Abstract
Insect peptidoglycan recognition protein-S (PGRP-S), a member of a family of innate immunity pattern recognition molecules conserved from insects to mammals, recognizes bacterial cell wall peptidoglycan and activates 2 antimicrobial defense systems, prophenoloxidase cascade and antimicrobial peptides through Toll receptor. We show that mouse PGRP-S is present in neutrophil tertiary granules and that PGRP-S–deficient (PGRP-S-/-) mice have increased susceptibility to intraperitoneal infection with gram-positive bacteria of low pathogenicity but not with more pathogenic gram-positive or gram-negative bacteria. PGRP-S-/- mice have normal inflammatory responses and production of tumor necrosis factor α (TNF-α) and interleukin 6 (IL-6). Neutrophils from PGRP-S-/- mice have normal phagocytic uptake of bacteria but are defective in intracellular killing and digestion of relatively nonpathogenic gram-positive bacteria. Therefore, mammalian PGRP-S functions in intracellular killing of bacteria. Thus, only bacterial recognition by PGRP-S, but not its effector function, is conserved from insects to mammals.
Introduction
The innate immune system recognizes micro-organisms through a series of pattern recognition receptors that are highly conserved in evolution and are specific for common motifs found in micro-organisms but not in higher eukaryotes.1-3 Peptidoglycan recognition proteins (PGRPs) are a novel family of innate immunity pattern recognition molecules that are highly conserved from insects to mammals.4-6
The first member of the PGRP family, now designated PGRP-S (for 19 kDa PGRP-short),5 was first described in 1996 as a protein present in hemolymph and cuticle of a silkworm (Bombyx mori).7 It binds gram-positive bacteria and peptidoglycan (PGN) and activates prophenoloxidase cascade.7 Prophenoloxidase cascade is an innate immunity mechanism in insects that generates antimicrobial products and surrounds micro-organisms with melanin.7,8 PGRP-S was then identified and cloned in a moth (Trichoplusia ni) as a protein that is up-regulated by a bacterial challenge,4 which led to the discovery of mouse and human PGRP-S orthologues.4
Sequencing of the Drosophila melanogaster genome has led to the discovery of a family of 12 highly diversified PGRP homologues.5 On the basis of the predicted structures of the gene products, Drosophila PGRPs were grouped into 2 classes: short PGRPs (PGRP-S), which are small extracellular proteins similar to the original PGRP, and long PGRPs (PGRP-L), which have long transcripts and are either intracellular or membrane-spanning proteins.5
Many of the Drosophila PGRPs are expressed in immune-competent organs, such as fat body, gut, and hemocytes, and their expression is up-regulated by injections of PGN,5 suggesting their role in insect immunity. Indeed, Drosophila PGRP-SA is required for the activation of Toll receptor pathway by gram-positive bacteria, which results in induction of antibacterial peptides and generation of effective immunity to gram-positive bacteria.9 Moreover, Drosophila PGRP-LC participates in the induction of antibacterial peptides in response to gram-negative and gram-positive bacteria10-12 and in phagocytosis of bacteria,12 and PGRP-LE participates both in the induction of antibacterial peptides and in the activation of prophenoloxidase cascade.13 Thus, insect PGRPs play an important role in innate immunity to bacteria.
Sequencing of the human genome has led to the discovery and cloning of 3 additional human PGRPs,6 designated PGRP-L, PGRP-Iα, and PGRP-Iβ, which, together with the previously discovered PGRP-S,4 define a new family of mammalian pattern recognition molecules. All mammalian and insect PGRPs have C-terminal PGRP domains that are highly conserved from insects to mammals (have up to 69% conserved identity and up to 83% conserved similarity between insects and mammals).6 The remaining N-terminal portions of PGRP molecules, however, have very little homology within the PGRP family and between insects and mammals.6
Mammalian PGRPs bind to bacteria and their PGN component.4,6,14 PGN is an essential cell wall component of virtually all bacteria; thus, it is an excellent target for recognition by the eukaryotic innate immune system. PGN induces strong antibacterial responses in insects8,15 and activates monocytes, macrophages, and B lymphocytes in mammals.16-21 Activation of mammalian monocytic cells by PGN is mediated by CD142,16-18,22-24 and TLR2,2,3,25-27 and leads to the production of numerous mediators of inflammation.16-20
Binding of PGRPs to PGN and bacteria suggests their direct role in recognition of bacteria. However, the consequences of this binding and, thus, the exact role of PGRPs in innate immunity to bacteria in mammals are not known. Because mammalian PGRP-S is highly expressed in bone marrow and polymorphonuclear leukocytes (PMNs) and has direct antibacterial effect in vitro,4,6,14,28 we hypothesized that PGRP-S plays a role in immunity to bacterial infections. The purpose of this study was to test this hypothesis by directly evaluating the role of PGRP-S in susceptibility to infection with gram-positive and gram-negative bacteria in PGRP-S–deficient mice.
Materials and methods
Generation, breeding, and characterization of PGRP-S–deficient(PGRP-S-/-) mice
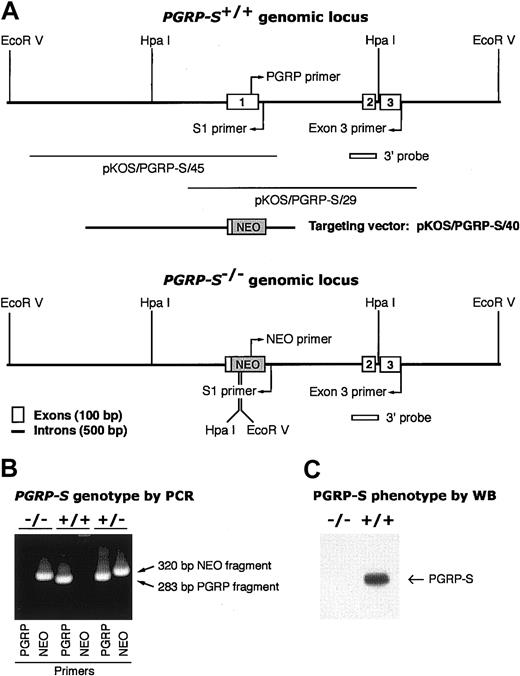
Screening of mouse strain 129/SvEvBrd genomic DNA library in the λ phage knock-out shuttle (λKOS)29 yielded 3 overlapping genomic clones (pKOS/PGRP-S/45, pKOS/PGRP-S/40, and pKOS/PGRP-S/29), which contained all 3 PGRP-S exons (Figure 1A). The contigs containing exon 1 and exons 2 and 3 were sequenced (GenBank accession nos. AY144360 and AY144361, respectively). The targeting vector was constructed from pKOS/PGRP-S/40 using a yeast-mediated recombination step that led to the deletion of nucleotides 8824 to 9147 of the PGRP-S exon 1 contig, and this region was replaced with an IRES LacZ/MC1-Neo (NEO) selection cassette29 (Figure 1A). Lex1 embryonic stem (ES) cells from 129/SvEvBrd mice were transfected with the targeting vector, and successful recombination of the targeting vector with the PGRP-S gene was identified in EcoRV or HpaI digests of 5 resistant clones by Southern blots with 3′ probe, which yielded, respectively, 28.5-kb and 9.2-kb fragments in the wild-type and 18.5-kb and 8.7-kb fragments in the mutant gene (data not shown). Positive ES clones were microinjected into C57BL/6 (albino) blastocysts and implanted into uteri of foster ICR mothers. Chimeric mice were screened for germ line transmission of the mutated allele in heterozygous PGRP-S+/- mice by Southern blot analysis of EcoRV or HpaI digests of tail DNA as described earlier and by polymerase chain reaction (PCR) analysis. Homozygous PGRP-S-/- mice were obtained by breeding heterozygous PGRP-S+/- mice and screening of their tail DNA by PCR analysis with PGRP14 or NEO (GCA GCG CAT CGC CTT CTA TC) sense primers and S1 antisense primer (GTC CTG CCT TGC AGT ATG C) (Figure 1B). PCR conditions were 35 cycles of 94°C for 30 seconds, 64°C for 30 seconds, and 72°C for 2 minutes. The identity of all PCR fragments was confirmed by sequencing.6
Construction of PGRP-S-/- mice. (A) Genomic organization of PGRP-S+/+ and PGRP-S-/- loci with 3 PGRP-S exons, genomic clones, targeting vector, and replacement of exon 1 with NEO cassette in PGRP-S-/- mice. Relevant restriction sites, primers, and screening probes are also shown. (B) Genotypes of PGRP-S-/-, PGRP-S+/+, and PGRP-S+/- mice determined by typing of genomic DNA by PCR using either PGRP-specific or NEO cassette–specific sense primer and intron-specific (S1) antisense primer. DNA from homozygous PGRP-S-/- mice yielded only 320-bp NEO PCR fragment and no PGRP fragment, demonstrating the replacement of PGRP-S exon 1 with NEO cassette in both chromosomes. DNA from wild-type PGRP-S+/+ mice yielded only 283-bp PGRP PCR fragment and no NEO fragment, whereas DNA from heterozygous PGRP-S+/- mice yielded both PGRP and NEO PCR fragments. (C) Phenotypes of PGRP-S-/- and PGRP-S+/+ mice determined by Western blot: lysates of 2 × 106 bone marrow cells/lane were separated by SDS-PAGE, and PGRP-S protein was detected by anti–PGRP-S antibodies on Western blots in lysates from PGRP-S+/+ but not from PGRP-S-/- mice.
Construction of PGRP-S-/- mice. (A) Genomic organization of PGRP-S+/+ and PGRP-S-/- loci with 3 PGRP-S exons, genomic clones, targeting vector, and replacement of exon 1 with NEO cassette in PGRP-S-/- mice. Relevant restriction sites, primers, and screening probes are also shown. (B) Genotypes of PGRP-S-/-, PGRP-S+/+, and PGRP-S+/- mice determined by typing of genomic DNA by PCR using either PGRP-specific or NEO cassette–specific sense primer and intron-specific (S1) antisense primer. DNA from homozygous PGRP-S-/- mice yielded only 320-bp NEO PCR fragment and no PGRP fragment, demonstrating the replacement of PGRP-S exon 1 with NEO cassette in both chromosomes. DNA from wild-type PGRP-S+/+ mice yielded only 283-bp PGRP PCR fragment and no NEO fragment, whereas DNA from heterozygous PGRP-S+/- mice yielded both PGRP and NEO PCR fragments. (C) Phenotypes of PGRP-S-/- and PGRP-S+/+ mice determined by Western blot: lysates of 2 × 106 bone marrow cells/lane were separated by SDS-PAGE, and PGRP-S protein was detected by anti–PGRP-S antibodies on Western blots in lysates from PGRP-S+/+ but not from PGRP-S-/- mice.
The expression of PGRP-S mRNA in bone marrow of wild-type and homozygous PGRP-S-/- mice was determined by reverse transcriptase (RT)–PCR with sense PGRP (exon 1) and antisense exon 3 primers and mouse glyceraldehyde phosphate dehydrogenase (GAPDH)–specific primers (as loading control).14 The expression of PGRP-S protein in bone marrow of wild-type and homozygous PGRP-S-/- mice was determined by Western blot with anti–PGRP-S antibodies.14
PGRP-S+/+ and PGRP-S-/- mice were bred and kept in sterile isolator cages with sterile food and sterile acidified drinking water, and they were handled sterilely under a laminar flow hood. A parallel breeding colony was maintained in a conventional environment. Because there were no differences in the development and susceptibility to infections between mice kept under sterile and conventional conditions, most experiments were performed on mice kept under conventional conditions. In all experiments PGRP-S+/+ and PGRP-S-/- mice were 6 to 10 weeks old and age- and sex-matched littermates derived from the same parents.
Materials
Bacillus subtilis ATCC 6633, Micrococcus luteus ATCC 4698, Staphylococcus aureus Rb (a clinical isolate) or strain 845, Escherichia coli K12 or K- mutant (a PMN-sensitive strain,30 obtained from Dr Alan Cross, University of Maryland, Baltimore), and Proteus vulgaris ATCC 13315 were grown in Luria-Bertani (LB) broth at 37°C with shaking or (for colony counts) on LB agar. Similar results were obtained with S aureus Rb and 845 and with E coli K12 and K- mutant in infection and phagocytosis experiments, and S aureus Rb and E coli K12 were used for most reported experiments. Lactobacillus acidophilus ATCC 4356 was grown in Lactobacilli MRS broth (Difco; Becton Dickinson, Sparks, MD) or agar as described above. Recombinant mouse PGRP-S was cloned, expressed in Sf-9 cells, purified, and analyzed as described.4,14 All reagents were from Sigma (St Louis, MO), unless otherwise indicated.
Survival following infection
Groups of PGRP-S+/+ and PGRP-S-/- mice were injected by intraperitoneal or intravenous route with the numbers and species of bacteria indicated in “Results,” and survival was monitored every 6 hours for the first 48 hours and every 12 hours for 10 days. The significance of differences in the survival of PGRP-S+/+ and PGRP-S-/- mice were calculated using the Chi-square test.
Bacterial counts in organs following infection
Groups of PGRP-S+/+ and PGRP-S-/- mice were injected intraperitoneally or intravenously with the numbers and species of bacteria indicated in “Results.” At times indicated in “Results,” mice were anesthetized and exsanguinated, their peritoneal cavities were washed with 1 mL saline, and their organs (liver, spleen, kidneys) were homogenized in 2 mL sterile water using Polytron (Brinkmann Instruments, Westbury, NY). Bacterial loads in tissue fluids and organs were determined by dilutions and colony counts on agar plates. Numbers of leukocytes in peritoneal fluid were also counted, and percentages of PMNs and mononuclear cells were determined on Wright-stained smears. The results are expressed as means of log10 numbers of bacteria per organ or per milliliter of blood. The significance of differences was calculated using the Mann-Whitney U test.
In vitro killing of bacteria by PMNs and macrophages
Peritoneal PMNs were elicited by intraperitoneal injection of 1 mL LB broth with 0.75 mg agar/mL, and the cells were harvested 16 to 20 hours later by washing the peritoneal cavities with Hanks balanced saline solution (BSS) without Ca2+ and Mg2+ with 1 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid). The total numbers of elicited peritoneal cells per mouse were similar for PGRP-S+/+ and PGRP-S-/- mice: 8.68 ± 1.42 × 106 and 7.34 ± 0.83 × 10,6 respectively (means ± SE, n = 30). These cells also contained similar percentages of PMNs (39%-48%), lymphocytes (33%-49%), and macrophages (8%-9%). Elicited peritoneal cells were pooled from 5 PGRP-S+/+ or PGRP-S-/- mice, and PMNs were obtained by centrifugation through 2 Histopaque layers.14 Unelicited bone marrow PMNs were obtained by flushing cells from femurs of 3 PGRP-S+/+ or PGRP-S-/- mice and centrifugation through Histopaque as described earlier. PMNs were 85% to 90% pure as determined by morphology on Wright-stained smears. Unelicited peritoneal macrophages were obtained by washing peritoneal cavities as described earlier, followed by 1-hour adhesion to tissue culture plates, washing, and removal of adherent macrophages with a plastic scraper. PMNs or macrophages were suspended to 5 × 106/mL in Hanks BSS with Ca2+ and Mg2+ with 1 mM HEPES with 12.5% autologous plasma (unless otherwise indicated), and B subtilis (3.3 × 106/mL), M luteus (15 × 106/mL), L acidophilus (5 × 106/mL), S aureus (1 × 106/mL), E coli (1 × 106/mL), or P vulgaris (1 × 106/mL) (all final concentrations). The phagocytes/bacteria mixtures were rotated at 10 rpm at 37°C. Samples were removed at 1 hour and 1.5 hours for PMNs or 2 hours for macrophages for B subtilis, M luteus, and L acidophilus and at 2 hours and 3 hours for PMNs or 4 hours for macrophages for S aureus, E coli, or P vulgaris; diluted with 0.25% NaCl; and plated for colony counts. Under these conditions, the percentages of killed bacteria were approximately 95% for B subtilis and M luteus, 90% for L acidophilus, 70% for S aureus, and 75% for E coli and P vulgaris, which gave the most sensitive determination of differences between killing capacity by PMNs for each bacterial species.31 In some experiments, PMNs were allowed to phagocytize bacteria for 10 minutes and were then centrifuged, washed, returned to the medium, and rotated at 37°C for 1 to 3 hours. The results are expressed as the numbers of viable bacteria recovered. To compare the results from different experiments for different bacteria, the numbers of viable bacteria in assays with PMNs (or macrophages) from wild-type mice (+/+) were taken as 100%, and the results in assays with PMNs (or macrophages) from PGRP-S-/- mice (-/-) were calculated as the percentage of wild-type [-/- = (100% × number of bacteria recovered in -/-) /(number of bacteria recovered in +/+)], and reported as means from the number of experiments indicated in “Results.” The significance of differences was calculated using the one-sample Student t test.
In the experiments on the effect of PGRP-S on in vitro killing of bacteria by PMNs, bacteria (B subtilis or M luteus) were first incubated with 50 μg recombinant PGRP-S/mL (or buffer control) for 10 minutes at 37°C, and then PMNs and serum were added to yield 10 μg PGRP-S/mL and the indicated concentrations of PMNs and bacteria. The killing assay was then performed as described earlier. The 10-minute preincubation (at 50 μg/mL) or 60- to 90-minute incubation (at 10 μg/mL) of PGRP-S with bacteria without PMNs did not change the numbers of viable bacteria.
In vitro ingestion of bacteria by PMNs
Elicited peritoneal cells (5 × 106/mL) were rotated with B subtilis or M luteus (5 × 107/mL) as described earlier. After 10 or 20 minutes, peritoneal cells were washed 4 times at 4°C with Dulbecco phosphate buffered saline (PBS) without Ca2+ and Mg2+ with 0.2% bovine serum albumin at 200g to remove extracellular bacteria,32 deposited onto microscope slides in a cytocentrifuge at 400g (Shandon, Sewickley, PA), fixed with ethanol, and stained with Diff-Quik (Dade Behring, Newark, DE).32 Numbers of PMNs and macrophages that ingested and did not ingest bacteria and the numbers of ingested bacteria per cell were counted under the microscope, and the phagocytic index (the measure of bacterial uptake) for PMNs and macrophages was calculated14,32 : phagocytic index = (percentage of cells containing at least one bacterium) × (mean number of bacteria per positive cell). The results are expressed as mean phagocytic indexes. The significance of differences was calculated using the Mann-Whitney U test. Representative cells were also photographed under immersion oil, magnification of 10 × 100.
Localization of PGRP-S in PMN granules
Bone marrow PMNs (pooled from at least 5 PGRP-S-/- or wild-type mice) were disrupted by nitrogen cavitation,33 nuclei and intact cells were removed by centrifugation, and postnuclear supernatant was fractionated on a 3-layer Percoll gradient.33 The supernatant (cytosol), 4 bands corresponding to the primary, secondary, tertiary, and secretory granules, and the sediment (membranes) were collected,33 lyophilized, dissolved in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and separated by SDS-PAGE.14 PGRP-S was detected on Western blots.14 The granules were identified by their location on the gradient33 and by the presence of the following granule markers33,34 detected on Western blots: myeloperoxidase (MPO) for primary (azurophil) granules (with rabbit anti-MPO antibodies from Calbiochem, La Jolla, CA), lactoferrin for secondary (specific) granules (with rabbit antilactoferrin antibodies from Sigma), and matrix metalloproteinase 9 (MMP-9 or gelatinase B) for tertiary (gelatinase) granules (with goat anti–MMP-9 antibodies from Santa Cruz Biotechnology, Santa Cruz, CA).
Oxidative burst
Bacterially induced oxidative burst in mouse peripheral blood PMNs was measured by nitro blue tetrazolium (NBT) reduction method with 2.5 × 107 bacteria/mL, exactly as described before for human PMNs.14 The results are expressed as the percentage of NBT-positive PMNs. The significance of differences was calculated using the 2-sample Student t test.
Antibacterial activity of PGRP-S
To measure bacteriostatic and bactericidal activity of PGRP-S, logarithmic phase bacteria (B subtilis, M luteus, L acidophilus, S aureus, E coli, or P vulgaris) were washed and incubated at 1.5 to 15 × 105 bacteria/mL for 2 hours at 37°C with rocking in buffer alone or with 50 μg/mL purified mouse recombinant PGRP-S in 10 mM PIPES (piperazine-N-N′ bis (20-ethanol sulfonic acid)) with 5 mM glucose,28 at pH 7.8, 7.0, and 6.2, without or with 140 mM NaCl or 200 mM KCl. The numbers of viable bacteria were determined by plating on agar plates. The results were statistically analyzed using 2-sample Student t test and Mann-Whitney U test.
Detection of cytokines
Mice were bled before or 1.5, 3, or 6 hours after intraperitoneal injection of 2 × 107 to 2 × 109 live B subtilis. Concentrations of tumor necrosis factor α (TNF-α) and interleukin 6 (IL-6) in their serum were determined using the L929 cytotoxicity assay19 and ELISA (enzyme-linked immunosorbent assay) R&D System20 (R&D Systems, Minneapolis, MN), respectively. The data were calculated using 4-parameter curve fitting with DeltaSoft3 ELISA software (Biometalics, Princeton, NJ) and were analyzed statistically using the Mann-Whitney U test.
Statistical analysis
GB-Stat PPC6.5.6 (Dynamic Microsystems, Silver Spring, MD) was used, and differences were considered significant at P ≤ .05.
Results
Generation and characterization of PGRP-S-/- mice
We generated PGRP-S-/- mice by targeted disruption of the PGRP-S gene, followed by germ line transmission of the mutant gene into chimeric, heterozygous, and homozygous PGRP-S-/- mice (Figure 1). The PGRP-S exon 1 DNA was detectable only in wild-type and heterozygous but not in PGRP-S-/- mice by PCR (Figure 1B). To study the expression of PGRP-S mRNA and protein, bone marrow was selected, because it expresses the highest level of PGRP-S mRNA in mice, humans, and cows.4,6,14,28 PGRP-S mRNA was detectable only in the bone marrow of wild-type but not of PGRP-S-/- mice by RT-PCR, with exon 1 sense and exon 3 antisense primers14 (not shown). PGRP-S protein was also detectable only in the bone marrow of wild-type but not of PGRP-S-/- mice by Western blot (Figure 1C). These results confirm the replacement of PGRP-S exon 1 with NEO cassette and the lack of expression of PGRP-S mRNA and protein in PGRP-S-/- mice.
PGRP-S-/- mice were viable and fertile, bred normally, and yielded the expected male-to-female ratios and similar litter size as the wild-type and heterozygous mice. They had the same weight as the wild-type and heterozygous mice and developed normally with no obvious defects. Their major internal organs had normal macroscopic appearance, and histologic appearance on hematoxylin/eosin-stained sections of spleen, lymph nodes, liver, bone marrow, and thymus was also normal. PGRP-S-/- mice had normal numbers of neutrophils, eosinophils, basophils, monocytes, and lymphocytes in peripheral blood and normal numbers of PMNs and mononuclear cells in the bone marrow (not shown).
Increased susceptibility of PGRP-S-/- mice to infection with B subtilis and M luteus
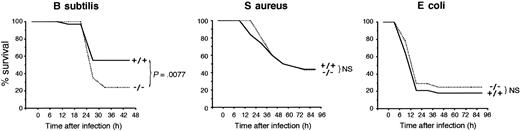
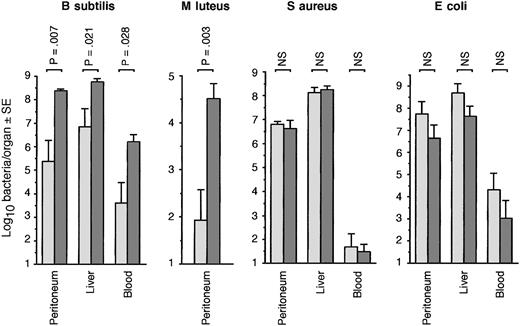
The mortality of PGRP-S-/- mice was twice as high as that of wild-type PGRP-S+/+ mice following intraperitoneal infection with B subtilis, and this difference was statistically significant (Figure 2). PGRP-S-/- mice also had significantly higher (100-1000 times higher) numbers of B subtilis in peritoneal cavity, blood, liver (Figure 3), spleen, and kidneys (not shown) than wild-type mice 24 to 36 hours after intraperitoneal infection. Significantly higher loads of B subtilis in internal organs were also found in PGRP-S-/- mice, when 2 times or 10 times less bacteria were injected (not shown). PGRP-S-/- mice also had higher susceptibility than wild-type mice to M luteus, manifested by significantly higher numbers of M luteus in peritoneal cavity 24 to 36 hours after intraperitoneal infection (Figure 3). However, unlike B subtilis, M luteus infection did not significantly spread to internal organs and did not cause mortality in either group of mice (not shown).
PGRP-S-/- mice have lower survival following intraperitoneal infection with B subtilis but not S aureus and E coli. Mice were injected intraperitoneally with 4 × 109B subtilis (n = 58/group; left panel), 1 × 109S aureus (n = 32/group; middle panel), or 3 × 108E coli (n = 28/group; right panel), and their survival was monitored for 10 days. The significance of differences in the survival of PGRP-S+/+ and PGRP-S-/- mice was calculated at 48 hours (B subtilis) or 96 hours (S aureus and E coli) after challenge using Chi-square test (no additional deaths occurred after that time). The results from 2 to 4 separate experiments were combined.
PGRP-S-/- mice have lower survival following intraperitoneal infection with B subtilis but not S aureus and E coli. Mice were injected intraperitoneally with 4 × 109B subtilis (n = 58/group; left panel), 1 × 109S aureus (n = 32/group; middle panel), or 3 × 108E coli (n = 28/group; right panel), and their survival was monitored for 10 days. The significance of differences in the survival of PGRP-S+/+ and PGRP-S-/- mice was calculated at 48 hours (B subtilis) or 96 hours (S aureus and E coli) after challenge using Chi-square test (no additional deaths occurred after that time). The results from 2 to 4 separate experiments were combined.
PGRP-S-/- mice have higher bacterial load following intraperitoneal infection withB subtilis and M luteus but not S aureus and E coli.PGRP-S+/+ (light gray bars) or PGRP-S-/- (dark gray bars) mice were injected intraperitoneally with 2 × 109B subtilis (n = 9/group), 15 × 109M luteus,4 × 108S aureus, or 1.5 × 108E coli (n = 8/group), and bacterial loads in tissue fluids and organs at 24 to 36 hours after challenge were determined. The significance of differences was calculated using the Mann-Whitney U test.
PGRP-S-/- mice have higher bacterial load following intraperitoneal infection withB subtilis and M luteus but not S aureus and E coli.PGRP-S+/+ (light gray bars) or PGRP-S-/- (dark gray bars) mice were injected intraperitoneally with 2 × 109B subtilis (n = 9/group), 15 × 109M luteus,4 × 108S aureus, or 1.5 × 108E coli (n = 8/group), and bacterial loads in tissue fluids and organs at 24 to 36 hours after challenge were determined. The significance of differences was calculated using the Mann-Whitney U test.
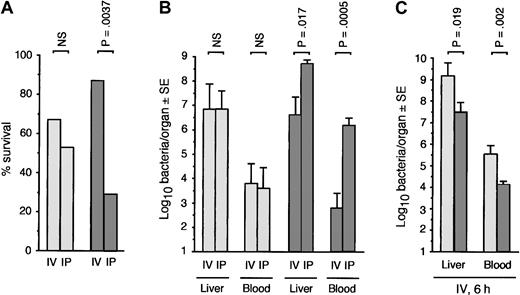
Increased susceptibility to infection of PGRP-S-/- mice with B subtilis and M luteus was evident only after intraperitoneal infection (Figures 2, 3) but not after intravenous infection (Figure 4). In fact, PGRP-S-/- mice were significantly more resistant to intravenous infection with B subtilis than wild-type mice. The survival of intraperitoneally infected PGRP-S-/- mice was significantly lower than of intravenously infected PGRP-S-/- mice, whereas there was no difference between the survival of intraperitoneally and intravenously infected wild-type mice (Figure 4). Also, in PGRP-S-/- mice, but not in wild-type mice, significantly higher numbers of B subtilis were recovered after intraperitoneal than intravenous infection (Figure 4). Moreover, the numbers of B subtilis in liver and blood following intravenous infection were significantly lower in PGRP-S-/- than in wild-type mice (Figure 4).
PGRP-S-/- mice are more resistant to intravenous than intraperitoneal infection with B subtilisPGRP-S+/+ (light gray bars) or PGRP-S-/- (dark gray bars) mice were injected intraperitoneally or intravenously with B subtilis, 4 × 109 (A, n = 16/group), or 2 × 109 (B-C, n = 8/group). Survival at 48 hours (A) was compared using the Chi-square test (no additional deaths occurred after that time). Bacterial loads in tissue fluids and organs at 30 hours (B) or 6 hours (C) after challenge were determined, and the significance of differences was calculated using the Mann-Whitney U test.
PGRP-S-/- mice are more resistant to intravenous than intraperitoneal infection with B subtilisPGRP-S+/+ (light gray bars) or PGRP-S-/- (dark gray bars) mice were injected intraperitoneally or intravenously with B subtilis, 4 × 109 (A, n = 16/group), or 2 × 109 (B-C, n = 8/group). Survival at 48 hours (A) was compared using the Chi-square test (no additional deaths occurred after that time). Bacterial loads in tissue fluids and organs at 30 hours (B) or 6 hours (C) after challenge were determined, and the significance of differences was calculated using the Mann-Whitney U test.
However, PGRP-S-/- and PGRP-S+/+ mice had similar susceptibility to infection with S aureus and E coli, as evidenced by similar survival rates following intraperitoneal (Figure 2) or intravenous (not shown) infection, and similar numbers of S aureus and E coli were recovered from internal organs following intraperitoneal (Figure 3) and intravenous (not shown) infection (note that lower numbers of S aureus and E coli than of B subtilis and M luteus were used in Figures 2 and 3, because of higher virulence of S aureus and E coli). Similar results were also obtained with 10 and 100 times lower doses of S aureus and E coli (not shown).
These results indicate that PGRP-S-/- mice have higher susceptibility than wild-type mice to intraperitoneal infection with 2 less pathogenic gram-positive bacteria (B subtilis and M luteus) but not with more pathogenic gram-positive (S aureus) and gram-negative (E coli) bacteria.
Deficient killing of B subtilis, M luteus, and L acidophilus by PMNs from PGRP-S-/- mice
Phagocytic cells (PMNs and macrophages) are the first line of defense against bacterial infections and are responsible for ingestion, killing, and digestion of bacteria.35 Because PGRP-S is highly expressed in bone marrow and PMNs, but is not expressed in lymphocytes, monocytes, and macrophages,4,6,14,28 we then focused on the role of PGRP-S in PMN-mediated defenses against infections.
Increased susceptibility of PGRP-S-/- mice to intraperitoneal infection with B subtilis and M luteus was not due to a deficiency in the total numbers of PMNs, because, as already indicated, there was no significant difference between PGRP-S-/- and wild-type mice in numbers of PMNs in their peripheral blood and bone marrow (not shown). It was also not due to a deficiency in the recruitment and retention of PMNs at the site of infection (peritoneal cavity): the numbers of inflammatory cells in the peritoneal cavity of wild-type and PGRP-S-/- mice, respectively, 6 hours and 30 hours after intraperitoneal infection with B subtilis and M luteus were similar (means ± SE of 8-11 mice/group): 4.9 ± 0.6 × 106 versus 5.1 ± 0.7 × 106(B subtilis, 6 hours), 4.8 ± 0.5 × 106 versus 4.9 ± 0.7 × 106 (M luteus, 6 hours), 9.3 ± 1.5 × 106 versus 9.7 ± 1.9 × 106 (B subtilis, 30 hours), and 8.3 ± 1.1 × 106 versus 9.1 ± 1.4 × 106 (M luteus, 30 hours). The percentages of PMNs and mononuclear cells in these inflammatory cells were also similar in wild-type and PGRP-S-/- mice (not shown). The differences between the wild-type and PGRP-S-/- mice in the numbers of B subtilis and M luteus in the peritoneal cavity and internal organs became apparent only after 6 hours, ie, after PMNs infiltrated the peritoneal cavity (data not shown). Therefore, our next experiments were designed to test whether PMNs from PGRP-S-/- mice were deficient in phagocytosis and killing of bacteria.
Both elicited peritoneal PMNs and unelicited bone marrow PMNs from PGRP-S-/- mice were deficient in killing of B subtilis, M luteus, and L acidophilus in vitro, as evidenced by significantly higher numbers of these bacteria recovered after 60 and 90 minutes of phagocytosis by PMNs from PGRP-S-/- than from wild-type mice (Figure 5A-B). Similar deficiency in killing of B subtilis, M luteus, and L acidophilus was obtained when PMNs were allowed to phagocytize bacteria for 10 minutes and were then separated from the extracellular bacteria and allowed to kill the ingested bacteria (data not shown). There was no difference in the numbers of extracellular bacteria, thus suggesting a deficiency in the intracellular killing of bacteria.
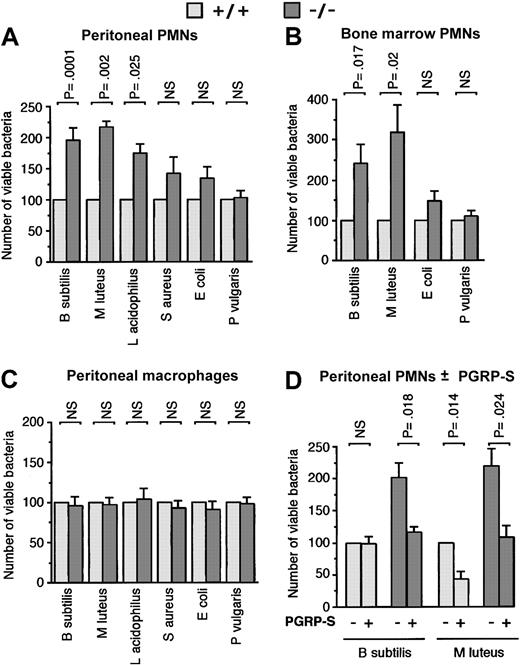
PMNs from PGRP-S-/- mice are deficient in killing of B subtilis, M luteus, and L acidophilus but not S aureus, E coli, and P vulgaris, and this defect is reversed by exogenous recombinant PGRP-S. Peritoneal (A) or bone marrow (B) PMNs or peritoneal macrophages (C) from PGRP-S+/+ (light gray bars) or PGRP-S-/- (dark gray bars) mice were incubated in vitro with the indicated bacteria, and the numbers of viable bacteria were determined as described in “Materials and methods.” (D) Bacteria were first incubated with 50 μg recombinant PGRP-S/mL (or buffer control) for 10 minutes, and then peritoneal PMNs were added to yield 10 μg PGRP-S/mL. The killing assay was then performed as in panel A. The results are means ± SE of the following number of experiments: (A) 21 (B subtilis), 4 (M luteus and L acidophilus), 10 (S aureus), 11 (E coli), and 8 (P vulgaris); (B) 12 (B subtilis and M luteus), 6 (E coli), and 12 (P vulgaris); (C) 6; and (D) 4. The significance of differences was calculated using the Student t test.
PMNs from PGRP-S-/- mice are deficient in killing of B subtilis, M luteus, and L acidophilus but not S aureus, E coli, and P vulgaris, and this defect is reversed by exogenous recombinant PGRP-S. Peritoneal (A) or bone marrow (B) PMNs or peritoneal macrophages (C) from PGRP-S+/+ (light gray bars) or PGRP-S-/- (dark gray bars) mice were incubated in vitro with the indicated bacteria, and the numbers of viable bacteria were determined as described in “Materials and methods.” (D) Bacteria were first incubated with 50 μg recombinant PGRP-S/mL (or buffer control) for 10 minutes, and then peritoneal PMNs were added to yield 10 μg PGRP-S/mL. The killing assay was then performed as in panel A. The results are means ± SE of the following number of experiments: (A) 21 (B subtilis), 4 (M luteus and L acidophilus), 10 (S aureus), 11 (E coli), and 8 (P vulgaris); (B) 12 (B subtilis and M luteus), 6 (E coli), and 12 (P vulgaris); (C) 6; and (D) 4. The significance of differences was calculated using the Student t test.
There was no significant difference in killing of S aureus, E coli, and P vulgaris by PMNs from PGRP-S-/- and wild-type mice in vitro (Figure 5A-B). There was also no significant difference between PGRP-S-/- and wild-type mice in killing of any of the bacteria by macrophages (Figure 5C), which is expected, because macrophages do not express PGRP-S14 and, therefore, should be similar in PGRP-S-/- and wild-type mice.
The deficient killing of B subtilis and M luteus by PMNs from PGRP-S-/- mice was reconstituted to the level of wild-type killing by addition of exogenous recombinant mouse PGRP-S (Figure 5D). Under these conditions, PGRP-S had no effect on the viability of extracellular bacteria throughout the duration of the assay, and it also had no effect on the numbers of bacteria ingested by PMNs (measured as described in Figure 6, not shown). However, the deficient killing of B subtilis by PMNs from PGRP-S-/- mice was not reversed by plasma from wild-type mice (data not shown), which indicates that plasma from wild-type mice does not contain sufficient PGRP-S to restore normal killing by PMNs. These results again suggest that the site of action of PGRP-S is within PMNs.
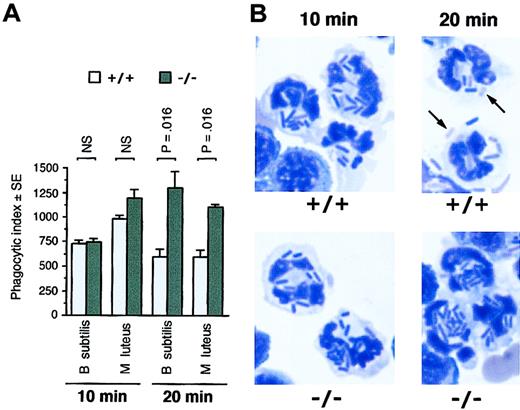
PMNs from PGRP-S-/- mice have normal uptake of B subtilis and M luteus but are defective in elimination of ingested bacteria. (A) Peritoneal exudate cells from PGRP-S+/+ or PGRP-S-/- mice were incubated in vitro with B subtilis or M luteus, and the numbers of bacteria ingested by PMNs (phagocytic index) were determined at 10 and 20 minutes. The results are means of 4 experiments, and the significance of differences was calculated using the Mann-Whitney U test. (B) Representative photographs of PMNs from PGRP-S+/+ or PGRP-S-/- mice after 10 and 20 minutes of phagocytosis of B subtilis (magnification, 10 × 100; Diff-Quik stain). The arrows point to poorly stained bacterial ghosts (partially digested bacteria), often seen in PMNs from PGRP-S+/+, but not PGRP-S-/-, mice after 20 minutes of phagocytosis.
PMNs from PGRP-S-/- mice have normal uptake of B subtilis and M luteus but are defective in elimination of ingested bacteria. (A) Peritoneal exudate cells from PGRP-S+/+ or PGRP-S-/- mice were incubated in vitro with B subtilis or M luteus, and the numbers of bacteria ingested by PMNs (phagocytic index) were determined at 10 and 20 minutes. The results are means of 4 experiments, and the significance of differences was calculated using the Mann-Whitney U test. (B) Representative photographs of PMNs from PGRP-S+/+ or PGRP-S-/- mice after 10 and 20 minutes of phagocytosis of B subtilis (magnification, 10 × 100; Diff-Quik stain). The arrows point to poorly stained bacterial ghosts (partially digested bacteria), often seen in PMNs from PGRP-S+/+, but not PGRP-S-/-, mice after 20 minutes of phagocytosis.
PMNs from PGRP-S-/- mice were not deficient in the ability to ingest B subtilis and M luteus, as evidenced by similar numbers of bacteria ingested by PMNs from wild-type and PGRP-S-/- mice after 10 minutes of phagocytosis (Figure 6A). After 20 minutes of phagocytosis, the numbers of ingested intracellular bacteria in PMNs from wild-type mice began to decrease because of killing and digestion (Figure 6A). The wild-type PMNs, after 20 minutes of phagocytosis, often had poorly staining bacterial ghosts, indicative of bacterial digestion (Figure 6B). By contrast, after 20 minutes of phagocytosis, the numbers of ingested intracellular bacteria in PMNs from PGRP-S-/- mice continued to increase, and at that time they were significantly higher than the numbers of bacteria in PMNs from wild-type mice (Figure 6A). The PGRP-S-/- PMNs had few poorly staining bacterial ghosts (Figure 6B, similar results were obtained for M luteus, not shown). These results again point to the intracellular site of action of PGRP-S.
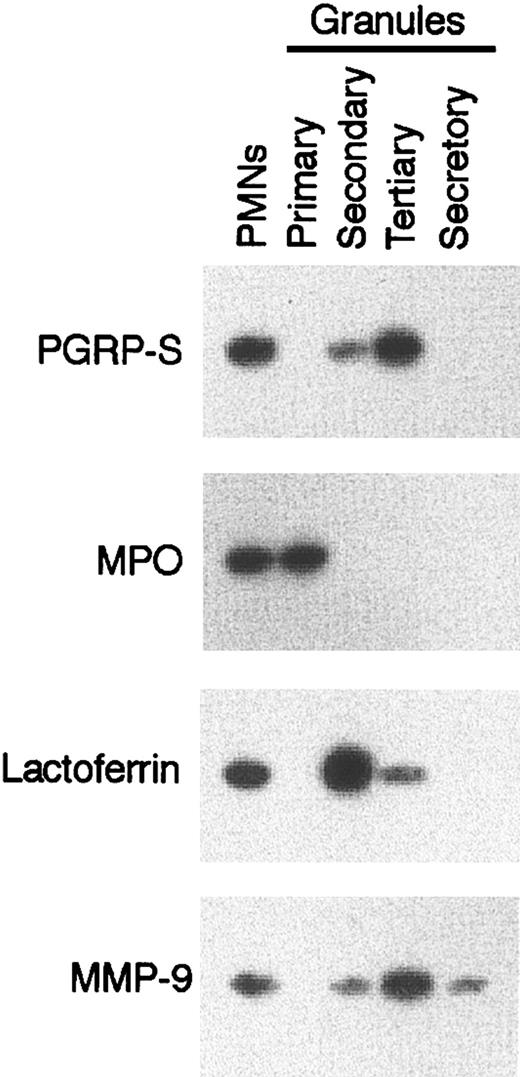
Our next experiments were performed to verify whether mouse PGRP-S, similar to bovine PGRP-S orthologue,28 is indeed stored intracellularly in granules of PMNs. Fractionation of mouse PMNs into 4 types of granules revealed that PGRP-S was present primarily in the tertiary (gelatinase) granules and in small amounts in secondary (specific) granules but was absent from the primary (azurophil) granules, secretory vesicles (Figure 7), membranes, and cytosol (not shown). PGRP-S was not detected in normal mouse serum.14 Therefore, altogether, these results indicate that PGRP-S is one of the intracellular granule proteins of PMNs and plays a role in intracellular killing of some relatively nonpathogenic gram-positive bacteria. PGRP-S has little effect on extracellular bacteria or on the rate of ingestion of bacteria by PMNs.
PGRP-S is present in PMNs in tertiary (gelatinase) granules. Lysates of bone marrow PMNs or their isolated granules were separated by SDS-PAGE, and PGRP-S protein was detected on Western blots. The following granule markers were also detected: myeloperoxidase (MPO) for primary (azurophil) granules, lactoferrin for secondary (specific) granules, and matrix metalloproteinase 9 (MMP-9 or gelatinase B) for tertiary (gelatinase) granules. The results are from 1 of 4 similar experiments.
PGRP-S is present in PMNs in tertiary (gelatinase) granules. Lysates of bone marrow PMNs or their isolated granules were separated by SDS-PAGE, and PGRP-S protein was detected on Western blots. The following granule markers were also detected: myeloperoxidase (MPO) for primary (azurophil) granules, lactoferrin for secondary (specific) granules, and matrix metalloproteinase 9 (MMP-9 or gelatinase B) for tertiary (gelatinase) granules. The results are from 1 of 4 similar experiments.
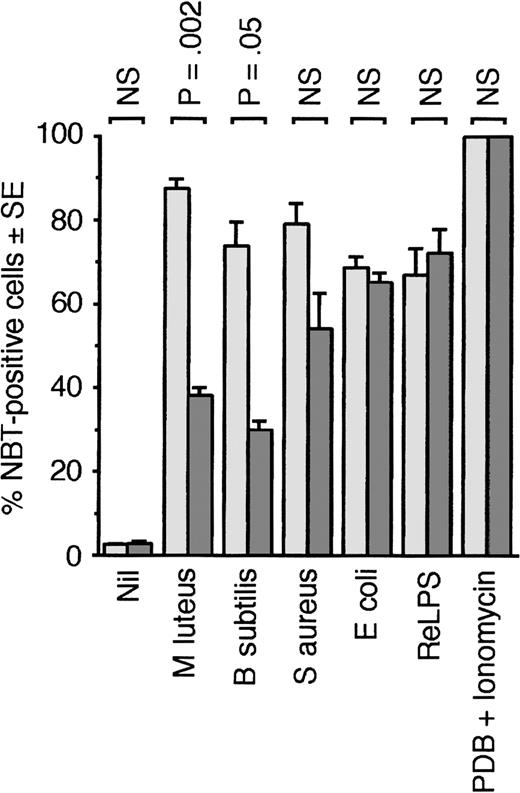
PGRP-S could either enhance generation or activity of other intracellular antibacterial mechanisms in PMNs or it could have a direct effect on bacterial survival inside phagocytic cells, or both. To test the first hypothesis, we compared generation of oxidative burst (which is a major mechanism of bacterial killing) in response to bacteria in PMNs from wild-type and PGRP-S-/- mice. PMNs from PGRP-S-/- mice were significantly less effective than PMNs from wild-type mice in generating oxidative burst in response to M luteus and B subtilis but not to S aureus, E coli, re-type lipopolysaccharide (ReLPS), and protein kinase C activator (PDB) plus Ca2+-ionophore (ionomycin) (Figure 8).
PMNs fromPGRP-S-/- mice are defective in generating oxidative burst in response to B subtilis andM luteus. Oxidative burst in blood PMNs from PGRP-S+/+ (light gray bars) or PGRP-S-/- (dark gray bars) mice in response to the indicated stimuli was measured by the NBT test. The results are means of 6 experiments, and the significance of differences was calculated using the Student t test.
PMNs fromPGRP-S-/- mice are defective in generating oxidative burst in response to B subtilis andM luteus. Oxidative burst in blood PMNs from PGRP-S+/+ (light gray bars) or PGRP-S-/- (dark gray bars) mice in response to the indicated stimuli was measured by the NBT test. The results are means of 6 experiments, and the significance of differences was calculated using the Student t test.
To test the second hypothesis, we measured the direct effect of PGRP-S on bacteria in vitro. Our previous results indicated that mouse recombinant PGRP-S inhibited growth of gram-positive but not gram-negative bacteria in vitro in LB broth, but it had no bactericidal effect.14 However, recent data indicated that bovine orthologue of PGRP-S was bactericidal in vitro for a number of gram-positive and gram-negative bacteria in the absence of salts.28 Therefore, we re-tested antibacterial activity of mouse PGRP-S at 50 μg/mL in 10 mM PIPES with 5 mM glucose,28 at pH 7.8, 7.0, and 6.2, without or with 140 mM NaCl or 200 mM KCl. We selected these conditions because NaCl inhibits antimicrobial activity of several neutrophil proteins (eg, defensins), and because the environment inside phagocytic vacuoles transiently changes from acidic to alkaline and from low K+ to high K+ concentration during different stages of phagocytosis and killing.35-39 However, under all of these conditions, mouse PGRP-S again showed only bacteriostatic activity for gram-positive bacteria and no bactericidal activity for B subtilis, M luteus, L acidophilus, S aureus, E coli, and P vulgaris (not shown).
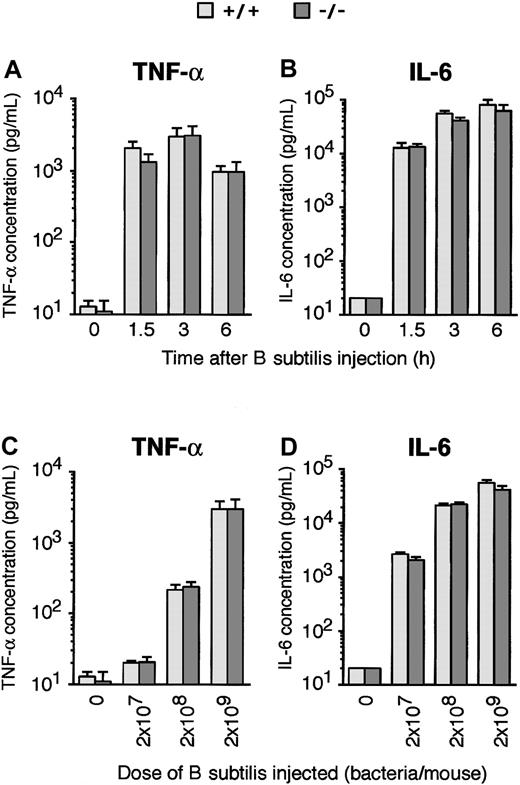
Normal induction of TNF-α and IL-6 in PGRP-S-/- mice by B subtilis
We also tested whether the induction of cytokines by bacteria in PGRP-S-/- mice was deficient, because early cytokine response in infection can profoundly influence the innate and acquired immunity and subsequent outcome of infection1-3 and because in Drosophila PGRP-SA (an orthologue of mammalian PGRP-S) is required for Toll receptor–mediated induction of antibacterial peptides by gram-positive bacteria.9 Therefore, it was also of interest to determine whether mammalian PGRP-S, by analogy to insects, was required for the activation of cytokines by gram-positive bacteria, which in mammals occurs through Toll-like receptors (primarily TLR2).2,3,25-27
However, both kinetics and dose response of in vivo induction of TNF-α and IL-6 production by B subtilis were the same in PGRP-S+/+ and PGRP-S-/- mice (Figure 9). These results suggest that the increased susceptibility to infection in PGRP-S-/- mice was not due to a defective initial cytokine response and demonstrate that PGRP-S is not required for normal stimulation of TNF-α and IL-6 secretion by B subtilis in vivo.
Similar induction of cytokines in PGRP-S-/- and PGRP-S-/- mice byB subtilis. Mice were injected intraperitoneally with 2 × 109 (A-B) or the indicated doses (C-D) of B subtilis, and the concentrations of TNF-α and IL-6 in their serum were assayed before (0 hour) and 1.5, 3, or 6 hours (A-B) or 3 hours (C-D) after the bacterial challenge. The results are geometric means ± SE of 8 mice per group. IL-6 at time or dose 0 was undetectable in any of the mice and is shown at the level of detection of the assay. There were no statistically significant differences between PGRP-S-/- and PGRP-S+/+ mice, calculated by Mann-Whitney U test.
Similar induction of cytokines in PGRP-S-/- and PGRP-S-/- mice byB subtilis. Mice were injected intraperitoneally with 2 × 109 (A-B) or the indicated doses (C-D) of B subtilis, and the concentrations of TNF-α and IL-6 in their serum were assayed before (0 hour) and 1.5, 3, or 6 hours (A-B) or 3 hours (C-D) after the bacterial challenge. The results are geometric means ± SE of 8 mice per group. IL-6 at time or dose 0 was undetectable in any of the mice and is shown at the level of detection of the assay. There were no statistically significant differences between PGRP-S-/- and PGRP-S+/+ mice, calculated by Mann-Whitney U test.
Discussion
Our results demonstrate that PGRP-S-/- mice have increased susceptibility to intraperitoneal infection with gram-positive bacteria of low pathogenicity (such as B subtilis and M luteus) and suggest that this is due to deficient intracellular killing and digestion of these bacteria by PMNs. PGRP-S is stored in tertiary granules of PMNs, and its primary site of antibacterial action is intracellular in PMNs, although purified PGRP-S is also bacteriostatic in vitro. PMNs from PGRP-S-/- mice have normal rate of phagocytic uptake of bacteria.
Phagocytosis and killing of bacteria are the main functions of PMNs. PMNs have several mechanisms to kill bacteria, broadly divided into oxygen-dependent mechanisms, which include several reactive oxygen intermediates that act in conjunction with myeloperoxidase and halide ions, and oxygen-independent mechanisms, which include nitrogen-derived toxins, granule proteases, lysozyme, and antimicrobial peptides (defensins, cathelicidins).35-38 Therefore, PGRP-S can be classified as another antibacterial protein in PMNs that has activity against gram-positive bacteria of low pathogenicity, such as B subtilis, M luteus, and L acidophilus.
PGRP-S deficiency has no effect on susceptibility to infection in vivo or on killing by PMNs in vitro of more pathogenic gram-positive bacteria (S aureus) and gram-negative bacteria (E coli and P vulgaris), which suggests that S aureus, E coli, and P vulgaris can evade antibacterial activity of PGRP-S. Efficient killing of bacteria usually requires concerted action of several killing mechanisms, which often act synergistically.35-38 Oxygen-dependent mechanisms have been long considered the most significant.35-38,40,41 However, different bacteria have different sensitivity to these mechanisms.35-38,40-45 Some pathogenic bacteria, such as S aureus, require oxygen-dependent mechanisms for killing36,42-44 ; some bacteria, such as E coli or P vulgaris, are more efficiently killed by the oxygen-dependent mechanisms but are still sensitive to oxygen-independent killing42,44,45 ; and other bacteria can be efficiently killed by either mechanisms.35-38,41,42 These differences, as well as lower binding of mouse PGRP-S to S aureus and gram-negative bacteria,14 could explain why PGRP-S deficiency has no effect on the susceptibility to several more pathogenic bacteria. This difference is also consistent with the notion that bacteria of low pathogenicity are much more easily killed or removed from the body by the immune system than the more pathogenic bacteria, which evolved to evade some of the killing mechanisms of the host. The effectiveness of bacterial killing also depends on several factors other than bacterial species, such as strain, bacteria-to-phagocyte ratio, opsonization, and assay conditions.32,35-38,40-45 Moreover, recent data indicate that granule proteases, especially cathepsin G and elastase, are of primary importance for microbial killing and that the role of the oxygen-dependent mechanisms is to trigger the release and activation of these proteases.39
How PGRP-S exerts its antibacterial activity is not known, because PGRP-S has no sequence homology to any known leukocyte-derived antimicrobial peptides. PGRP-S binds to bacteria and their PGN component with nanomolar affinity14 (and probably also to other polysaccharides, R.D., unpublished data, December 19, 2002), and at higher concentrations it most likely forms aggregates.14 The binding specificity of PGRP-S to PGN is similar to that of vertebrate lysozyme.14 However, PGRP-S has no lysozyme-like PGN-lytic or bacteriolytic activity.14 PGRP-S has some sequence homology to bacteriophage T3 and T7 lysozymes,4 which are amidases (they hydrolyze the bond between muramic acid and the peptide in PGN), but PGRP-S has no amidase activity either.4,14 Mouse PGRP-S is bacteriostatic, but not bactericidal14 (current data), whereas bovine PGRP-S orthologue is bactericidal.28 This difference between mouse and bovine PGRP-S is consistent with the presence of different types of granules and granule proteins in ruminants than in other mammals.46
Mammalian PGRP-S may work directly on bacteria through its antibacterial activity, or it may interact with, activate, or increase activity of another antibacterial system in PMNs. The latter hypothesis is supported by the lower induction of oxidative burst by B subtilis and M luteus in PGRP-S-/- mice.
The increased susceptibility of PGRP-S-/- mice to B subtilis and M luteus infection is manifested only on intraperitoneal but not intravenous challenge, which may be related to the differences in cells primarily responsible for removal of bacteria from these sites: infiltrating PMNs in peritoneal cavity following intraperitoneal infection versus macrophages (Kupfer cells in the liver and alveolar macrophages in the lungs) following intravenous infection. PGRP-S is expressed only in PMNs and bone marrow PMN precursors and is not expressed in monocytes and macrophages,4,6,14,28 and macrophages from PGRP-S-/- mice show normal in vitro killing of B subtilis and M luteus. Moreover, soluble PGRP-S inhibits uptake of bacteria by macrophages in vitro14 (and data not shown), whereas it does not inhibit the uptake of bacteria by PMNs. This inhibition of phagocytosis of bacteria by macrophages in wild-type mice is likely caused by extracellular PGRP-S, which can be released from PMNs by exocytosis at inflammatory sites. Indeed, we have shown that approximately half of intracellular PGRP-S is released from PMNs by exocytosis following intravenous or intraperitoneal injection of B subtilis or M luteus into mice (R.D., unpublished data, December 19, 2002). These results are consistent with the presence of PGRP-S in the gelatinase granules, because 40% of their contents is readily released from PMNs by exocytosis.34,35 This effect would be absent in PGRP-S-/- mice, which would result in higher immediate clearance of intravenously injected bacteria by alveolar macrophages in the lungs and by macrophage-like Kupfer cells in the liver. It should be noted that blood PMNs play only a minor role in mice, because of the very low numbers of PMNs in peripheral blood in mice (7%) compared with humans (65%). Differences in susceptibility to infection by different routes are often seen in knock-out mice, eg, cathepsin G–deficient mice have increased susceptibility to intravenous infection39 but not intraperitoneal infection47 with Saureus.
Our results demonstrate that the function of PGRP-S in mammals is different from that in insects. In insects, PGRP-S functions as a soluble protein in hemolymph and in cuticle that recognizes PGN and gram-positive bacteria, activates the prophenoloxidase cascade (that generates melanin),7,8 or has PGN-amidase activity (PGRP-SC1B).48 In Drosophila, PGRP-SA also activates Toll receptor pathway in immune cells, which induces production of antimicrobial peptides.9 By contrast, mammalian PGRP-S functions as an intracellular antibacterial protein in PMNs and has direct antibacterial effect, but it is not an amidase.4,14,48 Also in contrast to insects, we found no evidence of any interaction of mammalian PGRP-S with TLR or CD14 receptors. PGRP-S-/- mice have normal induction of TNF-α and IL-6 by B subtilis, which is a TLR2-mediated process.25-27,49 Also, PGRP-S had no effect on PGN- or bacteria-induced cytokine production or activation of several transcription factors or signal transduction molecules in mouse macrophage RAW264 and J774A cells or human (THP1) monocytic cells, or in primary mouse macrophages or human monocytes, or in cells transiently or stably transfected with CD14 and/or TLR2 (R.D. and D.G., unpublished data, December 19, 2002). PGRP-S did inhibit PGN-induced production of cytokines in mouse RAW264 macrophage cells, but this was an exception, only observed in the RAW264 cell line adapted to growth in serum-free medium.14 Moreover, CD14- or TLR2-mediated cell activation in both native cells, cell lines, or transfected cells does not require the presence of PGRP-S,22-27,49 PGRP-S and CD14 do not inhibit each other's binding to PGN,14 and monocytes and macrophages (the main CD14/TLR2-positive PGN-responsive cells) do not express PGRP-S.14 Also, in contrast to insect Toll receptor, which does not directly interact with bacteria,9,50 mammalian TLR receptors, such as TLR2 or TLR4, directly interact with microbial stimulants.51-54 Therefore, we show that mammalian PGRP-S has a different effector function than insect PGRP-S. Thus, only the bacterial recognition function of PGRP-S, but not its effector function, is conserved in evolution from insects to mammals.
Prepublished online as Blood First Edition Paper, March 20, 2003; DOI 10.1182/blood-2002-12-3853.
Supported by US Public Health Service grant AI2879 from National Institutes of Health (R.D.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Robert Rukavina for breeding and maintaining our mice.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal