| B | CD53 | NM000560 | 1.6 | 1.4 |
| Cell-surface molecules/adhesion marker | ||||
| C | CD44H | AX060538 | 0.5 | 0.7 |
| D | CD5 | X04391 | 0.7 | 0.7 |
| E | LFA1-α | AF114104 | 0.4 | 0.3 |
| Transcription factors | ||||
| F | GATA-3 | NM002051 | 1.5 | 7.6 |
| G | Stat5 | NM003152 | 1.8 | 16 |
| H | ISGF3-γ | NM006984 | 0.6 | 0.7 |
The regulation of gene expression in T cells upon G-CSF treatment in vivo was investigated in CD3+ and CD4+ T cells from healthy stem cell donors before and after G-CSF application. The results of cDNA array hybridizations are shown as mean factor of expression ratios (after vs before G-CSF) of regulated genes from 3 paired array experiments each for CD3+ and CD4+ T cells. Array analyses of CD3+ T cells were performed from individual donors before and after G-CSF application; CD4+ T cells had to be pooled for each array experiment from 3 stem cell donors in order to obtain sufficient starting material
Induction of GATA-3 and Th2 type at the mRNA level in donor T cells upon G-CSF treatment in vivo
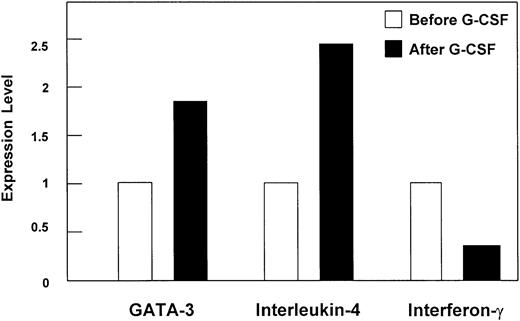
In order to confirm the induction of the master Th2 immune response regulator GATA-3 by an independent method, quantitative real-time RT-PCR was performed. An approximately 2-fold increase of GATA-3 mRNA expression was found in CD4+ T cells from a pool of 3 donors upon G-CSF treatment in vivo (mean value of 2 independent experiments; Figure 2) as well as in CD3+ T cells (data not shown). As representative cytokines for a Th1/Th2 immune response, IFN-γ and IL-4 gene expression, respectively, were quantified in CD3+ (data not shown) and CD4+ T cells of healthy donors before and after 4 days of G-CSF treatment using real-time RT-PCR. G-CSF stimulation resulted in a 2.5-fold increase of IL-4 and 3-fold decrease of IFN-γ (Figure 2) mRNA expression in CD4+ T cells.
Differential gene expression in CD4+ T cells upon G-CSF treatment in vivo quantified by real-time RT-PCR. Regulation of gene expression levels of GATA-3, interleukin-4, and interferon-γ in CD4+ T cells of stem cell donors upon G-CSF treatment in vivo analyzed by quantitative real-time RT-PCR. The mRNA expression was quantified by applying Taqman technology (Perkin Elmer, Wellesley, MA), and data are presented as mean (bars) of relative ratios of expression levels. Performed were 2 independent experiments of a pool of 3 stem cell donors each. SEM was below 10%.
Differential gene expression in CD4+ T cells upon G-CSF treatment in vivo quantified by real-time RT-PCR. Regulation of gene expression levels of GATA-3, interleukin-4, and interferon-γ in CD4+ T cells of stem cell donors upon G-CSF treatment in vivo analyzed by quantitative real-time RT-PCR. The mRNA expression was quantified by applying Taqman technology (Perkin Elmer, Wellesley, MA), and data are presented as mean (bars) of relative ratios of expression levels. Performed were 2 independent experiments of a pool of 3 stem cell donors each. SEM was below 10%.
Induction of GATA-3 and IL-4 secretion at the protein level in donor T cells upon G-CSF treatment in vivo
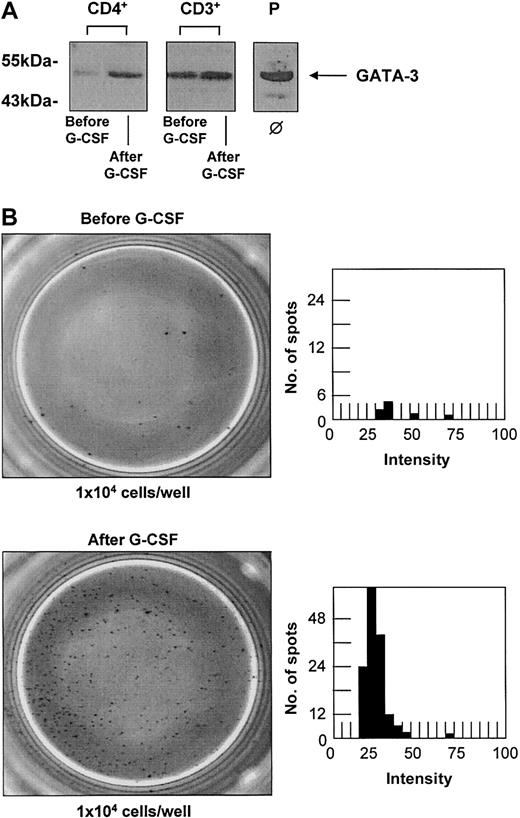
CD3+ and CD4+ T-cell populations were isolated before and after G-CSF treatment of the donors with a purity higher than 98%. Without any in vitro stimulation the same cell numbers of each T-cell population were directly lysed for further protein studies. Up-regulation of GATA-3 upon G-CSF stimulation in vivo could be confirmed at the protein level (Figure 3A) in 6 different donors with an increase up to 2.6-fold in CD3+ and 1.8-fold in CD4+ T cells. The effect of G-CSF on the secretion of IL-4 as a representative cytokine for a Th2 immune response was also analyzed at the protein level using an ELISPOT assay. As ELISPOT analysis has a higher sensitivity than the quantification of cytokine secretion using enzyme-linked immunosorbent assay (ELISA) techniques, no additional mitogeneic stimulation in vitro was necessary. The spontaneous production of IL-4 by CD3+ T cells was quantified ex vivo after G-CSF stimulation of a peripheral stem cell donor and compared with the spontaneous secretion of T cells from a healthy untreated donor. PHA-stimulated mononucleated cells were used as a positive control and cell-free medium was used as a negative background control. G-CSF treatment in vivo led to a 15- to 20-fold increase of spontaneous IL-4 secretion by CD3+ T cells (Figure 3B).
G-CSF treatment in vivo leads to an up-regulation of GATA-3 and increase of spontaneous IL-4 secretion in T cells. G-CSF treatment in vivo leads to an up-regulation of GATA-3 and an increase of spontaneous IL-4 secretion in T cells of healthy stem cell donors. (A) Western blot analysis of CD3+ and CD4+ T cells confirmed the up-regulation of GATA-3 upon G-CSF treatment in vivo at the protein level in 6 individual donors with increases up to 1.8-fold in CD3+ and 2.6-fold in CD4+ T cells (Western blots of representative donors are shown). The T-cell leukemia cell line MOLT-4 served as a positive control (P). ϕ indicates no in vitro stimulation. (B) The production of IL-4 as representative cytokine for a Th2 immune response was quantified in CD3+ T cells before (top panels) and after (bottom panels) G-CSF treatment by ELISPOT analysis. PHA-stimulated PBMCs served as a positive control, and cell-free medium served as a negative control. The number of spot-forming units is shown in dependence of their intensity determined by ELISPOT Reader Software. G-CSF treatment resulted in a 10- to 20-fold increase of spontaneous IL-4 secretion by CD3+ T cells in 3 different donors analyzed (1 representative donor is shown). Original magnification, × 20.
G-CSF treatment in vivo leads to an up-regulation of GATA-3 and increase of spontaneous IL-4 secretion in T cells. G-CSF treatment in vivo leads to an up-regulation of GATA-3 and an increase of spontaneous IL-4 secretion in T cells of healthy stem cell donors. (A) Western blot analysis of CD3+ and CD4+ T cells confirmed the up-regulation of GATA-3 upon G-CSF treatment in vivo at the protein level in 6 individual donors with increases up to 1.8-fold in CD3+ and 2.6-fold in CD4+ T cells (Western blots of representative donors are shown). The T-cell leukemia cell line MOLT-4 served as a positive control (P). ϕ indicates no in vitro stimulation. (B) The production of IL-4 as representative cytokine for a Th2 immune response was quantified in CD3+ T cells before (top panels) and after (bottom panels) G-CSF treatment by ELISPOT analysis. PHA-stimulated PBMCs served as a positive control, and cell-free medium served as a negative control. The number of spot-forming units is shown in dependence of their intensity determined by ELISPOT Reader Software. G-CSF treatment resulted in a 10- to 20-fold increase of spontaneous IL-4 secretion by CD3+ T cells in 3 different donors analyzed (1 representative donor is shown). Original magnification, × 20.
Direct induction of GATA-3 and Th2 type in T cells upon G-CSF exposure in vitro
A kinetic study was performed with highly purified CD3+ T cells (purity > 98%) in order to analyze direct effects of G-CSF on T cells; in particular, the secondary changes in production of GATA-3, IL-4, and IFN-γ were monitored. As demonstrated in 2 independent experiments, G-CSF stimulation in vitro led to a peak of IL-4 and GATA-3 mRNA production after 4 hours of culture, whereas IFN-γ mRNA expression decreased continuously starting as early as 2 hours after G-CSF stimulation of T cells (Figure 4). These results were confirmed at the protein level in 2 additional independent experiments applying the ELISPOT assay for the quantification of spontaneous IL-4 and IFN-γ secretion by CD3+ T cells upon G-CSF stimulation in vitro (data not shown). The secretion of IL-4 and IFN-γ by CD3+ T cells was biased significantly after 12 hours of in vitro stimulation with G-CSF: spontaneous IL-4 secretion increased nearly 2-fold, whereas a nearly 50% reduction of IFN-γ secretion was observed.
Kinetic analysis of secondary changes in IL-4, IFN-γ, and GATA-3 expression in CD3+ T cells upon G-CSF treatment in vitro. Fold change in gene expression levels of IL-4, IFN-γ, and GATA-3 in CD3+ T cells was determined by quantitative real-time RT-PCR upon G-CSF treatment in vitro at the indicated time points. The mRNA expression was quantified by applying Taqman technology. Data are expressed as mean of 2 independent experiments from 2 healthy donors (n = 2).
Kinetic analysis of secondary changes in IL-4, IFN-γ, and GATA-3 expression in CD3+ T cells upon G-CSF treatment in vitro. Fold change in gene expression levels of IL-4, IFN-γ, and GATA-3 in CD3+ T cells was determined by quantitative real-time RT-PCR upon G-CSF treatment in vitro at the indicated time points. The mRNA expression was quantified by applying Taqman technology. Data are expressed as mean of 2 independent experiments from 2 healthy donors (n = 2).
Discussion
The immune modulatory effect of G-CSF on T cells is believed to be mediated exclusively through other effector cells, for example, through monocytes by down-regulation of costimulatory molecules,7 increased IL-1013 production and decreased secretion of IL-12 and TNF-α,3,11 or selective mobilization of type 2 dendritic cells skewing T-cell differentiation toward a Th2 phenotype.15 As it is still unclear whether T cells actually do express the G-CSFR and thereby enable direct effects of G-CSF on the T-cell immune system, our study sought to clarify this issue at the single-cell level. While others failed to detect the G-CSFR on T lymphocytes in a bulk population by applying RT-PCR or flow cytometric analysis,9 we could clearly demonstrate by single-cell RT-PCR that both CD4+ and CD8+ T cells express the G-CSFR after G-CSF treatment. The single-cell approach described in “Materials and methods” combines high sensitivity for the detection of the G-CSFR with high specificity for T cells by coamplification of the TCR-α chain in the analyzed single cells. Kinetic studies and coculture experiments show that G-CSF directly induces the G-CSFR in T cells in a time-dependent manner. However, G-CSFR expression was not detected in T cells prior to G-CSF stimulation. Furthermore, repetitive stimulation of T cells with G-CSF in vitro was found to reinduce or sustain G-CSFR expression. These results might argue for functional G-CSFR expression below the detection limit of the single-cell assay and up-regulation of the receptor through an autoloop mechanism. Since both helper and cytotoxic T cells express the G-CSFR, preferential expression in a distinct functional T-cell subset is unlikely.
The functional alteration of T cells results in a reduced alloreactivity of G-CSF–mobilized grafts with an unexpected low incidence/severity of acute GVHD despite the relatively large numbers of mature T cells used for transplantation. However, the observed alteration might also be responsible for an increased risk of chronic GVHD, as recent studies report a higher incidence of chronic GVHD after G-CSF–mobilized PBSCT.5,6
This phenomenon was analyzed in greater detail by our cDNA array approach. CD4+ T cells showed signs of activation upon G-CSF stimulation with an mRNA increase of the surface molecules CD69 and CD53, which belong to the tetraspanin superfamily and transduce activation signals.19 In contrast, mRNA expression of costimulatory molecules was down-regulated. This might in fact reflect the recent activation state of T lymphocytes upon stimulation with G-CSF directly or via accessory cells in vivo. CD5, which up-regulates TCR signaling through crosslinking of rafts,20 was decreased by almost 30% in CD4+ T cells. CD44H, a CD28-independent costimulatory surface molecule, was also down-regulated upon G-CSF application, which may lead to a diminished TCR-mediated activation and proliferation.21 Hence, down-regulation of costimulatory molecules on T cells might contribute to the reduced alloreactivity observed in PBSCT. In addition, the α-chain of the adhesion molecule LFA-1 (CD11a), which is important not only as a costimulatory molecule for the enhancement of TCR-dependent T-cell proliferation, but also for homing of naive T cells to peripheral lymphoid tissues,22 was decreased 3-fold in CD4+ T cells. Since lymphocyte homing into secondary lymphoid organs as predominant sites of lymphocyte sensitization to novel antigens is initiated by the interaction of homing receptors with their ligands, the down-regulation of LFA-1α in donor T cells may affect the homing process of the graft and abrogate donor T-cell activation to host antigens leading to an amelioration of alloreactive immune responses. In a murine GVHD model, Li et al23 recently showed that transient blocking of homing receptors reduces the alloreactivity of donor T cells and delays or even prevents acute GVHD. Furthermore, gene expression profiling identified the regulation of some transcription factors in donor T cells upon G-CSF treatment: Stat5 mRNA, which is critical for cell proliferation and known to be regulated by cytokines including G-CSF via the G-CSFR,24 was up-regulated in CD4+ T cells (> 10-fold). Most importantly, the master regulatory factor for induction and stabilization of a Th2 immune response, GATA-3,25-29 was up-regulated in CD4+ T cells upon G-CSF stimulation as determined by both mRNA and protein levels: cDNA array analysis showed a more than 7-fold increase of GATA-3 in CD4+ T cells. Quantitative real-time RT-PCR and Western blot analysis confirmed the induction of this transcription factor in CD3+ and CD4+ T cells analyzed from pools of stem cell donors in independent experiments. The molecular mechanisms by which cytokine signaling and antigen stimulation via the TCR might control Th1 and Th2 effector cell differentiation are still poorly understood. Recently, GATA-3 was identified as a T-cell–specific transcription factor selectively expressed in Th2 cells, but not Th1 cells.25 GATA-3 controls T-helper cell differentiation, directs to Th2 commitment,25-29 and stabilizes this phenotype26-28 ; however, much less is known about the regulation of GATA-3 expression itself. GATA-3 activation in CD4+ T cells seems to lead to an induction of chromatin remodeling of the intergenic regulatory region for the IL-4/IL-13/IL-5 gene cluster,27 directly activates the IL-5 promoter,25 and exhibits enhancer activity for IL-4 gene expression.29 To further characterize the Th1/Th2 phenotype, we investigated IFN-γ and IL-4 production in donor T cells before and after G-CSF stimulation ex vivo. Th1/Th2 immune responses were unbalanced upon G-CSF application with a remarkable down-regulation of IFN-γ and up-regulation of IL-4 production. Single-cell analysis at the protein level by ELISPOT demonstrated a more than 10-fold increase of spontaneous IL-4 secretion of T cells in healthy peripheral stem cell donors after G-CSF application. In addition to activating a Th2 program, GATA-3 directly represses the opposing Th1 immune response most likely by interfering with the IL-12 signal transduction pathway.28 Interestingly, G-CSF may additionally limit the IFN-γ signaling in T cells by suppressing the gene expression of ISGF3-γ subunit/p48 in CD4+ donor T cells. Together with Stat1 and Stat2, p48 forms the transcriptional complex,30 a key regulator for IFN-γ signaling as it is essential for the induction of interferon-stimulated response elements.31 In vitro studies stimulating highly purified T cells with G-CSF support the idea of direct interaction of G-CSF on G-CSFR–positive T cells inducing Th2 phenotype with an increase of IL-4 and a decrease of IFN-γ secretion. Nevertheless, the mode of action of G-CSF (direct and/or indirect) on donor T cells in vivo cannot be discriminated by this approach. One can envision that G-CSF might directly initiate the Th2-like immune response and that host alloantigens further promote the ongoing Th2 polarization via activated accessory cells (eg, dendritic cells), leading to reduced severity of acute GVHD.
In conclusion, our studies provide new mechanistic insights into the immune modulatory effects of G-CSF on the T-cell immune system. Our results suggest that G-CSF acts as a strong immune regulator in T cells and directly modulates T-cell immune responses via its receptor on T cells. The regulation of several immune modulatory genes, most importantly LFA1-α, ISGF3-γ, and GATA-3, was identified in T cells upon G-CSF stimulation in vivo. The regulation of these genes is likely to be responsible for the reduced reactivity and/or alloreactivity of G-CSF–stimulated T cells. Therefore, G-CSF is an interesting candidate for a specific immune modulation in transplantation, especially acute GVHD and other pathologic conditions associated with Th1/Th2 imbalance, such as bone marrow failure syndromes and Th1-mediated autoimmune diseases.
Prepublished online as Blood First Edition Paper, April 3, 2003; DOI 10.1182/blood-2002-04-1200.
Supported by the Deutsche Forschungsgemeinschaft, SFB 265, and the Dr-Wilhelm-Kempe Stiftung, Springe, Germany.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank James P. Di Santo from the Pasteur Institute in Paris not only for the discussion of the role of G-CSF in lymphocyte biology, but also for natural language processing.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal