Abstract
Rearrangement of the FLI-1 locus and ensuing overexpression of FLI-1 protein is an early event in Friend murine leukemia virus (F-MuLV)-induced erythroleukemia. When overexpressed in primary erythroblasts, FLI-1 converts erythropoietin (Epo)-induced terminal differentiation into a proliferative response. We found that SLAP, a gene encoding a recently described negative regulator of T-cell antigen receptor function during thymocyte development, is up-regulated both at the RNA and protein levels in FLI-1-transformed erythroblasts. Src-like adaptor protein (SLAP) was found in a specific complex with erythropoietin receptor (EpoR), a cytokine receptor essential to erythroid differentiation. Constitutive expression of SLAP severely impairs hemoglobinization and late survival during Epo-induced terminal differentiation of erythroblasts. This impairment is associated with the specific inhibition of several critical Epo-dependent signaling events, including signal transducer and activator of transcription 5 (STAT5) activation and up-regulation of the expression of the antiapoptotic BCL-X gene. Our data support a model by which FLI-1 inhibits normal erythroid differentiation through the deregulation of genes encoding adaptors/effectors that modify the signaling output of cytokine receptors normally required for terminal differentiation. (Blood. 2003; 102:4555-4562)
Introduction
FLI-1 is a transcription factor of the ETS protein family. The common feature of ETS proteins is an 85-amino-acid domain (the ETS domain), which is responsible for their binding to specific DNA sequences.1 In addition, FLI-1 possesses other functional domains that are involved in its transcriptional regulatory properties.2,3 Inactivation of FLI-1 demonstrates its crucial role in mouse development as FLI-1-/- embryos die at E11.5 because of loss of vascular integrity. FLI-1-/--deficient embryos also show a defect in the terminal differentiation of megakaryocytes.4,5
The FLI-1 locus is a common proviral integration site for Friend murine leukemia virus (F-MuLV), a retrovirus that induces a multistep erythroleukemia in susceptible newborn mice.6,7 The emergence of proliferating erythroid progenitors in the spleen of infected mice coincides with the rearrangement of the FLI-1 locus because of integration of an F-MuLV provirus and up-regulation of FLI-1 expression. These erythroblasts are not initially immortalized, and additional genetic events, including the loss of function of wild-type p53, are required for tumor development.8 Studies conducted with different erythroid cell systems have demonstrated that FLI-1 is clearly implicated in erythroid transformation.9,10 To analyze the intrinsic consequences of FLI-1 expression in erythroid progenitors, we made use of primary avian erythroblasts. In the absence of erythropoietin (Epo), these cells rapidly die by apoptosis, whereas in the presence of Epo they terminally differentiate in a synchronous manner.11 We have shown that ectopic expression of FLI-1 inhibits apoptosis of primary erythroblasts maintained in culture in the absence of Epo, a property associated with the ability of FLI-1 to up-regulate BCL-2 gene expression.3 Moreover, FLI-1 prevents terminal differentiation normally obtained in response to Epo and converts the Epo-induced differentiation signal into a proliferation response.10
In the present work, by expression profiling we cloned the avian ortholog of SLAP, a gene encoding an adaptor molecule and showed its differential expression between control and FLI-1-transformed cells. We further demonstrate that enforced expression of src-like adaptor protein (SLAP) in erythroblasts at levels similar to those found in FLI-1-transformed cells specifically interferes with late events in Epo-induced erythroid differentiation. SLAP was found to be physically associated with erythropoietin receptor (EpoR) and to interfere with a subset of EpoR downstream signaling events.
Materials and methods
Cloning of chicken SLAP cDNA
RNA was obtained following lysis in guanidine isothiocyanate and phenol extraction from ts-v-Sea/SFCV-0 control and ts-v-Sea/SFCV-FLI-1 erythroblasts maintained in the absence of Epo for 10 hours at 42°C. PolyA-containing RNA was prepared, and double-stranded cDNA was synthesized using cDNA Timer Saver Kit (Pharmacia, Uppsala, Sweden). Representational difference analysis was performed as previously described.12
Rapid amplification of 5′ cDNA end was performed on RNA from FLI-1-transformed erythroblasts using the 5′/3′ rapid amplification of cDNA end (RACE) kit (Boehringer Mannheim, Mannheim, Germany), according to the manufacturer's instructions, using 3 specific primers: rev1, 5′-CTTCCAAATAACTGGTATTA-3′; rev2, 5′ AATCACTACTCAGAAGTTGC 3′; and rev3, 5′-CACTAACAACCACAGCATAAT-3′. A single RACE product of 500 bp corresponding to the 5′ part of ckSLAP cDNA was obtained. Primers 5′ ACCTTGAAGGTATAGATGTC 3′ from the 5′ untranslated region and 5′ TCTAAGTGCTCAACTTCCATT 3′ from the 3′ untranslated region were used to amplify the full-length ckSLAP cDNA by using cDNAs from FLI-1-transformed erythroblasts, sequenced (GenBank AY278230) and cloned into pCR2.1 TOPO.
DNA constructs
pRCAS-A, pRCAS-mEpoR, pCRNCM-mEpoR, pSFCV, pSFCV-FLI-1 (1-452) encoding a hemagglutinin A (HA)-tagged version of wild-type (wt) hFLI-1 and RCAS-hBCL-2 have been described previously.3,10,11 pSFCV-ckSLAP, pEFBos-ckSLAP, and pMSCV-eGFP-mSLAP were obtained by standard molecular biology techniques. pMSCV-eGFP was a gift from Dr H. Singh, University of Chicago. PEFBos-mSLAP and pΔODLO(NFAT)3 were from Dr A. Weiss (University of California, San Francisco).
Generation of recombinant retroviruses
Infectious stocks of avian retroviruses were generated as previously described.11 To generate infectious viral stocks of MSCV-eGFP (murine stem cell virus-enhanced green fluorescent protein) vectors, 5 × 106 cells of the Phoenix (Ampho) packaging cell line grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum were transfected by using the calcium phosphate (CaPi) coprecipitation method. Thirty-six hours after transfection, supernatants were collected, mixed with 6 μg/mL polybrene, and used to infect cells of the gpE86 packaging cell line.13 Infection of gpE86 was repeated every 12 hours for 2 days. The brightest eGFP-positive cells were sorted with the use of fluorescence-activated cell sorting (FACS) and expanded in culture. This sorting procedure was repeated until 100% of the cells were found brightly GFP positive.
Cell culture, retroviral infection, differentiation, and survival assays
ts-v-Sea primary avian erythroblast clones expressing the proteins of interest were obtained from bone marrow cells and analyzed for differentiation and survival as previously described.11 The EI/11 mouse erythroblast cell line was cultivated in serum-free conditions in a 1:1 mixture of Iscove modified Dulbecco medium (IMDM) and StemPro-34 SFM (serum-free medium; Invitrogen, Cergy Pontoise, France) supplemented with detoxified bovine serum albumin (BSA) (3 mg/mL), 2 mM glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin, human recombinant Epo (2 U/mL; Janssen-Cilag, Levallois Perret, France), murine recombinant stem cell factor (SCF; 100 ng/mL), and dexamethasone (10-6 M, Sigma, St Louis, MO). For differentiation induction, E1/11 cells were washed twice in phosphate-buffered saline (PBS) and seeded at 1.5 × 106 cells/mL in the same medium containing hEpo (5 U/mL), insulin (0.01 U/mL), and iron-saturated human transferrin (1 mg/mL; Sigma). Differentiating erythroblasts were maintained at densities between 1.5 and 6 × 106 cells/mL by either daily dilution or partial medium change. Retroviral transduction of EI/11 erythroblasts was carried out by cocultivating 0.5 × 106 cells with irradiated (1200 cGy) gpE86 virus-producing cells (1.5 × 106 cells/60-mm Petri dish) in the presence of 6 μg/mL polybrene. Forty-eight hours later, erythroblasts were collected and expanded in EI/11 proliferation medium. eGFP-positive erythroblasts were FACS sorted and further grown. Hemoglobin measurement and terminal dUTP nick-end labeling (TUNEL) assay were carried out as previously described.11
Culture and transfection of 293T cells
293T cells were cultivated in DMEM supplemented with 10% fetal calf serum. Cells (1 × 106 cells/60-mm Petri dish) were transfected by using the CaPi coprecipitation method with 2.5 μg pCRNCM-EpoR and 2.5 μg pcDNA3.1-mSLAP as indicated. The total amount of DNA was kept constant by the addition of the corresponding insert-free vectors.
RNA expression analysis
Total RNA was processed for Northern blot analysis as previously described.10 For reverse transcriptase polymerase chain reaction (RT-PCR) analysis, cDNAs were synthesized by using the first-strand cDNA synthesis kit (Amersham, Les Ulis, France) with the use of the manufacturer's Not I-d(T)18 primers. The generated cDNA products were amplified by using Taq polymerase (Eurobio, Les Ulis, France) and the following amplimers: chk-SLAP 5′ amplimer, 5′-TTCCTTGTCAGTACGGCACA-3′; chk-SLAP 3′ amplimer, 5′-TCTAAGTGCTCAACTTCCATT-3′; chk-S17 5′ amplimer, 5′-TACACCCGTCTGGGCAACGAC-3′; chk-S17 3′ amplimer, 5′-CCGCTGGATGCGCTTCATCAG-3′; chkBcl-X5′ amplimer, 5′-GGACCATGGACTCATTGAGGG-3′; chkBcl-X3′ amplimer, 5′-TGAGCTAGGATGCAGGGAGCC-3′; chk-S172 5′ amplimer, 5′-TACACCCGTCTGGGCAACGAC-3′; and chk-S172 3′ amplimer, 5′-CGCGATCTTGTTGCGCAGC-3′. Amplification conditions were 10 minutes at 94°C, followed by 30 amplification cycles (1 minute at 94°C, 1 minute at 57°C for ckSLAP or 60°C for ckS17, ckBclX, and ckS172 , 1 minute at 72°C). The amplification was followed by a 10-minute elongation step at 72°C. mBcl-XL and mβ-actin PCR products were amplified by using the following amplimers: mBcl-Xl5′ amplimer, 5′-TGGAGTCAGTTTAGTGATGTCG-3′; mBcl-Xl3′ amplimer, 5′-CCAGCAGAACCACACCAGCC-3′; mβ-Actin 5′ amplimer, 5′-GTGGGCCGCCCTAGGCACCAG-3′; mβ-Actin 3′ amplimer, 5′-CTCTTTGATGTCACGCACGATTTC-3′. Amplification conditions were 5 minutes at 94°C followed by 35 amplification cycles (30 seconds at 94°C, 30 seconds at 59°C, 1 minute at 72°C) followed by a 10-minute elongation step. PCR products were analyzed on a 2% agarose gel containing ethidium bromide.
Immunoblot analysis
Cells were processed for Western blot analysis as previously described.3 Rabbit antibodies to EpoR (sc-697), extracellular-regulated kinase 2 (ERK2; sc-154), signal transducer and activator of transcription 5 (STAT5; sc-835), and the B-cell lymphoma protein 2 (BCL-2) monoclonal antibody (sc-509) were from Santa Cruz Biotechnology (Santa Cruz, CA). The monoclonal antibody (05-495) specific for tyrosine-phosphorylated STAT5A and STAT5B and the 4G10 antiphosphotyrosine monoclonal antibody were from Upstate Biotechnology (Lake Placid, NY). The anti-Flag monoclonal antibody was from Sigma (F-3165). The phospho-ERK-specific antibody (9101) was from Cell Signaling Technology (Beverly, MA). The rabbit anti-ckSLAP antiserum, specific to the carboxy terminus of chicken SLAP, was obtained by injection of a keyhole limpet hemocyanin (KLH) conjugate of a peptide corresponding to the last 15 amino acids of ckSLAP (CKQKSTLQLPPTYYE). Protein/antibody complexes were revealed by using horseradish peroxidase-conjugated secondary antibodies (Amersham), and chemiluminescence was detected by using the enhanced chemiluminescence (ECL) kit (Amersham).
Electroporation and luciferase assays
Jurkat cells (107) were electroporated (250 V, 960 μF) with the indicated amounts of pΔODLO(NFAT)3-Luc reporter and pEF-Bos-SLAP, the total amount of expression plasmid was kept constant to 5 μg. After 24 hours, cells were treated with either the indicated amounts of anti-CD3 antibody (no. 553057; Pharmingen, San Diego, CA) cross-linked with the use of goat antimouse antibody (no. 1031-01, Southern Biotech, Clinisciences, Montrouge, France) or by treatment with phorbol 12-myristate 13-acetate (PMA; 20 ng/mL) and ionomycin (1 μg/mL). After 8 hours, cells were lysed, and lysates were assayed for luciferase activity by using the luciferase assay system kit (Promega, Madison, WI). Results were normalized with respect to the β-galactosidase activity encoded by 500 ng cotransfected pRSV-LacZ, using the Galacto-Star kit (Tropix, Applied Biosystems, Foster City, CA).
Immunoprecipitation assays
Cells were lysed in 20 mM Tris (tris(hydroxymethyl)aminomethane)-HCl pH 8, 137 mM NaCl, 10 mM EDTA (ethylenediaminetetraacetic acid), 1% NP40, 10% glycerol, 1% aprotinin, 100 μg/mL phenylmethylsulfonyl fluoride, 10 μg/mL leupeptin and centrifuged at 14 000 rpm for 15 minutes. Supernatants were collected, immunoprecipitation was carried out by using the indicated antibodies, and antigen/antibody complexes were insolubilized by using Protein A Sepharose (Amersham). After 2 washes using lysis buffer, immunoprecipitated proteins were dissolved in Laemmli sample buffer and subjected to polyacrylamide gel electrophoresis/sodium dodecyl sulfate (PAGE/SDS). Coimmunoprecipitated proteins were revealed by Western blot with the use of the indicated antibodies.
Electrophoretic mobility shift assay
Whole cell extracts were prepared by lysing 50 × 106 erythroblasts in 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) pH 7.9, 50 mM KCl, 300 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 1 mM Na3VO4, 10 mM 4-nitrophenylphosphate, 1% aprotinin, 100 μg/mL phenylmethylsulfonyl fluoride, and 10 μg/mL leupeptin. After 3 cycles of freezing and thawing, extracts were centrifuged at 15 000 rpm for 10 minutes at 4°C. Cellular proteins (15 μg) were incubated with either a STAT5-specific double-stranded oligonucleotide DNA probe (plus strand, 5′-AGATTTCTAGGAATTCAAATC-3′) or an ETS-specific oligonucleotide probe (plus strand, 5′-ATAAACAGGAAGTGGT-3′). Protein-DNA complexes were separated from the free probe by electrophoresis in a polyacrylamide gel in 0.25 × Tris borate EDTA (TBE) buffer for 90 minutes.
Results
RDA and cloning of ckSLAP
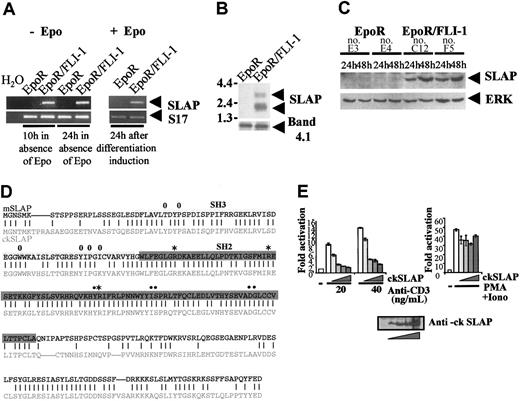
To identify genes specifically deregulated in FLI-1-transformed erythroblasts, we performed representational difference analysis (RDA) applied to cDNA12 with the use of RNA obtained from control primary erythroblasts and FLI-1-transformed erythroblasts (“Materials and methods”). To identify FLI-1 up-regulated genes, RNA from control cells was used as driver and RNA from FLI-1-transformed erythroblasts as tester. The differentially expressed PCR products were cloned, and a library was constructed. A full description of the transcripts specific to FLI-1-transformed erythroblasts will be presented elsewhere. We report here the characterization of clone no. 50, which was strongly differentially expressed. Indeed, RT-PCR analysis (Figure 1A) shows up-regulation of the corresponding transcript in EpoR/FLI-1 erythroblasts as compared with control cells. This comparison was confirmed by Northern blot analysis in which 2 transcripts of 2 and 3 kbp hybridizing with a probe to clone no. 50 were expressed at high levels in EpoR/FLI-1 erythroblasts (Figure 1B). Up-regulation of these transcripts in EpoR/FLI-1 cells occurred irrespective of the absence or presence of Epo (Figure 1A, left and right panels, respectively). Clone no. 50 was sequenced, and the open reading frame was found to encode a polypeptide highly homologous to the 149 carboxyterminal residues of mouse SLAP (mSLAP; Figure 1D). SLAP and the recently identified SLAP2 proteins form a novel family of adaptors that contain SH2 (Src homology domain 2) and SH3 domains. To confirm that clone no. 50 indeed encodes ckSLAP, the 5′ portion of the cDNA was obtained by 5′ RACE PCR. Primers were designed to amplify a full-length cDNA by RT-PCR by using RNA from a pool of 4 independent EpoR/FLI-1 clones. The derived amino acid sequence encoded by the open reading frame (ORF) of the full-length cDNA is represented in Figure 1D. It was found to encode a protein of 282 amino acids, showing an overall 83% similarity to mSLAP and to contain all characteristic domains of this adaptor. These domains include a short amino-terminal peptide containing a myristoylation site followed by SH3 and SH2 domains (90.4% and 75% identity to mSLAP, respectively) and a C-terminal part (48.1% identity to mSLAP) which comprises a 25-amino-acid domain unique to SLAP. To analyze whether the differential expression of SLAP translated at the protein level, we generated an antibody directed against the C-terminal part of ckSLAP. As shown in Figure 1C, this antibody specifically identified a 32- to 34-kDa protein in FLI-1-transformed erythroblasts. Thus, SLAP expression is specifically induced in FLI-1-transformed erythroblasts both at the RNA and protein levels.
Cloning and characterization of the differentially expressed ckSLAPgene. (A) RT-PCR analyses of SLAP expression. Control and FLI-1-transformed erythroblasts were maintained in the absence or in the presence of Epo as indicated. PCR was carried out with the use of primers for either ckSLAP (top blot) or the gene for ribosomal protein S17 as normalization control (bottom blot). The expected 520-bp SLAP and 129-bp S17 gene fragments are indicated. (B) Northern blot analysis. PolyA+ RNA from control and FLI-1-transformed erythroblasts were isolated from cells incubated for 10 hours in the absence of Epo. Hybridization was carried out with a probe corresponding to clone 50 (top blot) or to ckband4.1 gene as loading control (bottom blot). (C) Western blot analysis. Lysates from control and FLI-1-transformed clones cultivated in Epo for 24 and 48 hours were processed for Western blot by using an antibody to ckSLAP (top blot) or an antibody to ERK as loading control (bottom blot). (D) Alignment of mouse and chicken SLAP sequences. The deduced amino acid sequence of ckSLAP was aligned with that of mSLAP. Identities are shown as vertical lines. The SH3 domain is shaded in light gray and the SH2 domain in dark gray. The sequence corresponding to clone 50 corresponds to the last 144 amino acids. Basic amino acids conserved in the phosphotyrosine binding pocket of SH2 domains and those of the specificity determining region are highlighted by stars and dots, respectively. Hydrophobic residues involved in interactions of SH3 domains with proline-rich ligands are shown as circles. (E) ckSLAP inhibits TCR-induced activation of NFAT. Jurkat cells were cotransfected with 10 μg pΔODLO(NFAT)3-Luc reporter together with either 5 μg pEFBos control or an increasing amount (0.25, 0.5, 1, or 5 μg) of pEFBos-ckSLAP. Cells were then treated as indicated. Luciferase activity is presented as fold activation with respect to the basal activity of the reporter. The average values and standard deviations of 3 independent experiments are shown.
Cloning and characterization of the differentially expressed ckSLAPgene. (A) RT-PCR analyses of SLAP expression. Control and FLI-1-transformed erythroblasts were maintained in the absence or in the presence of Epo as indicated. PCR was carried out with the use of primers for either ckSLAP (top blot) or the gene for ribosomal protein S17 as normalization control (bottom blot). The expected 520-bp SLAP and 129-bp S17 gene fragments are indicated. (B) Northern blot analysis. PolyA+ RNA from control and FLI-1-transformed erythroblasts were isolated from cells incubated for 10 hours in the absence of Epo. Hybridization was carried out with a probe corresponding to clone 50 (top blot) or to ckband4.1 gene as loading control (bottom blot). (C) Western blot analysis. Lysates from control and FLI-1-transformed clones cultivated in Epo for 24 and 48 hours were processed for Western blot by using an antibody to ckSLAP (top blot) or an antibody to ERK as loading control (bottom blot). (D) Alignment of mouse and chicken SLAP sequences. The deduced amino acid sequence of ckSLAP was aligned with that of mSLAP. Identities are shown as vertical lines. The SH3 domain is shaded in light gray and the SH2 domain in dark gray. The sequence corresponding to clone 50 corresponds to the last 144 amino acids. Basic amino acids conserved in the phosphotyrosine binding pocket of SH2 domains and those of the specificity determining region are highlighted by stars and dots, respectively. Hydrophobic residues involved in interactions of SH3 domains with proline-rich ligands are shown as circles. (E) ckSLAP inhibits TCR-induced activation of NFAT. Jurkat cells were cotransfected with 10 μg pΔODLO(NFAT)3-Luc reporter together with either 5 μg pEFBos control or an increasing amount (0.25, 0.5, 1, or 5 μg) of pEFBos-ckSLAP. Cells were then treated as indicated. Luciferase activity is presented as fold activation with respect to the basal activity of the reporter. The average values and standard deviations of 3 independent experiments are shown.
Gene inactivation studies have shown that mSLAP is a negative regulator of T-cell antigen receptor (TCR) function at the CD4/CD8 double-positive stage of thymocyte development.14 In line with this finding, mSLAP overexpression in Jurkat T cells inhibits TCR signaling as assessed by its ability to interfere with the activation of nuclear factor of activated T cells (NFAT).14 To analyze whether ckSLAP is a functional homolog of mSLAP, we investigated whether ckSLAP also inhibited TCR-induced NFAT activation. For this, Jurkat cells were cotransfected with an NFAT-specific reporter plasmid, together with increasing doses of an expression vector encoding ckSLAP. By its own, ckSLAP had no detectable effect on the basal activity of the reporter. As shown in Figure 1E, TCR cross-linking by an anti-CD3 antibody induced activation of the NFAT-luciferase reporter gene. Under these conditions, ckSLAP expression inhibited NFAT activation in a dose-dependent manner (Figure 1E). As expected, ckSLAP did not interfere with NFAT activation induced in response to PMA/ionomycin stimulation, which bypasses early TCR signaling events (Figure 1E). Taken together, structural homology and functional assays indicate that the differentially expressed gene product identified in FLI-1-transformed erythroblasts encodes the chicken ortholog of mouse SLAP.
Constitutive expression of SLAP interferes with erythroid differentiation
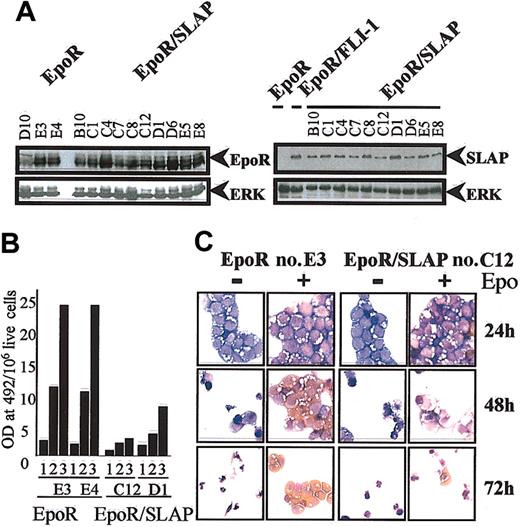
Because FLI-1 interferes with the response of erythroblasts to Epo and because SLAP is specifically up-regulated in FLI-1-transformed erythroblasts, we investigated whether ectopic expression of SLAP could also interfere with EpoR signaling in primary erythroblasts. We generated erythroblast clones coexpressing the mouse EPOR (EpoR) and ckSLAP by retroviral-mediated gene transfer and analyzed their ability to differentiate in response to Epo. Ten clones expressing either EpoR or both EpoR and ckSLAP were studied (Figure 2A and data not shown). Importantly, the level of exogenous SLAP protein expressed in these clones was comparable to the levels of endogenous SLAP observed in FLI-1-transformed erythroblasts (Figure 2A). EpoR and EpoR/SLAP erythroblasts were next studied for their response to Epo. As previously described, in response to Epo, control EpoR erythroblasts accumulated high levels of hemoglobin (Figure 2B) and acquired the oval morphology characteristic of mature erythrocytes (Figure 2C). The phenotype of EpoR/SLAP clones was essentially indistinguishable from that of EpoR clones for the first 24 hours of the differentiation process. In contrast, from 48 hours onward, unlike EpoR erythroblasts, which hemoglobinized and reduced their size, EpoR/SLAP cells accumulated little hemoglobin (Figure 2B) and differentiated into abnormally shaped cells or died (Figure 2C). No difference could be observed between EpoR and EpoR/SLAP erythroblasts maintained in the absence of Epo, as both died within 48 hours with the same kinetic (Figure 2C). These data show that SLAP specifically inhibits the response of erythroblasts to Epo, at the late stage of the differentiation process.
Constitutive SLAP expression interferes with Epo-induced differentiation. (A) Expression of exogenous proteins in EpoR and EpoR/SLAP erythroblast clones was analyzed by Western blotting by using either an anti-EpoR (left panel, top blot) or the anti-ckSLAP antibody (right panel, top blot). Expression of endogenous SLAP in a representative EpoR/FLI-1 erythroblast clone is shown in lane 2, right panel. (Bottom blots) ERK expression is shown as a loading control. (B) Quantitative determination of hemoglobin content in 2 EpoR and 2 EpoR/ckSLAP representative clones at days 1, 2, and 3 after differentiation induction in response to Epo. Normalized values (hemoglobin level per 106 live cells) are shown. (C) Cytocentrifugation analysis of EpoR and EpoR/ckSLAP representative clones maintained in absence (-) or presence (+) of Epo for 1, 2, and 3 days. Cells were stained with neutral benzidine (stains hemoglobin in brown) and histologic dyes (Diff-Quik). Original magnification × 60.
Constitutive SLAP expression interferes with Epo-induced differentiation. (A) Expression of exogenous proteins in EpoR and EpoR/SLAP erythroblast clones was analyzed by Western blotting by using either an anti-EpoR (left panel, top blot) or the anti-ckSLAP antibody (right panel, top blot). Expression of endogenous SLAP in a representative EpoR/FLI-1 erythroblast clone is shown in lane 2, right panel. (Bottom blots) ERK expression is shown as a loading control. (B) Quantitative determination of hemoglobin content in 2 EpoR and 2 EpoR/ckSLAP representative clones at days 1, 2, and 3 after differentiation induction in response to Epo. Normalized values (hemoglobin level per 106 live cells) are shown. (C) Cytocentrifugation analysis of EpoR and EpoR/ckSLAP representative clones maintained in absence (-) or presence (+) of Epo for 1, 2, and 3 days. Cells were stained with neutral benzidine (stains hemoglobin in brown) and histologic dyes (Diff-Quik). Original magnification × 60.
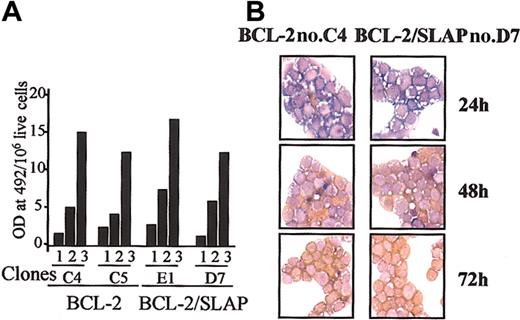
We next addressed whether SLAP-induced phenotype resulted from the ability of SLAP to interfere either with EpoR signaling or with the intrinsic erythroid differentiation program. To test this idea, we exploited the fact that massive expression of antiapoptotic molecules such as BCL-2 can completely bypass EpoR-induced signals to allow primary erythroblasts to terminally differentiate in the absence of Epo. We, therefore, analyzed the ability of SLAP to interfere with the Epo-independent differentiation of erythroblasts overexpressing BCL-2. Clones expressing either BCL-2 alone or both BCL-2 and SLAP were generated (data not shown) and analyzed for their ability to differentiate in the absence of Epo. As shown in Figure 3, the levels of hemoglobin accumulation over time (Figure 3A) and the appearance of mature, oval-shaped erythrocytes (Figure 3B) were comparable in BCL-2 and BCL-2/SLAP erythroblasts. The absence of SLAP-induced phenotype in BCL-2-overexpressing erythroblasts maintained in the absence of Epo suggests that expression of SLAP does not affect the intrinsic erythroid differentiation program per se but could rather inhibit EpoR-induced signals normally required for terminal differentiation.
SLAP does not interfere with intrinsic erythroid differentiation. (A) Quantitative determination of hemoglobin levels in representative BCL-2 and BCL-2/SLAP clones at days 1, 2, and 3 after differentiation induction in the absence of Epo. Normalized values (hemoglobin level per 106 live cells) are plotted. (B) Cytocentrifugation analysis of representative BCL-2 and BCL-2/SLAP erythroblast clones maintained in absence of Epo for 3 days. Cells were stained with neutral benzidine (stains hemoglobin in brown) and histologic dyes (Diff-Quik). Original magnification × 60.
SLAP does not interfere with intrinsic erythroid differentiation. (A) Quantitative determination of hemoglobin levels in representative BCL-2 and BCL-2/SLAP clones at days 1, 2, and 3 after differentiation induction in the absence of Epo. Normalized values (hemoglobin level per 106 live cells) are plotted. (B) Cytocentrifugation analysis of representative BCL-2 and BCL-2/SLAP erythroblast clones maintained in absence of Epo for 3 days. Cells were stained with neutral benzidine (stains hemoglobin in brown) and histologic dyes (Diff-Quik). Original magnification × 60.
SLAP interferes with the antiapoptotic properties of Epo
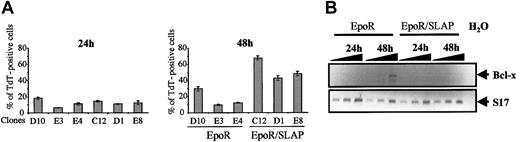
Epo is a cytokine endowed with antiapoptotic properties.15 We therefore investigated whether SLAP interfered with Epo-induced inhibition of apoptosis. Quantitative measurement of apoptosis by TUNEL assay revealed that 50% of EpoR/SLAP cells underwent apoptosis 48 hours after differentiation induction as compared with 15% of apoptotic cells in EpoR erythroblasts (Figure 4A). This increase in apoptosis was specific of the late stage of the differentiation process because no difference between control and SLAP erythroblasts could be detected 24 hours after differentiation induction (Figure 4A). A major antiapoptotic gene controlled by Epo stimulation is BCL-X.16 Up-regulation of BCL-X occurs late during the differentiation process, at the time of maximum hemoglobin synthesis,16 and gene inactivation studies have demonstrated a specific and nonredundant role for BCL-X for erythropoiesis.17 We, therefore, compared BCL-X gene expression in EpoR and EpoR/SLAP erythroblasts maintained under differentiation conditions by using semiquantitative RT-PCR. As shown in Figure 4B, BCL-X expression was up-regulated in differentiating EpoR cells. In contrast, BCL-X expression was inhibited in EpoR/SLAP erythroblasts (Figure 4B). Thus, ectopically expressed SLAP interfered with the response of primary erythroblasts to Epo, leading to impaired differentiation and apoptosis. At the molecular level, SLAP-expressing erythroblasts failed to up-regulate BCL-X expression, as normally seen during erythroid differentiation.
Constitutive SLAP expression interferes with Epo-induced survival and BCL-X induction. (A) Quantitative evaluation of apoptosis by TUNEL assay in EpoR and EpoR/SLAP erythroblast clones maintained in the presence of Epo for 24 hours (left panel) or 48 hours (right panel). The mean and standard deviation of 3 independent fields are shown. (B) RT-PCR analyses of BCL-x mRNA expression in EpoR and EpoR/SLAP erythroblasts that were maintained as in panel A. PCR was carried out by using primers specific for either the chicken BCL-x gene (top blot) or the S17 gene as normalization control (bottom blot). Aliquots of 0.5 μL, 1 μL, and 2 μL of each RT product were used. The expected 667-bp BCL-x fragment and 80-bp S17 gene fragment are indicated.
Constitutive SLAP expression interferes with Epo-induced survival and BCL-X induction. (A) Quantitative evaluation of apoptosis by TUNEL assay in EpoR and EpoR/SLAP erythroblast clones maintained in the presence of Epo for 24 hours (left panel) or 48 hours (right panel). The mean and standard deviation of 3 independent fields are shown. (B) RT-PCR analyses of BCL-x mRNA expression in EpoR and EpoR/SLAP erythroblasts that were maintained as in panel A. PCR was carried out by using primers specific for either the chicken BCL-x gene (top blot) or the S17 gene as normalization control (bottom blot). Aliquots of 0.5 μL, 1 μL, and 2 μL of each RT product were used. The expected 667-bp BCL-x fragment and 80-bp S17 gene fragment are indicated.
SLAP interferes with EpoR signaling
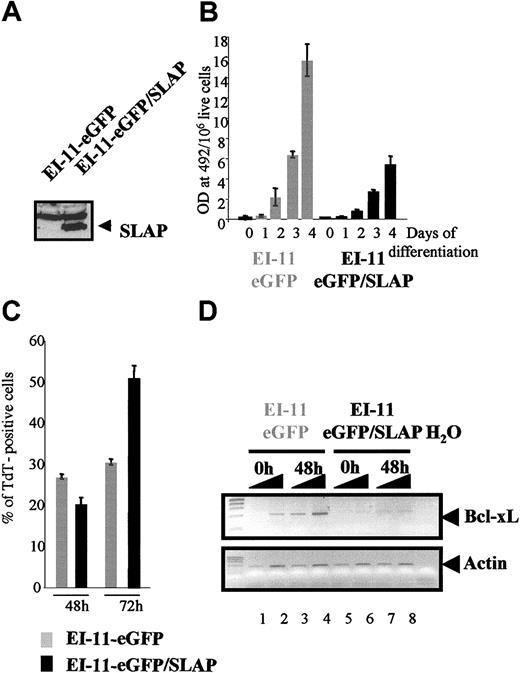
The results reported so far relied on the use of a heterologous system of avian erythroblasts in which EpoR is expressed as an exogenous protein. To verify that the conclusions drawn are not a peculiarity of the cell system, we studied the consequences of SLAP expression in mouse EI-11 erythroblasts.18 EI-11 erythroblasts proliferate as immature cells in the presence of stem cell factor (SCF), Epo, and dexamethasone (Dex) and differentiate into fully mature erythrocytes in response to Epo, thereby recapitulating the normal cellular response of primary erythroblasts. MSCV-eGFP and MSCV-eGFP/mSLAP retroviral vectors were used to transduce EI-11 erythroblasts, and GFP-positive cells were sorted by FACS. Western blot analysis using an anti-Flag antibody confirmed the specific expression of mSLAP in the eGFP/SLAP population (Figure 5A). The Epo response of control and SLAP erythroblasts was next analyzed. As illustrated in Figure 5B, eGFP/SLAP erythroblasts showed an impaired ability to differentiate in response to Epo as cells accumulated lower levels of hemoglobin over time as compared with control eGFP erythroblasts. The inhibition of differentiation imposed by SLAP was accompanied both by an increase in apoptosis late during the differentiation process (Figure 5C) and by impaired up-regulation of BCL-XL expression (Figure 5D). Therefore, as in primary erythroblasts, SLAP overexpression interferes with Epo-dependent cell survival and differentiation of EI-11 cells.
SLAP interferes with Epo-induced differentiation of EI/11 erythroblasts. (A) Expression of exogenous SLAP protein in EI/11-eGFP/SLAP culture was analyzed by Western blotting using an anti-Flag antibody. (B) Hemoglobin content in control eGFP and eGFP/SLAP cells maintained under proliferation conditions (day 0) and after differentiation induction for 1, 2, 3, and 4 days in the presence of Epo. Values were normalized with respect to cell number and volume (hemoglobin level per 106 live cells). The mean value and standard deviation of triplicate measurements are shown. (C) Quantitative evaluation of apoptosis by TUNEL assay in eGFP and eGFP/SLAP cells maintained in differentiation conditions for 48 or 72 hours. (D) RT-PCR analysis of BCL-XL mRNA expression in control eGFP and eGFP/SLAP erythroblasts maintained under either proliferative conditions (0 hours) or induced to differentiate for 48 hours in the presence of Epo. The mean value and standard deviation of 3 independent fields are shown. (D) PCR was carried out using 0.5 μL and 1 μL RT reactions. Primers used were specific for either the BCL-XL gene (top blot) or the β-Actin gene as normalization control (bottom blot). The expected 610-bp BCL-xL fragment and 540-bp Actin gene fragment are indicated.
SLAP interferes with Epo-induced differentiation of EI/11 erythroblasts. (A) Expression of exogenous SLAP protein in EI/11-eGFP/SLAP culture was analyzed by Western blotting using an anti-Flag antibody. (B) Hemoglobin content in control eGFP and eGFP/SLAP cells maintained under proliferation conditions (day 0) and after differentiation induction for 1, 2, 3, and 4 days in the presence of Epo. Values were normalized with respect to cell number and volume (hemoglobin level per 106 live cells). The mean value and standard deviation of triplicate measurements are shown. (C) Quantitative evaluation of apoptosis by TUNEL assay in eGFP and eGFP/SLAP cells maintained in differentiation conditions for 48 or 72 hours. (D) RT-PCR analysis of BCL-XL mRNA expression in control eGFP and eGFP/SLAP erythroblasts maintained under either proliferative conditions (0 hours) or induced to differentiate for 48 hours in the presence of Epo. The mean value and standard deviation of 3 independent fields are shown. (D) PCR was carried out using 0.5 μL and 1 μL RT reactions. Primers used were specific for either the BCL-XL gene (top blot) or the β-Actin gene as normalization control (bottom blot). The expected 610-bp BCL-xL fragment and 540-bp Actin gene fragment are indicated.
One of the earliest events in EpoR signaling is the activation of its associated Janus kinase 2 (JAK2) protein tyrosine kinase followed by STAT5 tyrosine phosphorylation and induction of its binding to DNA.19 We, therefore, compared the ability of STAT5 to bind to DNA in response to Epo in eGFP and eGFP/SLAP erythroblasts. Cells were hormone deprived for 4 hours and stimulated with Epo for 10 or 30 minutes, respectively. STAT5 DNA binding was analyzed by electrophoretic mobility shift assay (EMSA) by using a STAT5-specific 32P-labeled double-stranded oligonucleotide probe. Binding to an ETS-specific 32P-labeled oligonucleotide probe was used as normalization control. As shown in Figure 6A, maximum STAT5 DNA binding was observed 10 minutes after Epo stimulation (compare lanes 2 and 3) and decreased thereafter (compare lanes 3 and 4). Competitive EMSA and super shift experiments using an anti-STAT5-specific antibody confirmed that this complex corresponded to the specific binding of STAT5 to the oligonucleotide probe (data not shown). In contrast, the ability of STAT5 to bind DNA was strongly inhibited in SLAP-expressing erythroblasts (Figure 6A, compare lanes 3 and 6). Quantification of the experiment showed a 10-fold inhibition of STAT5 DNA binding in eGFP/SLAP erythroblasts as compared with control cells. STAT5 tyrosine phosphorylation was also analyzed in the same protein extracts that were used in EMSA. As previously reported,20 STAT5 was rapidly and transiently phosphorylated in response to Epo in control eGFP cells (Figure 6Bi). STAT5 activation was attenuated in SLAP erythroblasts as evidenced by its reduced tyrosine phosphorylation after Epo stimulation as compared with control cells (Figure 6Bi). Inhibition of STAT5 was not due to general quenching of signal transduction by SLAP because no significant difference in Epo-induced ERK1/2 activation was observed between eGFP and eGFGP/SLAP erythroblasts (Figure 6Biii).
SLAP interferes with STAT5 activation. (A) Electrophoretic mobility shift assay analysis. (Top blot) Cell extracts were tested for STAT5 DNA binding by using a STAT5-specific [32P]-labeled probe. (Bottom blot) The same extracts were incubated with a [32P]-labeled ETS probe, as loading control. The STAT5-containing specific complex is indicated by an arrow. The arrowhead points to a nonspecific complex. (B) Phosphorylation of STAT5 (i) and ERK (iii) was analyzed by Western blotting with the use of a STAT5 A/B phosphotyrosine-specific antibody or a phospho-ERK antibody. Immunoblotting with antibodies to STAT5 (ii) and ERK2 (iv) was used to quantify the total amount of STAT5 and ERK. (C) SLAP interferes with STAT5 phosphorylation during differentiation. Control eGFP and eGFP/SLAP EI-11 erythroblasts were induced to differentiate in response to Epo. Lysates were collected at 0, 24, 48, and 72 hours and analyzed by Western blot using the STAT5 A/B phosphotyrosine-specific antibody (top blot) or a STAT5 antibody to assess the total amount of STAT5 (bottom blot).
SLAP interferes with STAT5 activation. (A) Electrophoretic mobility shift assay analysis. (Top blot) Cell extracts were tested for STAT5 DNA binding by using a STAT5-specific [32P]-labeled probe. (Bottom blot) The same extracts were incubated with a [32P]-labeled ETS probe, as loading control. The STAT5-containing specific complex is indicated by an arrow. The arrowhead points to a nonspecific complex. (B) Phosphorylation of STAT5 (i) and ERK (iii) was analyzed by Western blotting with the use of a STAT5 A/B phosphotyrosine-specific antibody or a phospho-ERK antibody. Immunoblotting with antibodies to STAT5 (ii) and ERK2 (iv) was used to quantify the total amount of STAT5 and ERK. (C) SLAP interferes with STAT5 phosphorylation during differentiation. Control eGFP and eGFP/SLAP EI-11 erythroblasts were induced to differentiate in response to Epo. Lysates were collected at 0, 24, 48, and 72 hours and analyzed by Western blot using the STAT5 A/B phosphotyrosine-specific antibody (top blot) or a STAT5 antibody to assess the total amount of STAT5 (bottom blot).
We also analyzed STAT5 activation status during the differentiation process. As shown in Figure 6C, STAT5 activation was minimal when the cells were maintained under self-renewal conditions. Twenty-four hours after differentiation induction, STAT5 tyrosine phosphorylation was clearly enhanced and began to decline thereafter, to reach undetectable levels 3 days after differentiation induction. STAT5 tyrosine phosphorylation was attenuated in eGFP/SLAP erythroblasts 24 hours after differentiation-induction (Figure 6C, compare lanes 2 and 6). Interestingly, STAT5 inhibition was transient in SLAP-expressing erythroblasts, as the same level of STAT5 tyrosine phosphorylation was observed in both control and eGFP/SLAP cells at 48 hours after differentiation-induction (Figure 6C, compare lanes 3 and 7).
SLAP physically associates with EpoR
As SLAP has been described to associate with different signaling molecules, we investigated whether SLAP can be found in a physical complex with EpoR by using a coimmunoprecipitation approach. Lysates obtained from eGFP control and eGFP/SLAP erythroblasts were immunoprecipitated with an anti-EpoR antibody. Coimmunoprecipitated SLAP protein was investigated by Western blotting analysis with the use of the anti-Flag antibody. As shown in Figure 7A, a band of 32 to 34 kDa that comigrated with Flag-SLAP (Figure 7A, lanes 4 and 6) was specifically recovered in the EpoR immunoprecipitates from the EI-11 eGFP/SLAP cell lysates (Figure 7A, compare lane 1 with lane 2). Of note, the same amount of immunoprecipitated-EpoR protein was found in control EI-11 eGFP and EI-11 eGFP/SLAP cells (Figure 7A, bottom blot). To further analyze whether SLAP can directly interact with EpoR, coimmunoprecipitation analyses were performed by using 293T cells transfected with expression plasmids for these proteins. As shown in Figure 7B, the Flag-tagged SLAP protein was detected in the anti-EpoR immunoprecipitate (left blot). This coimmunoprecipitation is specific because SLAP was not immunoprecipitated by the anti-EpoR antibody when expressed alone (Figure 7B, compare lanes 3 and 4). Conversely, EpoR was recovered in the anti-SLAP immunoprecipitate (Figure 7B, lanes 5 and 6). In contrast, similar analyses failed to detect an interaction of SLAP with either JAK2 or STAT5 in EpoR-expressing 293T cells (data not shown). We conclude from these experiments that SLAP specifically interacts with EpoR. With the use of this reconstituted system we also found that expression of SLAP resulted in inhibition of STAT5 Epo-induced phosphorylation (data not shown).
SLAP interacts with EpoR. (A) eGFP and eGFP/SLAP EI-11 erythroblasts were lysed, and cell lysates were immunoprecipitated (lanes 1 and 2) with an EpoR antibody and immunoprecipitated proteins subject to Western blotting analysis using either anti-Flag (top blot) or anti-EpoR (bottom blot) antibodies. Immunoprecipitated EpoR (bottom blot) and coimmunoprecipitated SLAP (top blot) are indicated by arrowheads. Whole cell lysates from eGFP (lane 3) and eGFP/SLAP (lane 4) EI-11 cells and from 293T cells transfected either with plasmids encoding EpoR (lane 5) or Flag-SLAP (lane 6) as well as nontransfected control (lane 7) were analyzed by Western blot to identify EpoR (arrowhead in bottom blot) and SLAP proteins (arrowhead in top blot). (B) 293T cells were transfected with expression plasmids for EpoR, Flag-SLAP, or cotransfected with both expression plasmids as indicated. (Top blots) Cell lysates were immunoprecipitated with EpoR-specific (left blot) or SLAP-specific (right blot) antibodies, and coimmunoprecipitated proteins were identified by Western blot using anti-Flag or anti-EpoR antibodies as indicated. Coimmunoprecipitated proteins are indicated by arrowheads. (Bottom blot) The same immunoprecipitates were analyzed by Western blot by using the same antibody as that used in the immunoprecipitation step. Immunoprecipitated proteins are indicated by arrowheads.
SLAP interacts with EpoR. (A) eGFP and eGFP/SLAP EI-11 erythroblasts were lysed, and cell lysates were immunoprecipitated (lanes 1 and 2) with an EpoR antibody and immunoprecipitated proteins subject to Western blotting analysis using either anti-Flag (top blot) or anti-EpoR (bottom blot) antibodies. Immunoprecipitated EpoR (bottom blot) and coimmunoprecipitated SLAP (top blot) are indicated by arrowheads. Whole cell lysates from eGFP (lane 3) and eGFP/SLAP (lane 4) EI-11 cells and from 293T cells transfected either with plasmids encoding EpoR (lane 5) or Flag-SLAP (lane 6) as well as nontransfected control (lane 7) were analyzed by Western blot to identify EpoR (arrowhead in bottom blot) and SLAP proteins (arrowhead in top blot). (B) 293T cells were transfected with expression plasmids for EpoR, Flag-SLAP, or cotransfected with both expression plasmids as indicated. (Top blots) Cell lysates were immunoprecipitated with EpoR-specific (left blot) or SLAP-specific (right blot) antibodies, and coimmunoprecipitated proteins were identified by Western blot using anti-Flag or anti-EpoR antibodies as indicated. Coimmunoprecipitated proteins are indicated by arrowheads. (Bottom blot) The same immunoprecipitates were analyzed by Western blot by using the same antibody as that used in the immunoprecipitation step. Immunoprecipitated proteins are indicated by arrowheads.
Discussion
In line with the critical role of FLI-1 in F-MuLV-induced erythroleukemia, the enforced expression of FLI-1 profoundly modifies the normal response of erythroblasts to Epo. Overexpression of FLI-1 in the HB60 mouse erythroblastic cell line and primary avian erythroblasts results in the inhibition of Epo-induced differentiation and in the rerouting of the Epo-induced differentiation signal into a proliferative response.9,10 Similarly, enforced FLI-1 expression in the EI-11 erythroblast cell line18 and G1E-ER erythroblasts21 impairs their Epo-induced terminal differentiation (C.T.Q. and J.G., unpublished observations, January 2001). Taken together, these observations suggest that one way FLI-1 could interfere with the response of erythroblasts to Epo is through deregulation of the expression of adaptors or effectors able to directly modify the EpoR signaling pathway. In this report, we show that FLI-1-transformed cells up-regulate the SLAP adaptor molecule. SLAP was found to bind EpoR and to interfere with specific aspects of EpoR signaling during differentiation, eventually altering normal cellular response of erythroblasts to Epo.
Expression of exogenous SLAP at levels similar to those found in FLI-1-transformed primary erythroblasts failed to affect the early stages of Epo-induced differentiation, but specifically interfered with cell survival, hemoglobinization, and morphologic differentiation at later times. Epo/EpoR signaling is essential to red blood cell development because disruption of the genes encoding Epo or EpoR in mice suppresses definitive erythropoiesis.22,23 Although the fetal livers of these mice contain normal numbers of erythroid burst-forming units (BFU-Es) and erythroid colony-forming units (CFU-Es), terminal differentiation fails to occur because of a block at the erythroblast stage. Binding of Epo to its receptor induces multiple signaling pathways, the first detectable event being the autophosphorylation and activation of the receptor-associated JAK2 tyrosine kinase. Once activated, JAK2 transduces signals by activation of downstream targets either directly or through phosphorylation of specific tyrosine residues in the cytoplasmic tail of EpoR. This transduction results in the recruitment and often redundant activation of effectors and adaptors, including STAT5, phosphatidyl inositol-3 kinase (PI3K), growth factor-bound protein 2 (Grb2), Grb2-associated binder-1 and -2 (Gab1/2), Src homology 2 domain-containing protein 1 and 2 (SHP1/2), and SH2-containing inositol-5-phosphatase (SHIP).19 We found that, whereas SLAP interferes with Epo-induced survival and differentiation properties of both avian and murine erythroblasts, it failed to affect the implementation of the intrinsic differentiation program of these cells (Figure 3). These data favor a model whereby SLAP inhibition of terminal differentiation specifically results from its ability to impair EpoR signaling event(s) involved in erythroblast survival/differentiation rather than from its ability to interfere with the erythroid differentiation program per se. Consistent with this model, we found that (1) SLAP physically associates with EpoR both in erythroblasts and following its coexpression with EpoR in nonerythroid cells; (2) SLAP expression interferes with early, EpoR proximal signaling events such as STAT5 activation; and (3) SLAP interferes with the up-regulation of BCL-XL gene expression, an event which normally relies on the cooperation of Epo and GATA-1.24
BCL-X is a gene essential to erythroid differentiation, as its inactivation by homologous recombination leads to hemolytic anemia because of impaired survival of late-differentiating erythroblasts.17,25 Consistent with this finding, BCL-XL expression is strongly increased late during the differentiation process, at the time of globin gene induction and hemoglobinization, whereas the expression of other BCL-2 family members remain relatively constant.16,26 We observed that up-regulation of BCL-XL is impaired in SLAP-expressing erythroblasts, which strongly suggests that this default in BCL-XL induction contributes significantly to the impaired survival and differentiation of these cells.
The significance of the inhibition of STAT5 tyrosine phosphorylation and DNA binding in SLAP-expressing erythroblasts is more difficult to assess mainly because the role of STAT5 in erythroid differentiation itself is debated. On the one hand, gene inactivation studies have shown that STAT5 function is not required during normal mouse erythropoiesis.27 On the other hand, STAT5-deficient mice are anemic during their fetal life and most of their adult life.28,29 This property is paralleled by enhanced susceptibility of STAT5-/- erythroblasts to apoptosis ex vivo and an impaired response of STAT5-/- mice to erythropoietic stress.28,29 Interestingly, we found that STAT5 activation is increased during the differentiation process, reaching maximum levels at 24 hours after differentiation induction and decreasing afterward. Our results show that SLAP interferes with STAT5 activation normally observed at 24 hours of the differentiation process, suggesting that attenuation of STAT5 activation in SLAP erythroblasts could contribute to the effects of SLAP on terminal differentiation.
Enforced expression of SLAP does not impair EpoR processing and/or signaling in a general manner, as only some of the signaling events located downstream of the EpoR are affected. Indeed, enforced expression of SLAP failed to significantly inhibit Epo-induced EpoR tyrosine phosphorylation (I.L. and C.T.Q., unpublished observations, June 2002) and activation of the ERK1/2 pathway. Available evidence indicates that Epo-induced activation of ERK involves several redundant pathways, distinct from those implicated in STAT5 activation.30,31 This suggests that SLAP may (1) specifically inhibit the phosphorylation of EpoR on tyrosine residue(s) critical for STAT5 activation such as Y343,29 (2) interfere with the recruitment of inactive STAT5 to the phosphorylated EpoR, or (3) inhibit STAT5 phosphorylation by EpoR-associated JAK2. Comparison of the pattern of known and to be discovered EpoR-associated proteins between control and SLAP-expressing erythroblasts should allow a better understanding of the molecular mechanism underlying SLAP properties.
SLAP is a negative regulator of TCR and platelet-derived growth factor receptor (PDGF-R) signaling,14,32 and inactivation of SLAP enhances positive selection during mouse thymocyte development.33 In activated Jurkat cells, SLAP is complexed with tyrosine-phosphorylated CD3, ZAP70, lymphocyte cell kinase (LCK), SLP76, and VAV14 and associates with tyrosine-phosphorylated PDGF-R itself following PDGF stimulation of fibroblasts.32 Our results describe for the first time the association of SLAP with a cytokine receptor, EpoR. In contrast to the PDGF-R/SLAP complex, which is dependent on PDGF-R tyrosine-phosphorylation, the recruitment of SLAP to the EpoR was found both with the phosphorylated and unphosphorylated forms of EpoR (I.L. and C.T.Q., unpublished observations, June 2002). Disruption of SLAP results in marked up-regulation of TCR expression at the cell surface of thymocytes at the CD4+CD8+ stage of differentiation. Because SLAP has been shown to localize to endosomes,14 it has been suggested that SLAP could be involved in TCR down-regulation and degradation.33 Further studies are required to analyze whether the effects of SLAP on the response of erythroblasts to Epo involves an interference with EpoR trafficking.
Similar to SLAP-expressing erythroblasts, FLI-1-transformed progenitors also show inhibition of both Epo-induced STAT5 activation and of BCLXL up-regulation as compared with control erythroblasts (C.T.Q., unpublished observations, February 2002). It is, therefore, tempting to speculate that the deregulated expression of SLAP in FLI-1-transformed erythroblasts is at least in part responsible for the inhibition of STAT5 and BCL-X activation in these cells, a hypothesis that remains to be tested. Our results, therefore, suggest that FLI-1-transforming properties rely (1) on its ability to deregulate the expression of antiapoptotic genes to confer a survival advantage to immature erythroblasts in the absence or in the presence of suboptimal amounts of antiapoptotic cytokines or stroma-derived signals3 and (2) on the activation of adaptors like SLAP to inhibit signaling pathway(s) critical to erythroblasts' maturation and/or survival, late in the differentiation process.
Prepublished online as Blood First Edition Paper, August 28, 2003; DOI 10.1182/blood-2003-06-2077.
Supported by funds from CNRS, Institut National de la Santé et de la Recherche Médicale (INSERM), Institut Curie, Ligue Nationale Contre le Cancer (LNCC; équipe labellisée), and European Union (HPRNCT-2000-00083). I.L. was supported by a predoctoral fellowship from LNCC.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr O. Acuto, S. Roche, H. Singh, and A. Weiss for reagents; M. Pironin and E. Deiner for expert technical assistance; and Janssen-Cilag for its generous gift of recombinant Epo.

![Figure 6. SLAP interferes with STAT5 activation. (A) Electrophoretic mobility shift assay analysis. (Top blot) Cell extracts were tested for STAT5 DNA binding by using a STAT5-specific [32P]-labeled probe. (Bottom blot) The same extracts were incubated with a [32P]-labeled ETS probe, as loading control. The STAT5-containing specific complex is indicated by an arrow. The arrowhead points to a nonspecific complex. (B) Phosphorylation of STAT5 (i) and ERK (iii) was analyzed by Western blotting with the use of a STAT5 A/B phosphotyrosine-specific antibody or a phospho-ERK antibody. Immunoblotting with antibodies to STAT5 (ii) and ERK2 (iv) was used to quantify the total amount of STAT5 and ERK. (C) SLAP interferes with STAT5 phosphorylation during differentiation. Control eGFP and eGFP/SLAP EI-11 erythroblasts were induced to differentiate in response to Epo. Lysates were collected at 0, 24, 48, and 72 hours and analyzed by Western blot using the STAT5 A/B phosphotyrosine-specific antibody (top blot) or a STAT5 antibody to assess the total amount of STAT5 (bottom blot).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/13/10.1182_blood-2003-06-2077/6/m_h82435379006.jpeg?Expires=1769128826&Signature=J29Hq4UXVGP~OXeiuj7iKC7b3yqPdbukocaKiReirlcr4LyPf93cV0~nK~RULxunsw5MuICHVBNCeb1GbhGtcj6uMoEN7a5tSZvLAyJirkLyCq4wwbvHJtXofGborm6x17EMKnLMgeIDmGbM0pMAHcNpu6~DkP~lOxVFpsvY9l~HRXhY73Gn~ujfepatbGikPl3A7ZZfuigEZNKKnwaCQ9hcd4XJS6hMB6xMMphjPg8Zuw9-gD6RtHEVL4JFPEqMSDqvLxa27Clrk69aHQ3amHLhRXRtA6Y7QFZyhW9P-B6yKajH7zldvYT85H5XQXxr5cPw2chp3pAPBReY80Jn5A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal