Abstract
Interferon consensus sequence binding protein/interferon regulatory factor 8 (ICSBP/IRF-8) is a transcription factor that controls myeloid cell development. ICSBP-/- mice develop a chronic myelogenous leukemia (CML)-like syndrome. Several observations on patients and mouse models have implicated ICSBP in the pathogenesis of CML. In this paper, we investigated whether ICSBP modulates the growth-promoting activity of Bcr/Abl, the causal oncoprotein for CML. When transformed with p210 Bcr/Abl, ICSBP-/- myeloid progenitor cells lost growth factor dependence and grew in the absence of granulocyte-macrophage colony-stimulating factor. When ICSBP was ectopically expressed, Bcr/Abl-transformed cells underwent complete growth arrest and differentiated into mature, functional macrophages without inhibiting the kinase activity of Bcr/Abl. Providing a mechanistic basis for the growth arrest, ICSBP markedly repressed c-Myc messenger RNA (mRNA)-expression, a downstream target of Bcr/Abl. A further analysis with the ICSBP/estrogen receptor chimera showed that ICSBP repression of c-Myc is indirect and is mediated by another gene(s). We identified Blimp-1 and METS/PE1, potent c-Myc repressors, as direct targets of ICSBP activated in these cells. Consistent with this, ectopic Blimp-1 repressed c-Myc expression and inhibited cell growth. These results indicate that ICSBP inhibits growth of Bcr/Abl-transformed myeloid progenitor cells by activating several genes that interfere with the c-Myc pathway. (Blood. 2003;102:4547-4554)
Introduction
The p210 Bcr/Abl oncoprotein is encoded by the Bcr/Abl fusion gene generated by t(9;22) chromosomal translocation, which is essential and sufficient for the development of a clonal myeloproliferative disorder, chronic myelogenous leukemia (CML), in humans.1 CML begins with a chronic phase in which myeloid precursors and mature cells (especially granulocytes) expand in the bone marrow, blood, and extramedullary tissues. After 3 to 5 years, the disease progresses into a fatal blast crisis, characterized by the accumulation of immature precursor cells.2,3 Mainly through the deregulated Abl kinase activity, Bcr/Abl alters adhesion to stroma cells and extracellular matrix, activates mitogenic signaling, and reduces apoptosis, causing malignant transformation of hematopoietic cells.4 It has been shown that Bcr/Abl activates multiple signaling pathways, including the Myc, Ras, mitogen-activated protein (MAP) kinase, phosphatidylinositol 3 (PI3) kinase, Janus kinase (JAK), and signal transducers and activators of transcription (STAT) pathways. So far, allogeneic stem cell transplantation has been the only curative therapy for CML (up to 70% cure rates in chronic phase).5 However, because of the limitations of donor availability, the age of patients, and accompanied complications, only 5% of newly diagnosed patients are cured by allogeneic stem cell transplantation.6 Treatment with interferon α (IFNα) in chronic phase has prolonged survival of CML patients by selectively suppressing Bcr/Abl-positive cells, yet complete cytogenetic remission is achieved in only 10% to 20% of patients.5 Recently, the Bcr/Abl kinase inhibitor imatinib mesylate (STI571) was shown to be very effective even in IFNα-resistant patients.6,7 However, there are patients resistant to the drug, found especially among those in the late stage of chronic phase and in blast crisis, presumably due to secondary genetic changes including point mutations in or amplification of the Bcr/Abl gene.
Several lines of evidence have indicated the role of a transcription factor interferon consensus sequence binding protein (ICSBP)/interferon regulatory factor 8 (IRF-8) in the pathogenesis of this leukemia and its role in regulating the activity of Bcr/Abl: (1) ICSBP-/- mice develop a CML-like syndrome8 ; (2) ICSBP transcripts are absent in CML patients, and IFNα used for the therapy of CML induces ICSBP expression in vivo9 ; (3) ICSBP expression correlates with pretreatment risk features and cytogenetic response to IFNα in CML10 ; (4) coexpression of ICSBP can ameliorate p210 Bcr/Abl-mediated myeloid leukemia in mice11 ; and (5) ICSBP induces potent immunity against p210 Bcr/Abl-induced leukemia in mice.12 Despite the circumstantial evidence, however, whether ICSBP directly affects growth and differentiation in Bcr/Abl-transformed myeloid progenitor cells has yet to be elucidated.
ICSBP is a hematopoietic cell-specific transcription factor belonging to the IRF family.13 ICSBP messenger RNA (mRNA) is detected in lineage marker (-) cells14 and a purified stem cell population, Sca-1 (+) AA4.1+ c-Kit+ lineage marker-/low cells.15 While ICSBP expression is high in macrophages, B cells, and some dendritic cells (DCs), it is very low in granulocytes and unstimulated T cells. ICSBP mRNA is inducible by IFNγ through the IFNγ activation site (GAS) in the ICSBP promoter. ICSBP controls myeloid cell development by stimulating macrophage differentiation while inhibiting granulocyte differentiation, in both cases inhibiting cell growth.14,16 More recently, our laboratory and others found essential roles of ICSBP in development and function of DCs.17-20 ICSBP-/- mice are unable to produce interleukin-12 (IL-12) and IFNγ, defective in mounting a Th1 response, and immunodeficient against various pathogens. It was shown that ICSBP is a positive regulator of apoptosis in myeloid cells.21
By analyzing p210 Bcr/Abl-transformed ICSBP-/- myeloid progenitor cells we show that ICSBP can trigger growth arrest and macrophage differentiation even in the presence of Bcr/Abl. Furthermore, ICSBP repressed c-Myc gene expression, one of the critical mitogenic pathways activated by Bcr/Abl. We identified 2 target genes, B-lymphocyte-induced maturation protein-1 (Blimp-1) and mitogenic Ets transcriptional suppressor (METS/PE1), that are activated by ICSBP and repress c-Myc expression. Our results show that ICSBP antagonizes transcriptional pathways critical for Bcr/Abl-mediated growth promotion.
Materials and methods
Cells, reagents, and mice
Establishment and culture condition of an ICSBP-/- myeloid progenitor cell line Tot2 were described previously.16 Murine granulocyte-macrophage colony-stimulating factor (GM-CSF; Biosource International, Camarillo, CA), stem cell factor (SCF), IL-6, IL-3 (PeproTech, Rocky Hill, NJ), and macrophage colony-stimulating factor (M-CSF; BioSource International) were used at 2.5, 50, 10, 6, and 10 ng/mL, respectively. Morphologic differentiation was monitored by Wright-Giemsa staining (Sigma, St Louis, MO) of cytospin preparations. Imatinib mesylate (STI571) was a generous gift from Dr Richard J. Maraia (NICHD, Bethesda, MD). Lipopolysaccharide (LPS) from Escherichia coli (Sigma) was used at 200 ng/mL. IFNγ, a generous gift from Dr Atsuko Masumi (National Institute of Infectious Diseases, Tokyo, Japan), was added at 200 U/mL. β-Estradiol (Sigma) was dissolved in ethanol and used at 1 μM. The final concentration of ethanol was 0.1%, which did not affect growth and differentiation of Tot2p210 cells. Cycloheximide (Sigma; 10 μg/mL) was added 10 minutes before addition of estradiol and inhibited more than 95% of protein synthesis in Tot2 cells. ICSBP-/- and ICSBP+/+ mice8 on a C57BL/6 background were 6 to 10 weeks old and were housed under specific pathogen-free conditions. Bone marrow cells from 2 to 3 mice were collected, and mononuclear cells were prepared by density gradient centrifugation on Histopaque-1083 (Sigma). All animal work conformed to the guidelines of the animal care and use committee of the National Institute of Child Health and Human Development (Bethesda, MD).
DNA constructs
The p210 Bcr/Abl complementary DNA (cDNA)22 was blunt ended and ligated into pMSCV-hyg (Clontech, Palo Alto, CA) to generate MSCV-p210. To construct the ICSBP/estrogen receptor (ICSBP/ER) chimera, the stop codon of ICSBP cDNA was removed by polymerase chain reaction (PCR) using Pfu polymerase (Stratagene, La Jolla, CA) then ligated in-frame with the hormone-binding domain of human estrogen receptor cDNA23 in pcDNA3.1(+) (Invitrogen, Carlsbad, CA) and pMSCV-puro (Clontech) by using appropriate restriction enzymes. As control constructs, the hormone-binding domain of human estrogen receptor cDNA was subcloned into EcoRI sites of pcDNA3.1(+) and pMSCV-puro. In vitro-translated ICSBP/ER formed a complex with IRF-2 and PU.1 on IFN-stimulated response element (ISRE) and Ets-IRF composite element (EICE), respectively, in electrophoretic mobility shift assay, as did ICSBP itself (data not shown). Bicistronic pMSCV-CD8t vector24 was a generous gift from Dr Ricardo A. Feldman (University of Maryland School of Medicine, Bethesda, MD). Mouse Blimp-1 cDNA (XhoI-HpaI) was ligated into pMSCV-CD8t. The nucleotide sequence of all constructs containing PCR-derived DNA was confirmed by sequencing.
Retroviral transduction
Retroviral transduction into Tot2 and Tot2p210 cells was carried out as previously reported16 with slight modifications. Briefly, BOSC23 ecotropic retroviral packaging cells25 (American Type Culture Collection [ATCC], Manassas, VA) were transiently transfected with pMSCV retroviral vectors26 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Retroviral supernatant was collected at 48 hours. Cells (2 × 105 per one well of 24-well plate) were transduced by spinoculation (2500 rpm, 33°C, 2 × 1 hour) in 2 mL of retroviral cocktail containing 75% viral supernatant, 2.5 ng/mL GM-CSF (for Tot2 cells), and 4 μg/mL polybrene (Sigma). Virus titers measured on NIH3T3 cells or 32D cells were more than 1 × 106 infectious particles/mL, and transduction efficiency was 10% to 25% except MSCV-Blimp-1, which showed only 3% transduction efficiency in Tot2p210 cells, presumably because of rapid growth inhibition by Blimp-1. Transduced cells were selected either by adding puromycin (2 μg/mL) or by immunomagnetic cell sorting using magnetic-activated cell separation (MACS) CD8 microbeads (Miltenyi Biotec, Auburn, CA) according to manufacturer's protocol on day 2. More than 95% of untransduced cells were eliminated by puromycin within 2 days. The purity of sorted cells was more than 90%. Lineage marker-negative (lin-) cells were prepared from bone marrow cell suspension, as described previously,14 by immunomagnetic cell depletion using murine progenitor enrichment cocktail (StemCell Technologies, Vancouver, BC, Canada). Retroviral transduction of lin- cells was performed as described previously.14 Briefly, lin- cells were cultured in the presence of SCF, IL-6, and IL-3 for 1 day and were transduced with MSCV retroviruses by spinoculation on day 1 and day 2. On day 3, cells were washed and reinoculated in the presence of SCF and M-CSF. Transduced cells were selected by puromycin (0.5 μg/mL) and were further cultured until day 10. Transduction efficiency was 35% to 70%.
Semiquantitative and quantitative polymerase chain reaction with reverse transcription
Total RNA was prepared using Trizol (Invitrogen), and first strand complementary DNA (cDNA) was synthesized with Superscript II (Invitrogen) according to the manufacturer's instruction. The seminquantitative polymerase chain reaction (PCR) was performed as follows: 3 minutes of denaturing at 94°C and 30 cycles of amplification at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 45 seconds, followed by 7 minutes of extension at 72°C using Taq DNA polymerase (Promega, Madison, WI). To achieve semiquantitative measurement, the amount of cDNA appropriate for the reaction was determined before experiments. cDNA derived from 1 ng of total RNA was used for β-actin, 2 ng for myeloperoxidase (MPO) and c-Myc and 50 ng for other genes. The following primers were used: c-Myc (sense: 5′-gaa cta tga cct cga cta cga c-3′, antisense: 5′-atc gga cga ggt aca gga ttt g-3′); Blimp-1 (sense: 5′-gtt tcc tat ggt tcc gag ggc c-3′, antisense: 5′-tgt tct gac gtg gct gca gtt c-3′); and METS (sense: 5′-cac ctc tag aca gcc att ctc c-3′, antisense: 5′-ttg aac acc gaa cag gct tga g-3′). Primers for other genes were as described elsewhere.16 PCR products were resolved on 1.8% agarose gel electrophoresis and stained with ethidium bromide. Bands were visualized in an UV transilluminator Eagle-Eye system (Stratagene). Quantitative PCR was performed in triplicate by the real-time fluorescence detection method with the SYBR green PCR master kit (Applied Biosystems, Foster City, CA) using an ABI PRISM 7000 sequence detection system (Applied Biosystems) according to manufacturer's protocol. cDNAs derived from 2 ng of total RNA were used for PCR. The primer sets used were: Blimp-1 (sense: 5′-ttg agc acc atg aac aac atc a-3′, antisense: 5′-gac ggg ata caa cct agg gaa ga-3′); METS (sense: 5′-ggc cct ctg tgt cca tta gc-3′, antisense: 5′-tca ctg tct tcg gtc ccc tct a-3′); c-Myc (sense: 5′-acg aca aga ggc gga cac a-3′, antisense: 5′-gct gcg ctt cag ctc gtt-3′); and glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) (sense: 5′-gtg ttc cta ccc cca atg t-3′, antisense: 5′-tgt cat cat act tgg cag gtt tc-3′). SYBR green was incorporated into the reaction mixture to permit product measurement. Each of the primer sets gave a unique product. Transcript levels were normalized by GAPDH levels.
Cell cycle analysis
Cells were fixed in cold 70% ethanol, treated with 100 ng/mL RNaseA (Sigma), and stained with 50 μg/mL propidium iodide. Stained cells were analyzed on FACSCalibur (Becton Dickinson, San Jose, CA). Cell cycle profiles were obtained by using CellQuest and ModFitLD V2.0 softwares (Becton Dickinson).
Immunoblot analysis
Cell pellets were directly dissolved and sonicated in Laemmli buffer. Proteins from 5 × 105 cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% or 4%-20% gradient) and transferred to a polyvinylidene fluoride membrane. The anti-ICSBP antibody was rabbit antiserum raised against the C-terminal amino acids of ICSBP and used at 1:2000 dilution. Anti-c-Abl mouse monoclonal antibody (Ab-3; Oncogene, San Diego, CA) was used at 1:300 dilution and antiphosphotyrosine mouse monoclonal antibody (4G10; Upstate Biotech, Lake Placid, NY) was used at 1:1000. Second antibodies were horseradish peroxidase-conjugated donkey antirabbit and sheep antimouse immunoglobulin (Amersham, Piscataway, NJ). Protein expression was detected by enhanced chemiluminescence (NEN, Boston, MA).
RNA blot analysis
Total RNA (5 μg for c-Myc or 10 μg for METS) was electrophoresed on a 1% agarose formaldehyde gel, transferred onto a nylon membrane, and hybridized with 32P-labeled probe prepared by random priming using ReadyToGo (Amersham-Pharmacia, Piscataway, NJ). Probes were PCR amplicons using the primers described above. Templates for the PCR reaction were a plasmid containing mouse c-Myc cDNA (a generous gift from Dr Steven R. Bauer, Food and Drug Administration, Bethesda, MD) and cDNA synthesized from estradiol-treated Tot2p210 cells transduced with ICSBP/ER (for METS). The blots were quantified by an image analyzer FLA-3000 (FUJIFILM, Stanford, CT) and visualized by autoradiography following exposure for 5 days (c-Myc) or 10 days (METS).
Electrophoretic mobility shift assay
The preparation of nuclear extracts and electrophoretic mobility shift assay (EMSA) to detect DNA binding activity for the Blimp-1 binding site (the plasmacytoma repressor factor [PRF] site, 5′-cgc gta cag aaa ggg aaa gga cta g-3′) in the c-Myc promoter were as described27 with slight modifications. Briefly, 10 μg of nuclear extracts and 32P-labeled oligonucleotide probe were incubated in 10 μL reaction mixture containing NaCl (80 mM), 10% glycerol, Tris (20 mM, pH 7.5), EDTA (ethylenediaminetetraacetic acid; 0.2 mM), DTT (dithiothreitol; 0.1 mM), and poly(dI-dC) · (dI-dC) (2 μg) for 30 minutes at 4°C. Anti-Blimp-1 serum27 or control normal rabbit serum was added 30 minutes prior to addition of probe. Complexes were resolved by electrophoresis on a 6% acrylamide-bisacrylamide (37.5:1) gel run in 0.5 × Tris-borate-EDTA buffer at 4°C.
Results
Transformation of ICSBP-/- myeloid progenitor cells by p210 Bcr/Abl
To establish an experimental system for studying the role of ICSBP in the modulation of Bcr/Abl activity, we transduced a retrovirus carrying the p210 Bcr/Abl gene (MSCV-p210) into a GM-CSF-dependent ICSBP-/- myeloid progenitor cell line. This cell line, Tot2, possesses a bipotential myeloid capability to give rise to either macrophages or granulocytes, depending on whether cells were transduced with ICSBP or treated with G-CSF, respectively.16 Two days after Bcr/Abl transduction, GM-CSF was withdrawn and cell growth was monitored. While parental Tot2 cells as well as cells transduced with control MSCV died within a few days after GM-CSF deprivation (data not shown), a significant number of cells remained viable when cells were transduced with p210 Bcr/Abl, and these cells continued to grow in the absence of GM-CSF. Hereafter, p210 Bcr/Abl-transduced Tot2 cells (Tot2p210) were examined in the absence of GM-CSF. Morphologically, Tot2p210 cells remained immature as the parental Tot2 cells, both of which showed a large prominent nucleus and a small basophilic cytoplasm (Figure 1A). Tot2p210 cells grew at a rate similar to parental Tot2. Likewise, they displayed a cell cycle profile similar to Tot2 cells (Figure 1B). Immunoblot analysis confirmed the Bcr/Abl expression in Tot2p210 cells, in which numerous cellular proteins were phosphorylated at tyrosine (Figure 1C).
Transformation of ICSBP-/- myeloid progenitor cells by p210 Bcr/Abl. (A) Wright-Giemsa stain of parental Tot2 cells and Tot2 cells transduced with p210 Bcr/Abl retrovirus (Tot2p210 cells) (original magnification × 600). Tot2 cells were cultured in media containing GM-CSF, whereas Tot2p210 cells were cultured without GM-CSF. (B) Cell cycle analysis. Tot2 and Tot2p210 cells were analyzed for DNA content. (C) Immunoblot analysis for p210 Bcr/Abl and phosphotyrosine-containing proteins. (D) Effect of a Bcr/Abl kinase inhibitor, imatinib mesylate (STI571), on the growth and survival of Tot2 and Tot2p210 cells. Imatinib mesylate was added at indicated concentrations and viable cell number was monitored.
Transformation of ICSBP-/- myeloid progenitor cells by p210 Bcr/Abl. (A) Wright-Giemsa stain of parental Tot2 cells and Tot2 cells transduced with p210 Bcr/Abl retrovirus (Tot2p210 cells) (original magnification × 600). Tot2 cells were cultured in media containing GM-CSF, whereas Tot2p210 cells were cultured without GM-CSF. (B) Cell cycle analysis. Tot2 and Tot2p210 cells were analyzed for DNA content. (C) Immunoblot analysis for p210 Bcr/Abl and phosphotyrosine-containing proteins. (D) Effect of a Bcr/Abl kinase inhibitor, imatinib mesylate (STI571), on the growth and survival of Tot2 and Tot2p210 cells. Imatinib mesylate was added at indicated concentrations and viable cell number was monitored.
Imatinib mesylate (STI571) is a highly selective inhibitor of the protein tyrosine kinase family including Bcr/Abl protein, the platelet-derived growth factor receptor, and the c-kit receptor.7 This inhibitor strongly inhibited the growth and survival of Tot2p210 cells in a dose-dependent manner (Figure 1D). At a dose of 2.0 μM, all Tot2p210 cells died within a few days. In contrast, this inhibitor did not affect GM-CSF-dependent growth of parental Tot2 cells. Thus, p210 Bcr/Abl transformed ICSBP-/- myeloid progenitor cells through its kinase activity and rendered these cells growth factor independent. Consistent with these results, the transforming activity of Bcr/Abl has been reported for factor-dependent cell lines such as 32Dcl.3 and BaF3 cells. We also noted that G-CSF-mediated granulocytic differentiation of Tot2 cells was abolished upon Bcr/Abl expression (data not shown).
Induction of growth arrest and macrophage differentiation following ICSBP expression
To examine the effect of ICSBP on p210 Bcr/Abl-transformed cells, we transduced control (MSCV-puro) or ICSBP retrovirus (MSCV-ICSBP-puro) into Tot2p210 cells. Transduced cells were selected by puromycin and monitored for their growth and differentiation. As shown in Figure 2, ICSBP induced macrophage differentiation in Tot2p210 cells, similar to Tot2 cells run as a control. Wright-Giemsa stain of ICSBP-transduced Tot2p210 cells on day 6 showed a significantly enlarged, less basophilic cytoplasm with numerous vacuoles and a smaller, condensed nucleus, all of which are features of mature macrophages (Figure 2A). In contrast, cells transduced with control virus did not show these morphologic changes.
ICSBP induction of macrophage differentiation in Bcr/Abl-transformed ICSBP-/- myeloid progenitor cells. (A) Wright-Giemsa stain of Tot2 and Tot2p210 cells transduced with the control or ICSBP retrovirus on day 6 after transduction (original magnification × 600). (B) Semiquantitative reverse transcriptase-PCR (RT-PCR) analysis for macrophage differentiation-related genes. Expression of scavenger receptor (SR) and myeloperoxidase (MPO) transcripts was analyzed using RNA samples from indicated days after transduction. β-Actin was used as a control for equal loading. (C) Induction of LPS/IFNγ-responsive genes. Cells on 6 days after transduction were treated with 200 ng/mL LPS or 200 U/mL IFNγ for 6 hours. Indicated transcripts were analyzed by semiquantitative RT-PCR. iNOS indicates inducible nitric oxide synthase; and FcγRI, Fcγ receptor I.
ICSBP induction of macrophage differentiation in Bcr/Abl-transformed ICSBP-/- myeloid progenitor cells. (A) Wright-Giemsa stain of Tot2 and Tot2p210 cells transduced with the control or ICSBP retrovirus on day 6 after transduction (original magnification × 600). (B) Semiquantitative reverse transcriptase-PCR (RT-PCR) analysis for macrophage differentiation-related genes. Expression of scavenger receptor (SR) and myeloperoxidase (MPO) transcripts was analyzed using RNA samples from indicated days after transduction. β-Actin was used as a control for equal loading. (C) Induction of LPS/IFNγ-responsive genes. Cells on 6 days after transduction were treated with 200 ng/mL LPS or 200 U/mL IFNγ for 6 hours. Indicated transcripts were analyzed by semiquantitative RT-PCR. iNOS indicates inducible nitric oxide synthase; and FcγRI, Fcγ receptor I.
Macrophage differentiation was confirmed by expression of the macrophage-specific scavenger receptor gene (Figure 2B). In addition, expression of the myeloperoxidase (MPO) gene, known to be down-regulated during myeloid differentiation, was significantly decreased in ICSBP-transduced Tot2 and Totp210 cells, indicating their terminal maturation. Furthermore, ICSBP-transduced Tot2p210 cells responded to LPS to express the inducible nitric oxide synthase (iNOS) and IL-12 p40 genes, while no induction was detected in control virus-transduced cells (Figure 2C). Induction of these genes following ICSBP transduction was comparable between Tot2p210 and Tot2 cells. ICSBP-transduced cells also expressed the Fcγ receptor I (FcγRI) gene in response to IFNγ, while there was only a very modest induction in control cells. These results indicate that ICSBP can drive Bcr/Abl-transformed myeloid progenitor cells toward mature, functional macrophages.
Importantly, ICSBP introduction caused strong growth inhibition in Tot2p210 cells. In contrast, control virus did not have the effect, as cells grew exponentially (Figure 3A). Cell cycle analysis showed that ICSBP reduced the percentage of cells in S phase by day 4, causing almost complete cell cycle arrest at G0/G1 phase by day 6 both in Tot2 and Tot2p210 cells (Figure 3B).
Cell growth arrest by ICSBP in Bcr/Abl-transformed ICSBP-/- myeloid progenitor cells. (A) Growth rate of Tot2 and Tot2p210 cells transduced with the indicated viruses. Total cell yields are counted on the indicated days. (B) Cell cycle analysis. Cells transduced with control or ICSBP retrovirus were analyzed for DNA contents on the indicated days.
Cell growth arrest by ICSBP in Bcr/Abl-transformed ICSBP-/- myeloid progenitor cells. (A) Growth rate of Tot2 and Tot2p210 cells transduced with the indicated viruses. Total cell yields are counted on the indicated days. (B) Cell cycle analysis. Cells transduced with control or ICSBP retrovirus were analyzed for DNA contents on the indicated days.
We then sought to determine whether cell growth arrest and differentiation induced by ICSBP was due to an alteration of the kinase activity of Bcr/Abl in Tot2p210 cells. As shown in Figure 4, neither the expression of p210 Bcr/Abl nor tyrosine phosphorylation of cellular proteins was inhibited by ICSBP, indicating that ICSBP does not globally inhibit the kinase activity of p210 Bcr/Abl.
Expression of p210Bcr/Abl and phosphotyrosine-containing proteins. Immunoblot analyses were performed with Tot2p210 cells at 3 days after transduction of control or ICSBP retrovirus.
Expression of p210Bcr/Abl and phosphotyrosine-containing proteins. Immunoblot analyses were performed with Tot2p210 cells at 3 days after transduction of control or ICSBP retrovirus.
Down-regulation of c-Myc gene expression by ICSBP
During the course of this study we came to notice that c-Myc mRNA expression is reduced in Tot2 cells after ICSBP expression. The c-Myc gene encodes a basic helix-loop-helix leucine zipper (bHLHZip) transcription factor that regulates proliferation, differentiation, and apoptosis in many cell types including myeloid cells.28 c-Myc is shown to be required for the oncogenic activity of Bcr/Abl.29 Thus, it was of importance to assess whether ICSBP down-regulates c-Myc expression in the Bcr/Abl-transformed Tot2 cells. Data in Figure 5A show that c-Myc transcripts were expressed in p210 cells at a level similar to that of parental Tot2 cells. However, ICSBP transduction resulted in a marked reduction in c-Myc expression in both Tot2 and p210 cells, suggesting that ICSBP is capable of impeding Bcr/Abl activation of the c-Myc pathway. These results raised 2 possibilities: (1) ICSBP directly inhibits c-Myc transcription, or (2) ICSBP regulates c-Myc expression indirectly by activating another protein, which in turn represses c-Myc expression.
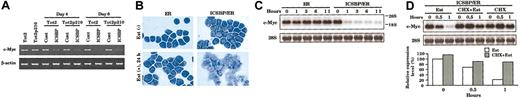
Down-regulation of c-Myc expression in ICSBP-transduced cells. (A) Semiquantitative RT-PCR analysis for c-Myc transcripts. (B) Wright-Giemsa stain of Tot2p210 cells transduced with retroviruses carrying hormone-binding domain of estrogen receptor (ER) or ICSBP/ER chimeric construct. Cells were either left untreated or treated with 1 μM β-estradiol (Est) for 24 hours (original magnification × 600). (C) RNA blot analysis for the c-Myc transcripts. Tot2p210 cells transduced with indicated viruses were treated with 1 μM β-estradiol. Total RNA (5 μg) from cells treated for the lengths of time was analyzed. The bottom panel indicates 28S ribosomal RNA. (D) Effect of cycloheximide (CHX) on ICSBP/ER-mediated repression of c-Myc expression. The top panel indicates RNA blot analysis; the bottom panel, quantification of the c-Myc transcripts. CHX (10 μg/mL) was added 10 minutes before addition of estradiol. The expression level in CHX + Est samples was normalized by the values in CHX alone-treated cells.
Down-regulation of c-Myc expression in ICSBP-transduced cells. (A) Semiquantitative RT-PCR analysis for c-Myc transcripts. (B) Wright-Giemsa stain of Tot2p210 cells transduced with retroviruses carrying hormone-binding domain of estrogen receptor (ER) or ICSBP/ER chimeric construct. Cells were either left untreated or treated with 1 μM β-estradiol (Est) for 24 hours (original magnification × 600). (C) RNA blot analysis for the c-Myc transcripts. Tot2p210 cells transduced with indicated viruses were treated with 1 μM β-estradiol. Total RNA (5 μg) from cells treated for the lengths of time was analyzed. The bottom panel indicates 28S ribosomal RNA. (D) Effect of cycloheximide (CHX) on ICSBP/ER-mediated repression of c-Myc expression. The top panel indicates RNA blot analysis; the bottom panel, quantification of the c-Myc transcripts. CHX (10 μg/mL) was added 10 minutes before addition of estradiol. The expression level in CHX + Est samples was normalized by the values in CHX alone-treated cells.
In order to effectively address these possibilities, we felt it necessary to establish another transduction system that allows us to study immediate early events after ICSBP expression. The system with ICSBP itself used above was not adequate for this purpose, since ICSBP was expressed in a nonsynchronous manner, making it difficult to study early changes. To this end, we employed an ICSBP/estrogen receptor (ICSBP/ER) chimeric system. In this system, ICSBP is induced in a relatively well synchronous manner enabling us to study early gene expression more precisely than with a system using a nonchimeric ICSBP. As shown in Figure 5B, in the absence of estradiol, ICSBP/ER-transduced Tot2p210 cells continued to grow without undergoing differentiation, similar to the ER-transduced cells. Upon estradiol treatment, however, ICSBP/ER-transduced cells ceased to grow and differentiated into macrophages in a synchronous manner. Wright-Giemsa stain of ICSBP/ER-introduced cells, after estradiol treatment for 24 hours, showed a cleaved nucleus and a moderately enlarged, less basophilic cytoplasm with some vacuoles, which corresponded to the intermediate stage of macrophage differentiation (Figure 5B). Cells transduced with ER alone did not show any sign of growth inhibition or differentiation. RNA blot analysis in Figure 5C demonstrated that c-Myc expression was strongly repressed within 1 hour after activation of ICSBP/ER and the levels remained very low for 11 hours. In contrast, c-Myc expression remained high in ER-transduced cells before and after estradiol addition (Figure 5C). Because c-Myc repression was observed within 1 hour after ICSBP activation, this repression is unlikely to be attributed to an event secondary to growth arrest/differentiation.
We next examined whether repression of c-Myc expression requires de novo protein synthesis. As shown in Figure 5D, although estradiol addition repressed c-Myc expression (5-fold reduction), pretreatment of cells with cycloheximide (CHX) obliterated the repression (< 20% reduction). Addition of CHX alone did not affect c-Myc expression, ruling out an induction of c-Myc by CHX itself.
Induction of Blimp-1 and METS/PE1 gene expression by ICSBP
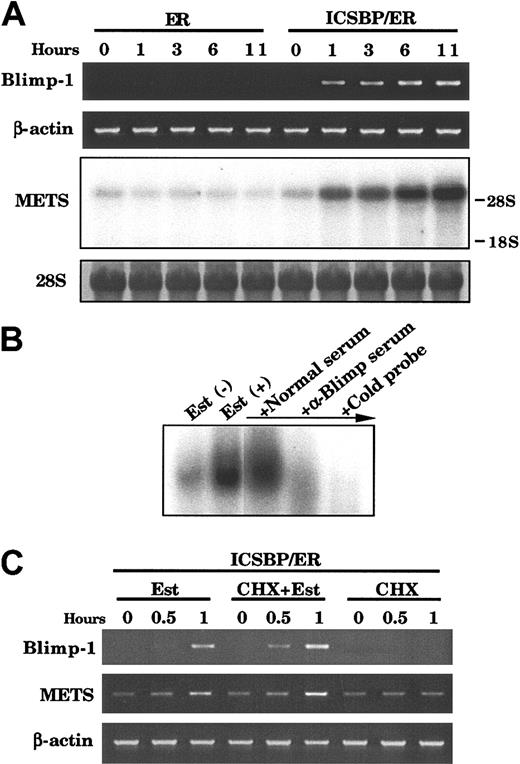
The observation that the repression of c-Myc required de novo protein synthesis suggests that ICSBP's effect is indirect and occurs through induction of a protein(s) that represses c-Myc expression. We searched for ICSBP-inducible genes that are capable of repressing c-Myc. We identified 2 known c-Myc repressors, Blimp-1 and METS/PE1, to be rapidly induced by ICSBP in Tot2 p210 cells. Blimp-1 carrying C2H2 zinc fingers is a transcriptional repressor that elicits potent antiproliferative effects on terminally differentiating B cells.27 METS/PE1 is an Ets family member that represses transcription and inhibits cell growth in myeloid cells.30 As shown in Figure 6A, both Blimp-1 and METS/PE1 transcripts were barely detectable or low in ICSBP/ER-transduced Tot2p210 cells before treatment, but were both induced within 1 hour after estradiol addition, with transcript levels increasing in the subsequent 11-hour period. Both transcripts were undetectable or low in ER-transduced cells before and after estradiol treatment (Figure 6A). EMSA demonstrated that in ICSBP/ER-transduced Tot2p210 cells, estradiol treatment induces the binding activity to the PRF site in the c-Myc promoter, the DNA element to which Blimp-1 binds.27 This activity was abrogated by the presence of anti-Blimp-1 serum but not by normal serum, suggesting that this activity contains Blimp-1 protein (Figure 6B).
Induction of Blimp-1 and METS/PE1 mRNA by ICSBP. (A) Expression of Blimp-1 and METS genes in ICSBP/ER-transduced Tot2p210 cells. RNA from cells treated with 1 μM β-estradiol for the indicated time was subjected to semiquantitative RT-PCR (for Blimp-1) and RNA blot analysis (for METS). (B) EMSA detection of DNA binding activity of Blimp-1. Nuclear extracts from ICSBP/ER-transduced Tot2p210 cells with or without estradiol treatment (10 hours) were analyzed with the PRF probe. In lanes 3 and 4, extracts from estradiol-treated cells were preincubated with anti-Blimp-1 serum, normal rabbit serum, or 100-fold excess unlabeled probe, prior to addition of labeled probe. (C) Effect of cycloheximide (CHX) on ICSBP/ER induction of Blimp-1 and METS transcripts. Cells were treated as in Figure 5D.
Induction of Blimp-1 and METS/PE1 mRNA by ICSBP. (A) Expression of Blimp-1 and METS genes in ICSBP/ER-transduced Tot2p210 cells. RNA from cells treated with 1 μM β-estradiol for the indicated time was subjected to semiquantitative RT-PCR (for Blimp-1) and RNA blot analysis (for METS). (B) EMSA detection of DNA binding activity of Blimp-1. Nuclear extracts from ICSBP/ER-transduced Tot2p210 cells with or without estradiol treatment (10 hours) were analyzed with the PRF probe. In lanes 3 and 4, extracts from estradiol-treated cells were preincubated with anti-Blimp-1 serum, normal rabbit serum, or 100-fold excess unlabeled probe, prior to addition of labeled probe. (C) Effect of cycloheximide (CHX) on ICSBP/ER induction of Blimp-1 and METS transcripts. Cells were treated as in Figure 5D.
To examine whether Blimp-1 and METS/PE1 are directly induced by ICSBP, the effect of CHX was tested (Figure 6C). Addition of CHX did not inhibit estradiol induction of either transcript and CHX alone did not induce these transcripts. These results indicate that Blimp-1 and METS/PE1 mRNA are directly induced by ICSBP. In addition to ICSBP/ER, nonchimeric ICSBP also induced expression of both mRNA in Tot2p210 as well as Tot2 cells (data not shown). It is of note that other genes reported to repress c-Myc, including CCCTC-binding factor,31 RFX1,32 and Myc intron-binding protein-133 were not induced by ICSBP/ER or ICSBP at a significant level (data not shown).
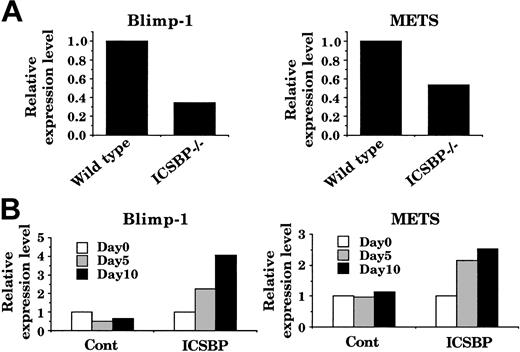
To explore additional evidence that supports ICSBP regulation of Blimp-1 and METS, we examined expression of these genes in freshly isolated bone marrow mononuclear cells in wild-type and ICSBP-/- mice by quantitative PCR. Consistent with the data with Tot2 cells, expression of Blimp-1 and METS transcripts were lower in ICSBP-/- cells than in wild-type cells (Figure 7A). We then tested whether introduction of ICSBP into freshly isolated ICSBP-/- hematopoietic cells can induce these genes. To this end, lineage marker-negative (lin-) bone marrow progenitor cells from ICSBP-/- mice were transduced with ICSBP retrovirus and were cultured in the presence of SCF and M-CSF. Transduction of ICSBP resulted in increased expression of both Blimp-1 and METS transcripts (Figure 7B). In contrast, no induction was observed in control virus-transduced ICSBP-/- cells. These results indicate that induction of Blimp-1 and METS genes by ICSBP is not a cell line-restricted phenomenon and support the notion that they are target genes of ICSBP.
ICSBP restoration of Blimp-1 and METS expression in ICSBP-/- bone marrow cells. (A) Blimp-1 and METS transcript levels in wild-type and ICSBP-/- bone marrow mononuclear cells. Transcript levels were quantified by real-time RT-PCR and were normalized by GAPDH levels. (B) Blimp-1 and METS transcript levels in ICSBP-transduced bone marrow lin- cells. Bone marrow lin- cells from ICSBP-/- mice were transduced with control MSCV-puro or MSCV-ICSBP-puro retrovirus on day 1 and day 2 as described in “Materials and methods.”
ICSBP restoration of Blimp-1 and METS expression in ICSBP-/- bone marrow cells. (A) Blimp-1 and METS transcript levels in wild-type and ICSBP-/- bone marrow mononuclear cells. Transcript levels were quantified by real-time RT-PCR and were normalized by GAPDH levels. (B) Blimp-1 and METS transcript levels in ICSBP-transduced bone marrow lin- cells. Bone marrow lin- cells from ICSBP-/- mice were transduced with control MSCV-puro or MSCV-ICSBP-puro retrovirus on day 1 and day 2 as described in “Materials and methods.”
To study the effect of Blimp-1 on c-Myc expression and cell growth, we transduced a bicistronic retrovirus that can express Blimp-1 and a truncated form of human CD8 (CD8t) into Tot2p210 cells. As a control, cells were transduced with an empty MSCV-CD8t virus. Transduced cells were sorted by antihuman CD8 antibody on day 2 and the level of c-Myc transcripts was quantified. Ectopic Blimp-1 repressed c-Myc gene expression more than 3-fold on day 2 (Figure 8B) and markedly inhibited Tot2p210 cell growth during day 2 to day 6 of culture (Figure 8C). Thus, ICSBP inhibits c-Myc expression and cell growth at least in part by activating c-Myc repressors.
Repression ofc-Mycand growth inhibition by Blimp-1. (A) Semiquantitative RT-PCR for ectopic Blimp-1. Tot2p210 cells were transduced with control MSCV-CD8t or MSCV-Blimp-1-CD8t. Transduced cells were purified by immunomagnetic cell sorting using anti-CD8 antibody on day 2. (B) c-Myc transcript level quantified by real-time RT-PCR. (C) Cell growth rate (fold increase in cell number) during day 2 to day 6. Data represent mean ± SD in triplicate.
Repression ofc-Mycand growth inhibition by Blimp-1. (A) Semiquantitative RT-PCR for ectopic Blimp-1. Tot2p210 cells were transduced with control MSCV-CD8t or MSCV-Blimp-1-CD8t. Transduced cells were purified by immunomagnetic cell sorting using anti-CD8 antibody on day 2. (B) c-Myc transcript level quantified by real-time RT-PCR. (C) Cell growth rate (fold increase in cell number) during day 2 to day 6. Data represent mean ± SD in triplicate.
Discussion
We found that introduction of ICSBP into Bcr/Abl-transformed ICSBP-/- myeloid progenitor cells causes marked growth arrest concomitant with the differentiation toward macrophages. These changes are similar to those seen in the parental Tot2 cells. That ICSBP's growth inhibitory and differentiation-promoting activities were not diminished following Bcr/Abl transformation shows that this oncoprotein does not significantly affect the function of ICSBP in these cells. Our results provide a potential mechanism by which ICSBP antagonizes the oncogenic activity of Bcr/Abl, reported in a series of clinical and experimental studies.9-12
The Bcr/Abl kinase inhibitor imatinib mesylate completely abolished the growth and survival of Tot2p210 cells. Thus, the cytokine-independent growth of these cells is still dependent on Bcr/Abl, rather than being maintained via secondary mutations that confer Bcr/Abl-independent growth.34 ICSBP did not inhibit the kinase activity of Bcr/Abl in Tot2p210 cells, indicating that ICSBP does not cause generalized repression of Bcr/Abl but has a more specific effect involving one or few of numerous pathways activated by Bcr/Abl.4 Consistent with this view, we found that ICSBP rapidly down-regulates c-Myc expression. It has been shown that c-Myc is required for the transforming activity of Bcr/Abl. The dominant-negative mutant form of Myc blocks transformation by Bcr/Abl or v-Abl.29 Efficient activation of c-Myc expression by Bcr/Abl requires the Src homology 2 (SH2) domain of Bcr/Abl, and overexpression of c-Myc partially rescues the transformation-defective Bcr/Abl SH2 mutant.35 Studies on v-Abl and Bcr/Abl suggested that the signal involves Ras/Raf, cyclin-dependent kinases, and E2F transcription factors that eventually activate the c-Myc promoter.36,37 A recent study showed the involvement of JAK2 as well.38
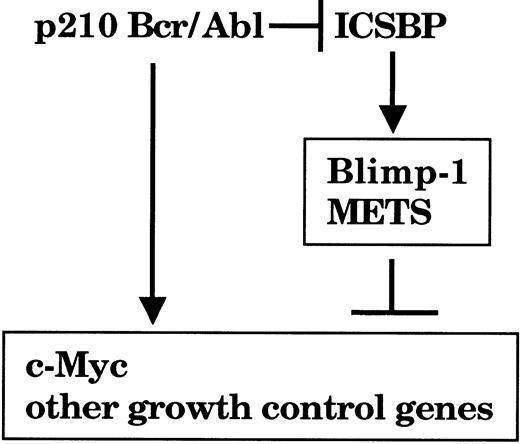
The observation that c-Myc repression by ICSBP required de novo protein synthesis led us to conclude that ICSBP does not directly act on c-Myc transcription but acts by inducing genes that repress the expression. In the present study we were able to identify 2 target genes activated by ICSBP, namely Blimp-1 and METS/PE1, both potent c-Myc repressors (Figure 9). It has been reported that Blimp-1 and METS directly repress c-Myc through the distinct DNA target elements PRF and Ets sites, respectively, on the c-Myc promoter.29,30 We detected an ICSBP-inducible DNA binding activity for the c-Myc promoter that contained Blimp-1 in Tot2p210 cells. In addition, ectopic Blimp-1 caused rapid repression of c-Myc gene expression, resulting in a marked growth inhibition of these cells. These results suggest that the growth inhibition by ICSBP is mediated at least in part by the c-Myc repressors. However, it is possible that additional factors induced by ICSBP may also contribute to the c-Myc repression and growth inhibition achieved by ICSBP. Induction of Blimp-1 and METS by ICSBP is intriguing, since they are expressed and active during macrophage differentiation, suggesting that their induction by ICSBP may be part of the developmental program specifying macrophage differentiation.30,39 Thus, upon induction by ICSBP, Blimp-1 and METS may activate and repress a series of their own target genes that are relevant not only to growth inhibition/c-Myc repression but also to macrophage differentiation/function.
A model for a growth inhibitory pathway activated by ICSBP in myeloid cells. Bcr/Abl activates c-Myc expression to stimulate cell growth ICSBP induces the Blimp-1 and METS genes, which contribute to repression of the c-Myc gene and presumably other growth control genes. Bcr/Abl may down-regulate ICSBP expression to achieve efficient mitogenic transformation in CML.
A model for a growth inhibitory pathway activated by ICSBP in myeloid cells. Bcr/Abl activates c-Myc expression to stimulate cell growth ICSBP induces the Blimp-1 and METS genes, which contribute to repression of the c-Myc gene and presumably other growth control genes. Bcr/Abl may down-regulate ICSBP expression to achieve efficient mitogenic transformation in CML.
Given the negative role of ICSBP in the Bcr/Abl function, our results are consistent with the notion that full oncogenic activity of Bcr/Abl depends on the down-regulation of ICSBP. This notion has gained increased acceptance based on recent observations. First, ICSBP mRNA expression is significantly reduced in CML patients and in a CML-like disease in mice caused by p210 Bcr/Abl.9,11 Second, it was recently reported that transduction of p210 Bcr/Abl retrovirus into umbilical cord blood cells decreases ICSBP mRNA expression in the CD34+ fraction.40 It would be of importance to study the mechanism by which Bcr/Abl interferes with ICSBP expression in hematopoietic cells.
It is likely that ICSBP negatively affects oncogenic activity of Bcr/Abl not only by inhibiting the c-Myc pathway but through additional mechanisms. Particularly important in this regard is the possible role that ICSBP plays in antigen presentation and antitumor immunity. It is well recognized that CML cells are sensitive to T-cell-mediated immunity.41,42 In allogeneic bone marrow transplantation, T-cell depletion from marrow cells results in a significant increase in relapse, especially in CML patients.43 Moreover, donor lymphocyte infusion, even without chemotherapy, is highly effective for relapsed CML patients who had received allogeneic transplantation.44 It was recently shown that IFNα induces a specific T-cell response in CML.45,46 For induction of a major histocompatibility complex (MHC)-restricted T-cell response, appropriate antigens such as peptides from cellular proteins including proteinase 3/myeloblastin as shown in the IFNα-induced T-cell immunity45,46 or those from the Bcr/Abl junction47 must be presented to T cells. Professional antigen-presenting cells, which include macrophages and DCs, serve this function. The importance of ICSBP in the development and function of macrophages in immune responses has been amply documented.14,16,48-51 In addition, we and others recently found that ICSBP is critical for the development and maturation of DCs.17-20 ICSBP is required for the expression of IL-12 p40 in macrophages and DCs, a cytokine required for IFNγ expression. Further, IFNα production in DCs is dependent on the expression of ICSBP, since ICSBP-/- mice lack plasmacytoid DCs that are the major IFNα-producing cells.19,20 This observation is significant, in view of the fact that IFNα is the main cytokine used for CML therapy. Moreover, ICSBP is essential for MHC class II expression in DCs,18 indicating that it plays a critical role in antigen presentation. It is therefore likely that ICSBP plays a key role in eliciting anti-Bcr/Abl immune responses. Based on this line of reasoning, it may be envisaged that macrophages/DCs from CML patients may have developmental and functional defects similar to ICSBP-/- cells. Given the ability of ICSBP to induce macrophage differentiation, even in the presence of Bcr/Abl activity, restoration of ICSBP expression may contribute to establishment of T-cell-mediated immunity against CML cells by generating functional Bcr/Abl-positive antigen-presenting cells. This may be achieved by treatment with IFNs or possibly more efficiently by direct ICSBP gene introduction as a gene therapy.
In conclusion, ICSBP can induce growth arrest and macrophage differentiation in p210 Bcr/Abl-transformed myeloid progenitor cells. ICSBP suppresses c-Myc expression at least in part by direct activation of Blimp-1 and METS, which may explain the mechanism of the growth arrest by ICSBP (Figure 9). The present work adds to the understanding of the role of ICSBP in the pathogenesis of CML and may help to develop strategies for CML therapy.
Prepublished online as Blood First Edition Paper, August 21, 2003; DOI 10.1182/blood-2003-01-0291.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Dr Steven R. Bauer for the c-Myc cDNA, Dr Ricardo A. Feldman for pMSCV-CD8t, Dr Richard J. Maraia for STI571, Pratima Thothakura for DNA constructions, and Dr Atsuko Masumi for IFNγ.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal