Abstract
Optimal methods of stem cell mobilization in multiple myeloma are undefined, and contaminating clonotypic cells could contribute to disease recurrence. A phase 2 trial of intravenous melphalan (60 mg/m2) and granulocyte colony-stimulating factor (G-CSF) (10 μg/kg/d) for mobilization was performed. To enhance reliability, contamination was assessed with 2 sensitive methods, immunoglobulin light and heavy chain variable region patient-specific limiting-dilution polymerase chain reaction (PCR). We evaluated 29 stem cell components (SCCs) from 15 patients; for 9 SCCs, only VL PCR was used because of light chain disease or technical problems with VH primers. For 20 SCCs, VL and VH PCR results were highly correlated (r2 = 0.93, P < .01), with 35% (7 of 20) having identical estimates. VH PCR gave significantly higher estimates for 8—and VL PCR for 5—SCCs, supporting the utility of using 2 methods. Estimated clonotypic contamination per SCC was 0.0009% (range, 0%-0.1%) or 0.5 × 104 clonotypic cells per kilogram (range, 0-41.2 × 104/kg), and contamination correlated with CD34+ cells collected (r2 = 0.42, P < .01). Melphalan-mobilized SCCs contain minimal clonotypic contamination.
Introduction
Multiple myeloma (MM) is an incurable hematologic malignancy. Autologous stem cell transplantation (SCT) prolongs survival compared with conventional chemotherapy1-3 although patients invariably relapse.3 Autologous SCT followed by reduced-intensity allogeneic SCT may improve response rates and survival4 but, even for such tandem SCT regimens, the contribution to disease relapse of autologous stem cell components (SCCs) contaminated by malignant plasma cells will need to be evaluated.5-9
The optimal mobilization strategy in myeloma might combine antimyeloma effect with collection of adequate, minimally contaminated stem cell components. Over the past 2 years we conducted a phase 2 trial employing intravenous melphalan (60 mg/m2) and granulocyte colony-stimulating factor (G-CSF) (10 μg/kg/d) for stem cell mobilization and reduction of disease. We assessed clonotypic contamination in SCCs by limiting-dilution polymerase chain reaction (PCR) assays using immunoglobulin (Ig) light and heavy chain (VL and VH, respectively) patient-specific primer sets (CDR1/CDR3),5,10-13 a strategy that combines a sensitive method and an independent check on estimates of contamination. In this report we describe the technical aspects of the assessment of tumor cell contamination of SCCs from patients with myeloma mobilized with melphalan.
Study design
Patients with chemoresponsive multiple myeloma (more than 50% reduction in M protein) were enrolled on this institutional review board (IRB)–approved protocol after providing written informed consent.14 Bone marrow and stem cell specimens were obtained for molecular studies, and data were recorded, as previously described.15 Stem cell collections began when peripheral blood leukocyte counts exceeded 5 × 109/L (5000/μL).
Clonal Ig V genes
Clonal Ig VL and VH genes were identified as previously described, using PCR primers for consensus VH and Cα or Cγ regions and for Vκ and Vλ subgroups.10-13,16,17 Sequences of the VDJ (VH) or VJ (VL) regions were evaluated. VL and VH CDR1/CDR3 primers were designed and optimized.12,17-20 Each primer pair was tested for specificity and sensitivity using buffy-coat DNA from healthy donors and DNA from the patient's marrow specimen.
Limiting-dilution PCR
Representative samples were obtained from each SCC without manipulation. To estimate the percent of clonotypic cells, limiting-dilution PCR with patient-specific CDR1/CDR3 primers was performed10-13,17-20 with DNA from 2 × 105 cells per SCC serially log-diluted with buffy-coat DNA. Specimens giving all negative results were assessed by reverse transcriptase (RT)–PCR, whereas specimens giving positive results underwent further PCR assays in which 5 to 10 identical PCR reactions were performed simultaneously with DNA at each dilution. The linearity of this approach has been validated,13,18 and the intra-assay variability was assessed using a control patient specimen with 1% clonotypic cells, repeating the PCR assays for 7 consecutive days. Each 25-μL PCR reaction contained cellular DNA in log dilutions, PCR buffer, 1.5 mM MgCl2, 0.2 mM of each deoxyribonucleoside triphosphate (dNTP), 0.4 μM of each primer, and 0.5 units of Taq (Invitrogen, Carlsbad, CA). Negative controls with and without buffy-coat DNA were amplified with each assay. PCR conditions were as previously described; annealing temperatures ranged from 50°C to 64°C, depending on primer pair.16,20
The percent of clonotypic cells per SCC was calculated using a Poisson analysis and MAXLIKE software (gift from F. Cremer and M. Moos, Universität Heidelberg, Heidelberg, Germany) as previously described.13 The amount of contamination was estimated by multiplying the percent clonotypic cells (using the higher of the VH or the VL estimate) by the total number of cells collected per kilogram.
Statistics
All analyses were performed with PRISM (GraphPad, San Diego, CA) and were 2-tailed, unless otherwise noted, using a P value of less than .05 to determine the significance level.
Results and discussion
Optimal methods for stem cell mobilization in myeloma and for assessment of stem cell components (SCCs) for contamination have not been defined. We evaluated 29 SCCs mobilized with melphalan from 15 patients, 12 with an IgG or IgA paraprotein and 3 with light chain disease (Table 1). In all cases the clonal Ig VL and VH genes were identified; they were distributed among heavy chain subgroups (VH2 = 1, VH3 = 8, and VH4 = 3) and light chain subgroups (Vκ1 = 7, Vκ4 = 1, Vλ1 = 1, Vλ2 = 3, and Vλ3 = 3). All genes showed evidence of somatic hypermutation.21,22
Data on 15 patients mobilized with melphalan (60 mg/m2) and G-CSF
Age, y (range) | 58 (33-73) |
| Sex | 8 men, 7 women |
| Myeloma protein | |
| IgG | κ = 3, λ = 3 |
| IgA | κ = 3, λ = 3 |
| Light chain only | κ = 1, λ = 1 |
| Nonsecretory | κ = 1 |
| Stage of disease, no. of patients | |
| II | 7 |
| III | 7 |
| Plasma cell leukemia | 1 |
| Days from melphalan to first collection (range) | 19 (13-22) |
| No. of leukaphereses (range) | 2 (1-5) |
| Total CD34+ cells collected × 106/kg (range) | 14.1 (3.5-40.0) |
| CD34+ cells × 106/kg infused for SCT (range) | 6.8 (3.1-10.7) |
| Days from SCT to ANC more than 0.5 × 109/L (range) | 9 (8-11) |
| Days from SCT to platelets more than 20 × 109/L untransfused (range) | 12 (10-16) |
| CD34+ cells in 13 paired—day 1, day 2—collections | |
| Day 1, % | 1.6 ± 1.4 |
| Day 1, no. × 106/kg | 3.2 ± 2.9 |
| Day 2, % | 1.9 ± 1.3 |
| Day 2, no. × 106/kg | 6.8 ± 5.6 |
| Day 1 vs day 2, paired t test, % | P = .11 |
| Day 1 vs day 2, paired t test, no. | P < .01 |
| Clonotypic cells* in 13 paired—day 1, day 2—collections | |
| Day 1, % | 0.009 ± 0.013 |
| Day 1, no. × 104/kg | 1.9 ± 2.7 |
| Day 2, % | 0.016 ± 0.033 |
| Day 2, no. × 104/kg | 4.0 ± 7.7 |
| Day 1 vs day 2, paired t test, % | P = .44 |
| Day 1 vs day 2, paired t test, no. | P = .37 |
| Clonotypic cells,* 29 collections; median (range) % | 0.0009 (0-0.1) |
| No. × 104/kg | 0.5 (0-41.2) |
Age, y (range) | 58 (33-73) |
| Sex | 8 men, 7 women |
| Myeloma protein | |
| IgG | κ = 3, λ = 3 |
| IgA | κ = 3, λ = 3 |
| Light chain only | κ = 1, λ = 1 |
| Nonsecretory | κ = 1 |
| Stage of disease, no. of patients | |
| II | 7 |
| III | 7 |
| Plasma cell leukemia | 1 |
| Days from melphalan to first collection (range) | 19 (13-22) |
| No. of leukaphereses (range) | 2 (1-5) |
| Total CD34+ cells collected × 106/kg (range) | 14.1 (3.5-40.0) |
| CD34+ cells × 106/kg infused for SCT (range) | 6.8 (3.1-10.7) |
| Days from SCT to ANC more than 0.5 × 109/L (range) | 9 (8-11) |
| Days from SCT to platelets more than 20 × 109/L untransfused (range) | 12 (10-16) |
| CD34+ cells in 13 paired—day 1, day 2—collections | |
| Day 1, % | 1.6 ± 1.4 |
| Day 1, no. × 106/kg | 3.2 ± 2.9 |
| Day 2, % | 1.9 ± 1.3 |
| Day 2, no. × 106/kg | 6.8 ± 5.6 |
| Day 1 vs day 2, paired t test, % | P = .11 |
| Day 1 vs day 2, paired t test, no. | P < .01 |
| Clonotypic cells* in 13 paired—day 1, day 2—collections | |
| Day 1, % | 0.009 ± 0.013 |
| Day 1, no. × 104/kg | 1.9 ± 2.7 |
| Day 2, % | 0.016 ± 0.033 |
| Day 2, no. × 104/kg | 4.0 ± 7.7 |
| Day 1 vs day 2, paired t test, % | P = .44 |
| Day 1 vs day 2, paired t test, no. | P = .37 |
| Clonotypic cells,* 29 collections; median (range) % | 0.0009 (0-0.1) |
| No. × 104/kg | 0.5 (0-41.2) |
ANC indicates absolute neutrophil count.
All 15 patients had stem cell collections on day 1; 13 had collections on day 2, and 1 had a third collection on day 3, giving a total of 29 collections, CD34 and clonotypic contamination data for the 13 paired collections (means ± SD) and clonotypic data for all 29 collections are shown. To compute estimates of contamination, the higher percentage obtained with VL or VH PCR was used.
Twenty-five of 27 primer sets passed specificity and sensitivity testing (15 VL and 10 VH). Two VH primer sets continued to amplify bands similar in size to the target amplicon despite using buffy-coat DNA from 3 healthy donors as substrate and repeated redesign of primers. (Most target amplicons were about 220 base pair [bp], ranging from 159 to 255 bp.) Thus, 10 VH primer sets passed testing for the 12 patients with IgG or IgA paraproteins. Sensitivity testing showed that all 25 specific primer sets had sensitivities 0.01% or below. The coefficient of variation (CV) for replicate patient-specific limiting-dilution PCR assays was 24%, acceptable and similar to CVs obtained for radioimmunoassays.23,24
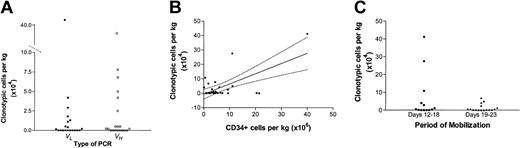
Results are available for all 29 SCCs. Thirty-five percent of the SCCs (7 of 20) successfully analyzed by both VL and VH PCR (n = 20) had identical estimates by both methods; in 5 of the concordant cases the estimate was zero or none detected (although RT-PCR assays for these were positive by both methods). VL and VH estimates differed for 65% (13 of 20) of the SCCs; in these instances, the higher value was used as the estimate of contamination. VH PCR was significantly higher for 8 (2.9 × 104 versus 0.4 × 104 cells per kilogram, P = .02 by 1-tailed paired t test), whereas VL PCR was higher for 5 SCCs (9.2 × 104 versus 7.5 × 104 cells per kilogram, P = .05). Results for these 20 SCCs were highly correlated (Figure 1A; r2 = 0.93, P < .01). For the remaining 9 SCCs, only VL PCR was used because of light chain disease (n = 5) or technical problems with VH primer sets (n = 4).
Estimates of clonotypic contamination in stem cell components collected after melphalan and G-CSF mobilization in 15 patients are shown as a function of different factors. (A) Estimates of clonotypic contamination in 20 stem cell components (SCCs) are shown as a function of the type of PCR assay used. The results with light and heavy chain (VL and VH) patient-specific PCR were correlated (r2 = 0.93, P < .01) and 35% (7 of 20) of the time were equal. Means were VL = 2.7 × 104 and VH = 3.3 × 104 clonotypic cells per kilogram per SCC. (B) Estimates of clonotypic contamination in 29 SCCs are shown as a function of CD34+ cells per kilogram in each SCC, calculated by flow cytometry. By simple linear regression, the line of best fit is y = (0.707)x – 0.669 with 95% confidence intervals as shown (r2 = 0.42, F = 19.1, P < .01). There is a relatively constant ratio of CD34+ to clonotypic cells, as previously suggested by Moos and colleagues.18 (C) Clonotypic contamination is shown in relationship to the timing of mobilization. Medians for days 12 to 18 and 19 to 23 were 0.5 × 104 and 0.35 × 104 clonotypic cells per kilogram. Differences due to timing of mobilization were not significant (Mann-Wilcoxon, P = .68).
Estimates of clonotypic contamination in stem cell components collected after melphalan and G-CSF mobilization in 15 patients are shown as a function of different factors. (A) Estimates of clonotypic contamination in 20 stem cell components (SCCs) are shown as a function of the type of PCR assay used. The results with light and heavy chain (VL and VH) patient-specific PCR were correlated (r2 = 0.93, P < .01) and 35% (7 of 20) of the time were equal. Means were VL = 2.7 × 104 and VH = 3.3 × 104 clonotypic cells per kilogram per SCC. (B) Estimates of clonotypic contamination in 29 SCCs are shown as a function of CD34+ cells per kilogram in each SCC, calculated by flow cytometry. By simple linear regression, the line of best fit is y = (0.707)x – 0.669 with 95% confidence intervals as shown (r2 = 0.42, F = 19.1, P < .01). There is a relatively constant ratio of CD34+ to clonotypic cells, as previously suggested by Moos and colleagues.18 (C) Clonotypic contamination is shown in relationship to the timing of mobilization. Medians for days 12 to 18 and 19 to 23 were 0.5 × 104 and 0.35 × 104 clonotypic cells per kilogram. Differences due to timing of mobilization were not significant (Mann-Wilcoxon, P = .68).
For the 29 SCCs examined, estimated clonotypic contamination was 0.0009% (range, 0%-0.1%) or 0.5 × 104 cells per kilogram (range, 0-41.2 × 104/kg). Some variability could be explained by the correlation between CD34+ cell mobilization and contamination (Figure 1B). There was no correlation between contamination and stage of disease (data not shown) or timing of mobilization (Figure 1C). In a study of SCC contamination that involved a similar cohort of patients mobilized with cyclophosphamide- or ifosfamide-containing regimens and G-CSF, a constant ratio of contaminating clonotypic cells to CD34+ cells collected was also observed using a similar PCR method.18 The investigators reported no difference in CD34+ cells obtained on the first or second days of collection,18 whereas with melphalan mobilization significantly more CD34+ cells were obtained on the second day of collection (Table 1).
In that study the investigators reported a median of 0.0066% contaminating cells per SCC (range, 0%-0.71%).18 We performed limiting-dilution PCR on 4 SCCs mobilized with cyclophosphamide and G-CSF from 3 chemoresponsive myeloma patients and found a median of 0.038% contaminating cells per SCC (range, 0.029%-0.046%), using the same technique that demonstrated a median of 0.0009% contamination of melphalan-mobilized SCCs. These data suggest that melphalanmobilized SCCs contain minimal clonotypic contamination and that the assessment of contamination is enhanced by use of Ig VL and VH patient-specific PCR.
In a phase 3 trial of SCT for myeloma, cyclophosphamidemobilized SCCs purged by CD34 selection were not superior to unpurged components.8 However, gene-marking studies have demonstrated that infused tumor cells in SCCs can contribute to relapse.25 Patients with myeloma mobilized with melphalan on this trial had minimally contaminated SCCs collected, and 40% of them experienced a further 50% disease reduction.14 All 15 patients we report had adequate SCCs collected, underwent autologous stem cell transplantation with a median of 6.8 million melphalanmobilized CD34+ cells per kilogram (range, 3.1-10.7 million cells per kg), and had routine and full hematologic recoveries (Table 1). We believe these results support the further assessment of melphalan for stem cell mobilization given its antimyeloma effect. Such studies can determine whether this mobilization strategy has a clinical impact on progression-free and overall survival.
Prepublished online as Blood First Edition Paper, March 20, 2003; DOI 10.1182/blood-2002-12-3674.
Supported by National Institutes of Health grant CA05826, the Graziano Fund, the Multiple Myeloma Research Foundation, the Mel Stottlemyre Myeloma Research Fund, the Donald Stein Myeloma Research Fund, and Amgen.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked ``advertisement'' in accordance with 18 U.S.C. section 1734.
We are grateful for the assistance of Suzanne Costello, Lilian Reich, Nancy Collins, Shanta Sharma, Lisa Drake, Joanne Santorsa, and the staffs of the Memorial Hospital Donor Room and Cytotherapy Laboratory.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal